Abstract
Frameshift mutations provide recognized mechanisms for changing the coding potential of an organism. Here, multiple frameshifts are identified in repetitive sequences within an Epstein-Barr virus unspliced early gene, LF3, which is associated with the viral replicative cycle and also transcriptionally expressed in many virally associated tumors. On the DNA strand encoding LF3, there are three open reading frames, only one of which contains an initiation codon. Most (>95%) of the gene consists of numerous (>20, varying with cell source) GC-rich copies of a 102-bp direct repeat (called IR 4) flanked by small unique sequences. LF3 may express a protein if its initiation and termination codons reside in the same reading frame, but this is not always the case. Frameshifting events, occurring in short runs of pyrimidines (mainly C residues) in the repeats, give rise to mutations which may provide a mechanism for escape of an LF3 function from host surveillance. Sequence studies link these frameshifts to DNA replication errors. Notably, the number of sites in LF3 at which such mutations can occur permits a very large amount of diversity in this gene. Our data also suggest a second degeneracy mechanism within the protein itself, which influences its stability and may reflect a host defense mechanism. LF3 thus provides a potentially important model for studying the quest for supremacy between a virus and its host.
Epstein-Barr virus (EBV), a human herpesvirus, is associated with a variety of human cancers as well as being a causative agent for infectious mononucleosis. Its genome (≈170 to >200 kbp) is characterized by unique DNA sequences interrupted by tandem repetitive sequences, internal repeats (IR) 1 to 4, and terminal repeats (TR). The repeats themselves are of various sizes, ranging from about 100 to >3,000 bp, but all reside within genes. Three of these repetitive elements (called IR 1, IR 3, and TR, respectively) are found in latent viral proteins, EBV nuclear antigens 5 and 1, and LMP2A and -B (membrane proteins). The other two repetitive elements, IR 2 (or NotI repeats) and IR 4 (PstI repeats), lie within genes that have been classified as expressing early (replication-associated) functions (23, 30). The corresponding transcripts, called BHLF1 (BamHI H leftward reading frame 1) and LF3 (leftward reading frame 3), are found in the polyribosomal fraction of EBV-infected cells and represent the most abundant viral transcripts synthesized during the EBV lytic cycle (5, 11, 15, 17, 30). Their open reading frames are almost entirely composed of the repetitive elements. Structurally, there are similarities between IR 2- and IR 4-containing genes, and both lie adjacent to viral origins of lytic replication (13). Their protein products, however, have different sequences, and antibodies to one do not cross-react with the other (28, 29).
Although EBV IRs resemble in size the short interspersed elements found in eukaryotic genomes, whereas short interspersed elements are generally noncoding and can act as retrotransposons, there is no evidence for retrotransposition of the EBV tandem repeats. They appear to remain physically as stable components of their respective viral genes. Nonetheless, like chromosomal repetitive elements in general (21), by offering the possibility of template-primer misalignments or generation of template hairpin structures during replication, the viral repeats provide opportunities for replication infidelity or transcriptional variation and mutations. Such events are postulated as relevant to viral evolution (14, 24, 41).
A remarkable feature of any microbial or mammalian cell is the incredible accuracy achieved in maintaining the fidelity of its genetic information. However, sequence alterations can and do occur due to RNA/DNA polymerase slippages during transcription or replication. In the RNA virus field, products have been observed with genetic alterations associated either with RNA polymerase slippages at tandem repetitive sequences or mRNA editing that produces frameshifts within a single open reading frame. Such mechanisms lead both to nucleotide deletions and insertions of G residues in paramyxoviruses (4, 16) or, in bovine parainfluenza virus 3, to the expression of all three reading frames in its P gene (32). An RNA polymerase stuttering or slippage model has been proposed to explain such events (40).
In the retrovirus field, infidelity of the reverse transcriptase frequently results in mutation events, many of which appear nonrandom (2). Further, many retroviruses use translational frameshifting on the ribosome at stem-loop structures for generating their pol gene products (31). In both Escherichia coli and Thermus thermophilus, frameshifting events at runs of A residues allow two components of DNA polymerase III to be expressed from a single gene. In the latter case, this mechanism has been attributed to slippage on the ribosome during translation (46), whereas in E. coli, the data suggest that the frameshifting has occurred during transcription (22).
In the DNA virus field, the fidelity of individual DNA replication complexes, whether mainly of cellular or mixed viral and cellular origin, is sufficiently high that records of polymerase slippages and frameshift alterations are rare. To expand or alter their limited coding potential, DNA viruses mainly appear to rely on the use of multiple, alternative splicing sites for processing transcripts that then allow different proteins to be expressed from a single gene, as observed, for example, with papovaviruses and adenoviruses. A classical case of infidelity, however, in the DNA virus field involves the lysozyme gene of bacteriophage T4. This has been attributed to stuttering of a replication complex at short runs of A residues at two sites in the gene (38). Polymerase slippages that alter mini- and microsatellite sequences in DNA are also well documented (6) and, as seen with the RAD50 gene in colorectal cancer and gastric carcinomas, may be related to tumorigenesis (19). Elsewhere, sequence slippages have been reported in clinical isolates involving herpes simplex virus type 2 (25).
Our attention became focused on slippage/frameshifting phenomena while studying the EBV LF3 (IR 4-containing) major transcript in virally associated cell lines and tumors, where alterations possibly explainable by such mechanisms appeared to occur (43). The literature on LF3 already pointed to sequence variations which, if accurate, could best be explained by the occurrence of mutation events (5, 11, 23, 30). A cartoon based in part on literature data is shown (Fig. 1). Further evidence for genetic diversity came from studying cell lines derived from a Burkitt's lymphoma patient, Daudi, which differed in their responsiveness to interferon alpha (12) as well as in their abilities to express a protein from the LF3 gene (44).
FIG. 1.
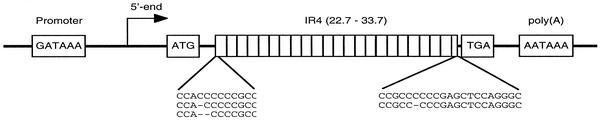
Schematic diagram of the structure of the LF3 early gene from EBV (not to scale). Within the LF3 coding region on the viral genome there are three open reading frames, only one with an initiation codon (ATG), and several termination codons, all in the same reading frame (see Fig. 2), the first of which (TGA) is shown. LF3 encodes an unspliced transcript with a single polyadenylation (AATAAA) signal. The gene has short unique sequences at its 3′ and 5′ ends (indicated by heavy solid lines) and contains numerous copies of 102-bp repeats (IR 4), as indicated. Within each repeat lie pyrimidine-rich sequences (two sets of which, at the 5′ and 3′ ends, are given) that are subject to frameshifting events during DNA replication, producing mutations, as noted. The number of repeats may vary (as indicated) according to the cell source, as do the oligopyrimidine sequences, and, depending upon the sequences at the latter slippage-prone regions, initiation and termination codons may or may not lie in the same reading frame.
These intriguing data led us to examine further the question of frameshifting with regard to LF3 in other viral cell lines and in primary tumors. Here, we describe stuttering or frameshifting events as a general property of the LF3 gene. We show that these events occur at the DNA level, that the altered sequences are conserved within the viral transcript, and that they significantly affect protein synthesis, structure, and stability. We suggest that the EBV LF3 gene may undergo frameshifts to accommodate and evade the immunological profile of its host while at the same time host mechanisms may be invoked to degrade the respective viral protein. As such, the data illustrate and in part define a hitherto unrecognized subtlety that exists between EBV and its host cell.
MATERIALS AND METHODS
Cell lines and cultivation.
B95-8, M-ABA, and M81 are cell lines immortalized with EBV from infectious mononucleosis (B95-8) or nasopharyngeal carcinoma (NPC) patients (M-ABA and M81). Raji, Daudi-ICRF, and mutant 100K (36) (from Ian Kerr, Imperial Cancer Research Fund), and Daudi-ATCC, P3HR-1, and BL74 are Burkitt's lymphoma-derived cell lines. Ramos is an EBV-negative Burkitt's lymphoma line. The origin of several Daudi cell lines is described elsewhere (20). All B cells were cultured in suspension in RPMI 1640 medium with 10% fetal calf serum under standard conditions. They could be induced to activate gene expression or produce virus as described previously (11).
Tumor materials.
North African NPC xenografts (C15, C17, and C18, from Pierre Busson) were maintained by propagation in nude mice (3). Primary Chinese NPC biopsies were obtained from Hong Kong, and Burkitt's lymphoma biopsies were obtained from Blantyre, Malawi. Southern blot data showed all tumors to be EBV positive.
RNA and DNA isolation for amplification protocols and sequencing.
Total RNA was isolated as a pellet from xenografts, cell lines, and fresh biopsy specimens by the guanidinium-cesium chloride method, as described previously (43), and polyadenylated RNA was selected on oligo(dT) mRNA purification columns (Pharmacia). Polyadenylated RNA (1 μg) or total RNA (5 μg) was treated with 10 U of DNase I (Boehringer) to remove any DNA in the RNA samples. DNA isolated from the supernatant of the total RNA preparation by standard procedures was used as a control for DNase I digests or as a substrate for PCR amplifications.
DNA sequencing.
PCR products were either sequenced directly from gel-isolated materials or cloned into a pBIISK vector (Stratagen) or M13mp19 and then sequenced with the ABI Prism dye terminator cycle sequencing kit as specified by the manufacturer (Applied Biosystems Inc.).
cDNA synthesis, RT-PCR, and PCR amplifications.
To obtain the 5′-end sequence of the LF3 transcripts from different EBV strains and tumor biopsies, DNase I-treated RNAs (1 μg of polyadenylated RNA or 5 μg of total RNA) were used for first-strand cDNA synthesis with a gene-specific primer, 5′-CTGCAGCCGGGTCCGGGGTT-3′ (EBV nucleotides 3470 to 3489) (30). Synthesized cDNA was diluted (to 100 μl) in l0 mM Tris-HCl (pH 8.0)-0.1 mM EDTA. PCR amplifications were carried out on 5 μl of cDNA over 36 cycles with 5′-GGTCCGGGGTTCCGGCCCTG-3′ (3479 to 3498) and 5′-CGCTCGGGGGGTGCACACCT-3′ (3684 to 3665) as primers, as described previously (43). Products were separated by electrophoresis on 1% agarose, and identities were verified by Southern blot hybridization and DNA sequencing. PCR amplification of DNA for sequencing purposes used the same primers and conditions as for cDNA.
RACE analysis.
For 3′ end sequence determination on LF3 transcripts, the rapid amplification of cDNA ends (RACE) protocol (9) was employed. mRNA (1 μg) was primed with 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′ and reversibly transcribed with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL). cDNA was amplified by PCR with a gene-specific primer, 5′-CCCGAGCTCCAGGGCCGGAA-3′ (1161 to 1142), and an adapter primer, 5′-GACTCGAGTCGACATCG-3′. Positive bands, identified by Southern blotting, were subcloned and sequenced.
Determination of IR 4 repeat copy numbers from cell lines and tumor biopsies.
Total chromosomal DNA, digested with BanI and DdeI, was separated by electrophoresis on a 1% agarose gel, transferred to nylon membranes, and hybridized with a 32P-labeled PstI fragment from Daudi cells (11). Product migration, relative to size markers, allowed copy numbers to be derived. Raji DNA, giving a fragment 2.5 kbp in size, with 24.7 copies of the repeat (30), served as an added control for this experiment; 0.7 of a repeat unit lacks a PstI site.
IR 4 repeat studies.
To obtain the sequence of the internal 102-bp repeat region, high-fidelity PCR was performed with primers 5′-CCCGAGCTCGAGGGCCGGAA-3′ and 5′-CTGCAGCCGGATCCGGGGTT-3′ (italic sequences are restriction enzyme sites used in cloning of the DNA products). PCR utilized heat-denatured total cellular DNA (1 μg) from uninduced or chemically induced Daudi-ICRF cells, 400 ng of each primer, 0.1 mM each of the deoxynucleoside triphosphates, and 2 U of HiFidelity DNA polymerase mix (ABgene). Amplification involved an initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 s at 95°C, 20 s at 60°C, and 40 s at 72°C. PCR products were digested with XhoI and BamHI restriction enzymes and cloned into M13mp19. Representative clones were sequenced with dye terminators.
Generation of antipolypeptide antisera.
Peptides from the three coding frames in the IR 4 repeat region were used: PP, H2N-PPPERGSGPSGPGGPPGRPPSSRAGTPDPAAAGH-COOH; PR, H2N-PRRSGAADPADPVGHPAAPRAPGPEPRTRLQPAT-COOH; and PA, H2N-PAGAGQRTQRTRWATRPPPELQGRNPGPGGSRPP-COOH. They were synthesized with a 433A solid-phase synthesizer (Applied Biosystems Inc., Poster City, Calif.). Peptide-keyhole limpet hemocyanin conjugates were emulsified at l mg/ml in Freund's complete adjuvant, and 2 ml was injected subcutaneously into New Zealand White rabbits. At 4 weeks, a second immunization was given with peptide-keyhole limpet hemocyanin in Freund's incomplete adjuvant. Immunization was repeated until a strong antibody titer (>l:1,000, checked by peptide enzyme-linked immunosorbent assay [ELISA]) was reached, and then the rabbits were bled. The resulting serum samples and preimmunization serum samples were checked for specificity and reactivity by peptide ELISA and immunoblot staining with chemically induced HH514 (P3HR-1-derived) cell extracts as described previously (27). No cross-reaction by ELISA was observed with antipeptide sera on the individual (see above) peptides.
Protein extraction and Western blotting analyses.
Confluent cells (30 ml), treated or not treated with chemical inducing agents (tetradecanoyl phorbol acetate and n-butyrate) (11), were harvested and pelleted. After three washes with ice-cold phosphate-buffered saline, cell pellets were resuspended in 0.6 ml of cytolysis buffer [20 mM Tris-HCl (pH 7.6), 50 mM KCl, 400 mM NaCl, 150 mM 2-mercaptoethanol, 1% (vol/vol) Triton X-100, and 20% glycerol]. Lysates were incubated on ice (20 min) and spun at 4°C (12 min), and supernatant containing cytoplasmic protein was transferred to a clean 1.5-ml microfuge tube and stored at −70°C prior to use. In all cases, samples from ≈2 × 106 cells were loaded for electrophoresis purposes. Western blots were performed as described previously (29).
Immunofluorescence analyses.
Healthy proliferating cells, with or without chemical induction, were pelleted, washed with ice-cold phosphate-buffered saline, and resuspended (in phosphate-buffered saline) to 2 × 106 cells/ml, and 20 μl was dropped onto glass slides coated with 3-aminopropyltriethoxysilane over an area of about 1 cm in diameter, dried at room temperature, and stored at −70°C (26). Cells were fixed with 4% paraformaldehyde (10 min) and blocked with 10% normal swine serum (20 min) and then incubated with rabbit anti-LF3 antibody (1:50 dilution) (29) or preimmune serum for 1 h. Cells were incubated with biotinylated swine anti-rabbit immunoglobulin (1:500 dilution; Dako) for 45 min, followed by Texas Red-conjugated streptavidin (1:200 dilution; Amersham) for 45 min. All antisera were diluted in 3% bovine serum albumin (Sigma) in phosphate-buffered saline, and intervening washes of slides were carried out twice with phosphate-buffered saline for 5 min. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and examined under a fluorescent microscope (Zeiss Axiophot).
RESULTS
Sequence variations at the 5′ and 3′ ends of LF3 occur initially at the DNA level and produce frameshift alterations.
LF3 is a highly structured GC-rich gene containing numerous 102-bp (34-amino-acid) repetitive (IR 4) sequences (Fig. 1). In its open reading frame, after a short (59 nucleotide) unique 5′ sequence, a repetitive region is encountered. Each repeat contains short runs of pyrimidines (mainly C residues), which, depending upon the source of the gene, have been reported to be TC3 (from M-ABA cells [1]), TC4 (from Raji and Daudi cells [references 30 and 11, respectively]), and C6 (from an NPC xenograft, C15 [43]). At the 3′ end of the gene, within the repetitive sequence, a similar run of oligopyrimidines exists, described as C5 in M-ABA and Raji cells, as C6 in Daudi cells and the C15 tumor, and as C7 in AG-876 B cells (5; the 5′ sequence was not reported in that paper). Notably, >95% of the gene is composed of repetitive sequence. The net result of the reported variations, assuming a constant 102-bp repeat, is that only in M-ABA cells does the sequence of LF3 include a classical open reading frame, where in-frame initiation and termination codons are encountered prior to a polyadenylation AATAAA (transcription maturation) signal in the gene. Transcripts of LF3 are not spliced but are linearly related to the DNA.
We began our studies by examining in greater detail the apparent frameshift mutations at the 5′ and 3′ ends of the LF3 gene, asking whether the sequence variations observed in cell lines such as those described above are reflected in the gene from primary EBV-associated tumors, whether sequence differences are tumor type specific, whether mechanistically the frameshifts have occurred during DNA replication or transcription (within a particular cell line or tumor), and how much diversity is observed. DNA (by PCR) and RNA (by RT-PCR) sequences over both 5′ and 3′ apparent frameshift variable regions were determined. Sequence data from relevant regions of the LF3 gene were obtained from both virus producer (P3HR-1 and BL74) and nonproducer (Raji, Daudi-ATCC, Daudi-ICRF, and 100K) human cell lines, from North Africa NPC xenografts (C15, C17, and C18), and from primary biopsies, two from Asian NPC tumors (NPC 1 and 2) and nine (BL001 to BL007, SD, and IB) from Malawian African Burkitt's lymphomas. Similar data were also obtained from a marmoset cell line, M81.
DNA sequence studies showed the unique regions of LF3 to contain invariable sequence. On the other hand, as given in Table 1, runs of pyrimidines at the 5′ end of the gene were variously found to be either C6, TC4, or TC3, and at the 3′ end, either C6 or C5. From a single source, the sequence data obtained on the mRNAs were identical with the DNA data. There appear to be two discrete patterns among the lymphomas and two among the NPCs, which overlap but are not entirely identical. However, the data are too limited to attach any significance to these results at present.
TABLE 1.
Oligopyrimidine sequences at the 5′ and 3′ frameshift variable regions in the LF3 gene and repeat copy numbers from different sourcesa
| EBV strain (repeat copy no.) | 5′-end sequence | 3′-end sequence | Pattern |
|---|---|---|---|
| C15∗ (22.7) | CCCCCC | CCCCCC | 1 |
| C17∗ (27.7) | CCCCCC | CCCCC | 2 |
| C18∗ (24.7) | CCCCCC | CCCCCC | 1 |
| NPC 1∗∗ (23.7) | TCCCC | CCCCCC | 3 |
| NPC 2∗∗ (22.7) | TCCCC | CCCCCC | 3 |
| M-ABA≠ (25.7) | TCCC | CCCCC | 6 |
| Raji≠ (24.7) | TCCCC | CCCCC | 4 |
| Daudi-ICRF/ATCC≠ (23.7) | TCCCC | CCCCCC | 3 |
| Daudi 100K≠ (ND) | TCCCC | CCCCCC | 3 |
| P3HR-1≠ (21.7, 25.7) | CCCCCC | CCCCCC | 1 |
| BL-74≠ (25.7) | TCCCC | CCCCCC | 3 |
| BL-001+ (28.7) | TCCCC | CCCCC | 4 |
| BL-002+ (24.7) | TCCCC | CCCCC | 4 |
| BL-003+ (24.7) | TCCCC | CCCCC | 4 |
| BL-004+ (24.7) | TCCCC | CCCCC | 4 |
| BL-005+ (29.7) | TCCCC | CCCCC | 4 |
| BL-006+ (33.7) | TCCCC | CCCCCC | 3 |
| BL-007+ (29.7) | TCCCC | CCCCC | 4 |
| BL-SD+ (28.7) | TCCCC | CCCCC | 4 |
| BL-IB+ (23.7) | TCCCC | CCCCC | 4 |
| M81 (ND) | TCCCC | CCCCCC | 3 |
All cells except M81, a marmoset line, were from human sources. ND, not done. ∗, xenografts from North African NPCs. ∗∗, Chinese NPC biopsies. ≠, cell lines from African Burkitt's lymphomas (BLs). +, BL biopsies. Combinations of sequences are designated, respectively: 5′-C6 with 3′-C6 or C5, 1 and 2; 5′ TC4 with 3′-C6 or C5, 3 and 4; 5′-TC3 with 3′-C6 or C5, 5 and 6. Only combination patterns 1 and 6 with nucleotide numbers (n = 12 or 9) divisible by 3 are predicted to have initiation and termination codons in the same reading frames in their transcripts and thus accommodate a protein (see Fig. 2).
Figure 2, given as a DNA sequence, corresponds to LF3 transcripts and predicted translation products from different cell sources (see Table 1), up to and including the poly(A) tail. It assumes that the repeats are 102 bp, as reported (11, 23, 30, 43), and that the first polyadenylation signal encountered in the sequence is used for the production of mature polyadenylated LF3 mRNA (43), as was subsequently found (see below). Thus, only sequence pattern combinations from 5′ and 3′ frameshifts that produce number combinations divisible by 3 (that is, n = 12 and 9, Table 1) should specify proteins. The combinations uncovered in this work are defined in Table 1 as patterns 1 to 6, where, as illustrated in Fig. 2, only 1 or 6 is predicted to generate a protein. In published studies on the LF3 translation products (29), proteins were observed with P3HR-1 and M-ABA cells, as predicted. A protein was also detected in BL74 tumor cells, which was not predicted on the sequence evidence presented here.
FIG. 2.
Predicted translation products from LF3 transcripts. The sequence shown is that corresponding to the stable mature LF3 RNAs from different sources (see Table 1), which generate protein data (as given) that are predicted from particular sequences obtained by combining those identified in the framshift variable regions at the 5′ and 3′ ends of the genome (bold type). Deletions generated by frameshifting events at the 5′ and 3′ ends are indicated (−). Given here (dashed line between arrowheads) are two complete and one partial (0.7) IR 4 repeat sequences as found in different LF3 genes. Also shown (underlined) are the PstI (CTGCAG) restriction sites which allow single repeat copies, the source of previous sequence data (5, 11, 23, 30, 43), to be excised from the gene, and the first termination codon (TGA, also see Fig. 1) and the polyadenylation (AATAAA) signal. Notably, multiple termination codons (in italics) are all in the same coding frame, and the single AATAAA is used for maturation of all LF3 transcripts. As indicated (patterns 2 to 5), the initiation (ATG) and termination codons may not lie in the same reading frame. Depending upon the reading frame translated (as determined by 5′- and 3′-end combination patterns, designated 1 to 6, see Table 1), the LF3 protein in two of three cases would contain aspartic acid-proline (DP) regions, which are predictably sensitive to enzyme degradation at acidic pH conditions.
RACE analyses show invariant 3′ ends for LF3 transcripts.
In the model that describes the LF3 open reading frame and frameshift effects (Fig. 2), it is assumed that the first AAUAAA in the transcript is used as the polyadenylation maturation signal for the RNA. In the case of Raji cells, although no protein was identified, it was suggested (30) that readthrough to the next polyadenylation signal containing an in-frame termination codon might occur to produce a protein. Since such a mechanism might allow a protein product to be made from other open reading frames in this region of the EBV genome, a RACE protocol (9) was used to determine the 3′ ends of LF3 transcripts in the cell lines and tumors studied here.
In every case examined (data not given), including Raji, with a ubiquitous internal sequence primer, the same-sized product was obtained, indicating that LF3 transcripts all terminate at the same position as that shown in Fig. 2. Sequencing of RACE products also showed that the first AAUAAA polyadenylation signal in the transcript, as noted, was used in every case and identified the poly(A) tail position shown. Thus, based on the two assumptions cited, were an LF3 protein identified in materials cited in patterns 2 to 5, as for example, in the case of BL74 cells (Table 1), mechanisms other than frameshifting events at the ends of the repetitive regions are necessary to permit protein expression.
Frameshifts in internal IR 4 repeats also occur to produce genetic variations.
It is generally assumed from sequence evidence that although point mutations may occur within the repeats in LF3 from different cell types, the size (102 bp) of the repetitive element is invariant (11, 23, 30, 43). There is, however, a single report of a 103-bp repeat (in AG876 B cells), involving expansion of a frequently observed C6 sequence to C7 (5), which alters the coding frame. The question of internal repeat variability could be addressed in detail were it possible to sequence across the repetitive elements, but this has to date defeated all attempts both by us and by others. This is probably due to the GC-rich (≈85%) repetitive element; in the LF3 transcript, this folds into a predictably highly stable secondary structure and behaves like a double-stranded RNA in enzymatic digests (12).
To explore this question, we adopted a cloning methodology previously used to search for DNA fidelity during replication (35). For this, we used cellular DNA from chemically induced or uninduced Daudi-ICRF cells as the work of others suggested that protein synthesis might require chemical inducing agents (29). Daudi-ICRF cells were chosen because preliminary data suggested that a protein could be produced in this cell line (44), although none had been predicted from simple sequence data (Table 1 and Fig. 2), nor was one detectable in another Daudi line, Daudi-ATCC (11). Of relevance, Daudi lines need not be identical. Historically, whereas their precise origins remain obscure, their natural history allows variation (20); four separate lines were produced over time from the tumor patient Daudi prior to and after chemotherapeutic response, followed by tumor relapse.
In our present study, PCR primers were chosen that should produce products which, when cloned into suitable M13 vectors, would yield blue plaques from a 102-bp repeat unit and white plaques from 101- or 103-bp repeats. For amplification, the Extensor Hi-Fidelity PCR enzyme (ABgene) was used because of its low error rate, in the range of 5 × 10−5 to 3 × 10−6 errors/base, depending on the nature of the template (information from the manufacturers). We obtained both blue and white plaques. Cloned DNA products were sequenced. Difficulties in designing suitable primers for PCR amplification of a complete repeat unit meant that amplification products were 90 and 91 bp, respectively. Only one variation was seen in the DNAs (Fig. 3). That is, whereas blue plaque isolates gave a product containing a C6 sequence from the repeat units, white plaque-derived materials had a C7 sequence. The ratio of C6 to C7 found in the Daudi 90- and 91-bp products was ≈7:1 in uninduced and ≈9:1 in chemically induced cells. This difference, probably not significant, implies that the frameshifts generally observed in LF3 are independent of chemical induction.
FIG. 3.
Slippage events in internal IR 4 repeats. Partial sequence profiles (see Fig. 2) in 102-bp (blue plaque) and 103-bp (white plaque) recombinant DNA repeats from Daudi-ICRF isolates. C6 and C7 sequences (bold type) were obtained from the same cell isolate and were the only sequence alterations found in the Daudi repeats, whether from chemically induced or uninduced cells.
Several clones were found to have multiple inserts. Interestingly, one of these contained two 90-bp (C6) sequences, but another had one 90-bp (C6) and one 91-bp (C7) IR 4 sequence. Whereas direct sequencing of 5′ and 3′ residues (Table 1 and Fig. 2) had failed to identify any C7 product from the Daudi cells, this cloning technique showed its existence. By this technique, however, we did not identify its precise internal location in the gene. Notably, however expansion from C6 to C7 in an internal repeat in Daudi cells would shift it from pattern 3 to a pattern 1 profile (Fig. 2) and permit protein expression.
Frameshifting is independent of the number of IR 4 repeats in the LF3 gene in different cells.
For eukaryotic cells, it has been observed that when replication frameshifts involve reiterated sequences, the number of potential misaligned intermediates and correct base pairs that can stabilize such misalignments increases as the length of the run increases (21). To ask whether repeat copy numbers might correlate with the observed slippage patterns of LF3, the numbers of IR 4 repeats from different EBV strains was determined, with a technique described previously (30). In each case examined (see Table 1), except for uncloned P3HR-1, where two copy number variants (n = 21.7 and n = 25.7) were found, each cell line had its own repeat copy number. These ranged from 21.7 to 33.7, indicating a potential difference of up to 408 amino acids in LF3 proteins from different sources. (The 0.7 represents the 3′-most incomplete repeat, shown in Fig. 2.) Unlike reports from studies on microsatellites (reviewed in reference 39), where replication errors resulted in copy number variations, we saw no correlation between repeat numbers and sequence specified, as best illustrated by the African Burkitt's lymphoma biopsies. Thus, whereas repeat copy numbers may be of relevance, no biological significance was apparent in this study.
LF3 protein expression.
Sequence variations might be of little consequence if LF3 were an untranslated gene whose function, if any, lay in its RNA (see reference 37 for discussion of this topic). However, as argument against this notion, not only is the primary transcript of LF3 polyadenylated, but proteins have been identified in several B-cell lines (29). Since determining frameshifts within all internal repeats is difficult by sequencing methodologies, protein structures of LF3 in individual cells or tumors cannot be predicted by such methods. Protein data may be highly relevant, however, to understanding the cellular selective pressures that lead to the frameshift mutational events. However, identification of proteins from the LF3 gene has also proved problematic. As noted earlier, for example, whereas in M-ABA cells one group (29) identified a protein, another (23) did not, and in our hands detection of proteins from a single cell type, such as M-ABA or P3HR-1, did not always give reproducible results (data not shown).
In the light of the gene sequence data (Table 1, Fig. 2 and 3), a polyclonal antibody generated from a recombinant clone that expressed 46 amino acids of the C-terminal part of the protein in M-ABA cells (N. Mueller-Lantzsch, personal communication), often used in experiments aimed at examining LF3 protein expression, may be suboptimal for protein assays in other cells. This antibody did, however, allow detection of a protein where none is predicted (Fig. 2) in the Burkitt's lymphoma-derived line BL74 (29) and in Daudi-ICRF cells (44), supporting further the notion of frameshifts within internal repeats.
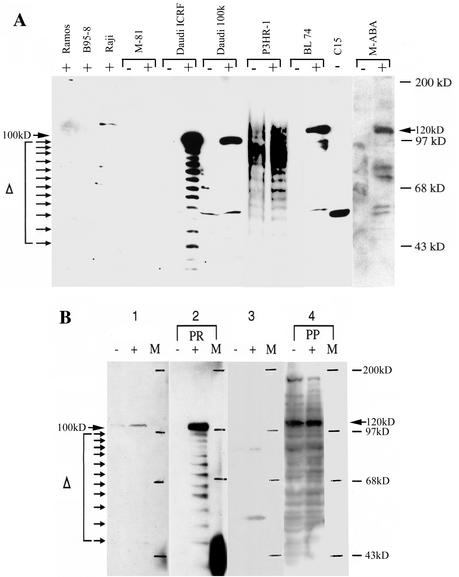
We used the antibody to probe Western immunoblots of proteins extracted from several other cell lines and controls, with and without chemical induction, and from the C15 NPC xenograft. Full-length proteins of expected sizes (Fig. 4A) were identified in Daudi-ICRF and its mutant variant Daudi-100K, in M-ABA cells, and in the EBV producer lines P3HR-1 and BL74. For C15, although the repeat copy number of LF3 is at the lower end of the range (22.7 copies, Table 1), the discrete protein band observed was only half its expected size. No proteins of expected sizes were observed for Raji and M81 cells. Since chemical induction was an important factor in allowing a protein to be identified (29; our data) but appeared to have little effect on frameshifting, this finding may merely reflect the increased mRNA levels found in induced cells (11).
FIG. 4.
Western blots of LF3 proteins. (A) LF3 polypeptides from cytoplasmic extracts of cells propagated in the absence (−) or presence (+) of the chemical inducing agents tetradecanoyl phorbol acetate and n-butyrate and identified with a rabbit polyclonal antibody from M-ABA cells (29). Sizes of protein markers are given on the right. Arrows indicate sizes predicted for full-length proteins from different sources or degraded residues with different numbers of repetitive (about 34 amino acids) sequences. No polypeptide ladders were observed in three cases, Daudi-100K, BL74, and C15, but in the last, the product was roughly half its expected size. No bands of the expected size were observed in Raji and M-81 cells (as predicted, Table 1 and Fig. 2). The data for M-ABA were taken from a separate Western blot. (B) Proteins from cytoplasmic extracts of Daudi-ICRF or P3HR-1 uninduced (−) and induced (+) cells (panels 2 and 4, respectively) stained with polypeptide antibody PR or PP, as noted, or with the corresponding preimmune serum samples (panels 1 and 3). Positions of protein size markers (lane M) are indicated on the right of each panel, and integrals suggesting loss of a repeat polypeptide are indicated on the left. In the case of Daudi cells, induction was required for protein detection, whereas this was not so with P3HR-1.
Figure 4A represents data from two Western blots which differed only in that whereas inexplicably in one experiment no protein was observed with M-ABA cells, in another experiment (as shown), in induced cells, full-length and shorter polypeptides were observed. Reproducibly, in the C15 NPC tumor, only a shorter (stable) polypeptide was ever observed. Since levels are low, our Western blot data (Fig. 4A) used cytoplasmic extracts from cells, which proved superior to whole cell extracts for protein analysis. In addition, they revealed polypeptide patterns not explicable by sequence frameshifts, but indicative of another level of degeneracy for LF3: in addition to full-length proteins, numerous smaller polypeptide bands with integrals of about 34 amino acids (equivalent to a 102-bp repeat) were also observed in materials from Daudi-ICRF, P3HR-1, and M-ABA cells, although inexplicably not from the chemically mutated Daudi-ICRF 100K line (36) or from BL74.
To evaluate protein expression further, antibodies were raised to peptides (designated PP, PR, and PA, from the first unique dipeptide in different reading frames, see Fig. 2) from the three internal repeat elements. With these antibodies and induced Daudi-ICRF cells, specific immunoreactive polypeptides were observed with PR (Fig. 4B) or PP but not with PA (not shown), induction being required (as in Fig. 4A) to observe the protein. With P3HR-1 cells, the PP antibody recognized a full-length discrete protein and smaller species in cells with or without induction (Fig. 4B). The data are consistent with findings given in Fig. 2 and 3. From both cells the polypeptide bands were notably strong. That the degraded bands were not so sharp for P3HR-1 as they were for Daudi-ICRF may reflect the fact that with regard to numbers of repetitive sequences, P3HR-1 is a mixed population of cells (Table 1). Again, the data support the hypothesis of internal frameshifts within the LF3 gene. The polypeptide degeneracy in products from some sources, which cannot be explained by frameshift mutations, raises the possibility that in the highly structured RNA from LF3 (12) either slippages are taking place on the ribosome during translation or that, in some cases, specific protein degradation is occurring.
Immunostaining of cells for LF3 protein give data consistent with genetic diversity.
To examine on a single cell basis whether chemical induction leads to large amounts of protein being generated in a small number of cells, or vice versa, immunostaining experiments were carried out. Such studies were also relevant for confirming the subcellular location of the LF3 translation product and whether it resides largely in the cytoplasm, as suggested by Western blot results, or might also be localized in the nucleus, in accordance with a role in DNA replication (13). For these studies, EBV-positive cell lines M-ABA, M81, and Burkitt's lymphoma-derived cell lines Raji, Daudi-ICRF, BL74, and P3HR-1 were used, both with and without induction, with Ramos (EBV negative) and B95-8 cells (with an LF3 deletion) as controls.
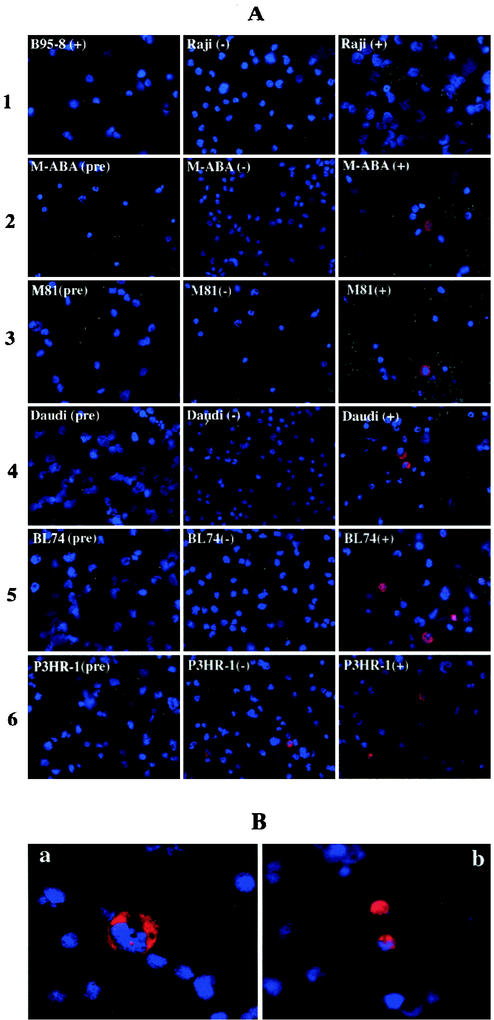
Data given in Fig. 5A show that whereas the low-virus-producer cell line M81 had appeared negative for expression of the LF3 protein on Western blots (Fig. 4A), on immunostaining following induction, small numbers of cells with low levels of fluorescence were observed. No expression was seen in Raji (or B95-8) cells. From immunostaining data, the LF3 protein appears in detectable amounts in a subpopulation of cells only (at most 10%), expression being generally enhanced upon chemical induction and more prominent in the cytoplasm than in the nucleus. An example of cytoplasmic (panel a) and nuclear (panel b) staining patterns in P3HR-1 cells is shown in Fig. 5B, where in the latter the stained cells appear to be in mitosis. Overall, the protein data accord with the genetic data.
FIG. 5.
Immunostaining of LF3 proteins. (A) Cells propagated without (−) or with (+) chemical inducing agents and stained with preimmune serum (vertical column 1) or with antibody (columns 2 and 3) from recombinant M-ABA polypeptide (29). Only in P3HR-1 (horizontal panels 6) were stained cells observed without chemical induction (see also Fig. 4). By this technique, all other isolates examined (except the B95-8 control and Raji), including M81, show small numbers of positively (mainly cytoplasmic) stained cells following chemical induction. Preimmune serum samples gave negative results. (B) Cytoplasmic and nuclear staining of LF3 protein in P3HR-1 induced cells, photographed by confocal microscopy. Panel a shows a cell with cytoplasmic staining, and panel b shows cells with nuclear staining. These data demonstrate that the protein can reside at two cellular sites and belongs to the early antigen-diffuse family of EBV proteins. Examination of larger fields of cells (not shown) indicated that cytoplasmic staining predominated. Similar conclusions were reached from Western blot data on Daudi-ICRF and M-ABA cell isolates.
DISCUSSION
Viruses, noted for the mechanisms that they adopt to increase their coding capacities and ensure survival, have also proved good models for events that can also occur in their hosts. The large DNA viruses (herpesviruses and poxviruses) are particularly adept in vivo at adapting to the immunological status of their host (1). We suggest that the latter may account for the frameshifting events we observe with the unusual LF3 gene of the oncogenic EBV. Such events lead to a demonstrably high degree of genetic variation.
LF3 contains only a small amount of unique sequence, the bulk of its information residing in multiple copies (Table 1) of a mainly 102-bp GC-rich repeat, designed IR 4. It is found transcriptionally expressed in a variety of virally infected cell lines and tumors (8, 11, 12, 43, 45). Its protein product has been postulated to be associated with productive expression of EBV (29), and in our studies, without the use of chemical inducing agents, protein was detected only in a cell line (P3HR-1) that can spontaneously generate virus. The frameshifts observed occur at short runs of pyrimidines (mainly cytidines) found throughout the gene. Sequence data, as illustrated in Fig. 2, support the need for frameshifting events to occur in order to generate a protein in some cells, and molecular (Fig. 3) and immunofluorescence (Fig. 5) data suggest that this occurs in a minority of cells only (10% or fewer) in most isolates. Notably, most EBV-infected cells do not spontaneously produce virus. LF3 is probably not unique to EBV in that a gene similar to it has been predicted to exist in the recently sequenced rhesus lymphocryptovirus (34), and it is worth keeping an open mind with regard to the possibility that among the many repetitive sequences in the human genome, one or more might well have properties similar to those found for LF3.
In normal eukaryotic cells, although DNA replication fidelity is exceedingly high, not all of the enzyme components involved in replication have proofreading-associated functions. Structural elements that promote stable frameshifts have been defined in part as those, like LF3, which contain a high frequency of reiterated (including homopolymer) sequences and tandem repeats (6). For LF3 as well, its high GC content (≈85%) could make it vulnerable and subject during replication to aberrant mispairing or replication complex stuttering mechanisms that contribute to slippage events (21, 24, 41). Coupling these data with the fact that malfunctions of mismatch repair processes occur in tumors (18), with hindsight it is probably not surprising that, consistent with molecular hybridization and translational data (11, 43, 45), variation in the sequence of this viral gene in different cell isolates occurs. We propose that sequence frameshifts in LF3, having occurred initially at the DNA level in short runs of pyrimidines, reflect DNA polymerase slippage and its failure of correction, rather than internal recombination events - since it seems unlikely that the latter would be sufficiently promiscuous to allow for all the variables in sequence observed or predicted (Table 1 and Fig. 2) and account for the apparent genetic diversity observed. The occurrence of frameshifting events in a gene consisting of repeats usually divisible by three theoretically allows expression of a unique protein in virally infected cells from any particular source, depending upon the repeat copy numbers in the gene and the sites at which mutations occur.
There are other unusual aspects to this gene. Western blots earlier identified proteins from LF3 that varied between 115 to 140 kDa, depending (presumably) on its repeat copy number in an individual cell (29). Two other things were noted in this cited work that are relevant to our findings and support protein diversity. One was that only a small fraction of human serum samples (4 of 57) that were positive for EBV early and late antigens also recognized the recombinant LF3 protein from M-ABA cells. The other was that protein derived from a recombinant construction containing the whole LF3 gene, expressed in E. coli, was rapidly degraded; only a protein considerably reduced in size (by removal of most of the repetitive sequences), as seen in the C15 NPC tumor (Fig. 4A), proved stable.
Our Western blot data (Fig. 4) with a single antibody (29) point to another aspect of diversity, epitomized by specific degradation of the protein, with integrals of about 34 amino acids in some (but not all) cells. In M-ABA, P3HR-1, and Daudi-ICRF cells, ladders extending from a predicted full-sized peptide to much smaller peptides (<40 kDa) were observed, whereas the chemically modified Daudi-100K and BL74 cells produced mainly a full-sized protein and the NPC xenograft C15 gave a truncated (about half the predicted size) stable protein; with polypeptide antibodies made from different repeat sequences (see Materials and Methods and Fig. 2), cytoplasmic extracts from Daudi-ICRF and P3HR-1 cells gave similar ladders. Notably here, in line with sequence data, one polypeptide antibody proved more responsive in the first case and another in the second (Fig. 4B).
Although slippages on the ribosome during translation, such as postulated for some RNA and RNA viruses (31, 42, 46) but not reported for transcripts from DNA viruses, might account for the protein diversity, it seems an unlikely mechanism to explain the degradation products observed. From different cell sources, the transcription products of LF3 generally represent point mutations only, and their sequences are not apparently influenced by the repeat copy number in a particular cell. An alternative explanation for the polypeptide ladders seen may be suggested by sequences within the repeats themselves, in that two of the three LF3 coding frames contain rare aspartic acid-proline (DP) dipeptides (Fig. 2), known to be susceptible to acid degradation. In vitro, the conditions for this specific degradation are quite drastic, low pH and high temperature (33), and not commensurate with the data shown in Fig. 4. Thus, the observed degradations do not seem wholly explainable by pH changes during cell culture or isolate work-up. They might occur, however, if the proteins were targeted to lysosomes, where they would encounter not only sustained low-pH conditions but also numerous acid hydrolases, such as those suggested to be used for specific enzymatic degradation of a salivary gland protein (10).
There is no evidence of a lysosome-targeting signal in LF3 proteins, and at this stage in our work, although we have evidence for the cytosolic location of the majority of the LF3 protein expressed (Fig. 5), we cannot comment on its specific localization. Notably the degradative process has specificity (as predicted, Fig. 2) and is not equally effective on all LF3 proteins (see those from the chemically modified 100K protein from Daudi-ICRF cells or the tumor-derived line BL74). The protein in the C15 xenograft may also have largely evaded the degradative pathway by losing most of its repeats. In the rhesus lymphoproliferative virus (34), the LF3 homologue identified should also have DP sequences in two out of its three coding frames, raising interesting questions about an apparently conserved protein function between these viruses.
In the work presented here, we have examined LF3 not because of its putative role in lytic DNA replication, but with regard to its notable genetic diversity. There may be a correlation between these topics. LF3 is strategically placed to play an important, possibly initiating role in lytic replication (8, 11, 13), being located adjacent to one of the two EBV lytic origins of replication, both of which contain AT-rich palindromic elements and a large homopurine-homopyrimidine tract that could adopt a classical triple-helical hinge and single-stranded DNA regions (11). Such structures conform to those found in replication origins of other DNA viruses (7), where susceptibility to mutations, above background levels, has been reported to occur during replication (35). As elsewhere (41), they allow polymerase pauses to occur during DNA replication, with dissociation of the enzyme and ultimate induction of slippage.
Our findings raise questions about selective pressures that give rise to the diversities that define LF3, both with regard to the gene and its protein, where a 'tug-of-war' between the virus and its host may take place. Chemical inducing agents, well known to support lytic replication of EBV, can influence the outcome. The frameshifting events themselves may be a survival stratagem of the virus, allowing it to produce a potentially immunogenic protein in some hosts but to evade the immune system and proliferate in others, that is, generate a viral defense mechanism. Destroying the viral protein in the absence of an effective immune mechanism by targeting it to a pathway where it can be degraded may reflect a host defense mechanism (1). The phenomena observed with LF3 merit further investigation not just with regard to virus-host interactions, but also for the light they might shed on cellular repetitive elements, often relegated to the status of junk DNA.
Acknowledgments
We thank colleagues (R. Broadhead, E. Molyneux, and E. Borgstein) in Paediatrics and Surgery at the Medical School in Blantyre, Malawi, for Burkitt's lymphoma biopsies and M. H. Ng (Queen Mary's Hospital, Hong Kong University) for NPC biopsies. We are grateful to G. Lindahl for comments on protein structure and N. Mueller-Lantzsch and C. M. Nuebling for the gift of an antibody.
REFERENCES
- 1.Alcami, A., R. Chazal, and J. W. Yewdell. 2002. Viruses in control of the immune system. EMBO Rep. 3:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, J. C., K. Bebenedk, and T. A. Kunkel. 1996. Analyzing the fidelity of reverse transcription and translation. Methods Enzymol. 275:523-537. [DOI] [PubMed] [Google Scholar]
- 3.Busson, P., G. Ganem, P. Flores, F. Mugneret, B. Clausse, B. Caillou, K. Brahan, et al. 1988. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinoma. Int. J. Cancer 42:599-606. [DOI] [PubMed] [Google Scholar]
- 4.Cattaneo, R., K. Kaelin, K. Baczko, and M. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. [DOI] [PubMed] [Google Scholar]
- 5.Dambaugh, T. R., and E. Kieff. 1982. Identification and nucleotide sequences of two similar tandem direct repeats in Epstein-Barr virus DNA. J. Virol. 44:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debrauwere, H., C. G. Gendrel, S. Lechat, and M. Dutreix. 1997. Differences and similarities between various tandem repeat sequences: minisatellites and microsatellites. Biochimie 79:577-586. [DOI] [PubMed] [Google Scholar]
- 7.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 35:6229-6263. [DOI] [PubMed] [Google Scholar]
- 8.Freese, U. K., G. Laux, J. Hudewentz, E. Schwarz, and G. W. Bornkamm. 1983. Two distant clusters of partially homologous small repeats of Epstein-Barr virus are transcribed upon induction of an abortive or lytic cycle of the virus. J. Virol. 48:731-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification with a single gene specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galli, J., and L. Wieslander. 1993. A repetitive secretory protein gene of a novel type in Chironomus tentans is specifically expressed in the salivary glands and exhibits extensive length polymorphism. J. Biol. Chem. 268:11888-11893. [PubMed] [Google Scholar]
- 11.Gao, Y., P. R. Smith, L. Karran, Q.-L. Lu, and B. E. Griffin. 1997. Induction of an exceptionally high-level, nontranslated, Epstein-Barr virus-encoded polyadenylated transcript in the Burkitt's lymphoma line Daudi. J. Virol. 71:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Y., S.-A. Xue, and B. E. Griffin. 1999. Sensitivity of an Epstein-Barr virus-positive tumor line, Daudi, to alpha in.rferon correlates with expression of a GC-rich viral transcript. Mol. Cell. Biol. 19:7305-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, J. M., W. Chaleeprom, J. Daleand, and A. Gibbs. 1995. Replication slippage in the evolution of potyviruses. J. Gen. Virol. 76:3229-3232. [DOI] [PubMed] [Google Scholar]
- 15.Hudewentz, J., H. Delius, U. K. Freese, U. Zimber, and G. W. Bornkamm. 1982. Two distant regions of the Epstein-Barr virus genome with sequence homologies have the same orientation and involve small tandem repeats. EMBO J. 1:262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacques, J. P., S. Hausmann, and D. Kolakofsky. 1994. Paramyxovirus mRNA editing leads to G deletions as well as insertions. EMBO J. 13:5496-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeang, K.-T., and S. D. Hayward. 1983. Organization of the Epstein-Barr virus DNA molecule. J. Virol. 48:135-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiricny, J. 1996. Genetic instability in cancer. Cancer Surveys 28:47-68. [PubMed] [Google Scholar]
- 19.Kim, N.-G, Y. R. Choi, M. J. Baek, Y. H. Kim, H. Kang, N. K. Kim, et al. 2001. Frameshift mutations at coding mononucleotide repeats of the hRAD50 gene in gastrointestional carcinomas with microsatellite instability. Cancer Res. 61:36-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, E., G. Klein, J. S. Nadkarni, J. J. Nadkarni, H. Wigzell, and P. Clifford. 1968. Surface IgM specificity on cells derived from a Burkitt lymphoma. Cancer Res. 28:1300-1310. [PubMed] [Google Scholar]
- 21.Kunkel, T. A. 1992. Biological asymmetries and the fidelity of eukaryotic DNA replication. BioAssays 14:303-308. [DOI] [PubMed] [Google Scholar]
- 22.Larsen, B., N. M. Wills, C. Nelson, J. F. Atkins, and R. F. Gesteland. 2000. Nonlinearity in genetic decoding: Homologous DNA replication genes use alternatives of transcriptional slipage or translational frameshifting. Proc. Natl. Acad. Sci., USA 97:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laux, G., U. K. Freese, and G. W. Bornkamm. 1985. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J. Virol. 56:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 25.Liljeqvist, J.-A., B. Svennerholm, and T. Bergstrom. 1999. Herpes simplex virus type 2 glycoprotein G-negative clinical isolates are generated by single frameshift mutations. J. Virol. 73:9796-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, Q. L., and T. A. Partridge. 1998. A new blocking method for application of murine monoclonal antibody to mouse tissue sections. J. Histochem. Cytochem. 46:977-983. [DOI] [PubMed] [Google Scholar]
- 27.Middeldorp, J. M., and P. Herbrink. 1988. Epstein-Barr virus specific marker molecules for early diagnosis of infectious mononucleosis. J. Virol. Methods 21:133-146. [DOI] [PubMed] [Google Scholar]
- 28.Nuebling, C. M., and N. Mueller-Lantzsch. 1989. Identification and characterization of an Epstein-Barr virus early antigen that is encoded by the NotI repeats. J. Virol. 63:4609-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuebling, C. M., and N. Mueller-Lantzsch. 1991. Identification of the gene product encoded by the PstI repeats (IR 4) of the Epstein-Barr Virus genome. Virol. 185:519-523. [DOI] [PubMed] [Google Scholar]
- 30.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339-346. [DOI] [PubMed] [Google Scholar]
- 31.Parkin, N. T., M. Chamorro, and H. E. Varmus. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 66:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelet, T., J. Curran, and D. Kolakofsky. 1991. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 10:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piszkiewicz, D., L. Landon, and E. L. Smith. 1970. Anomalous cleavage of aspartyl-proline peptide bonds during amino acid sequence determinations. Biochem. Biophys. Res. Commun. 40:1173-1178. [DOI] [PubMed] [Google Scholar]
- 34.Rivailler, P., H. Jiang, Y.-G. Cho, C. Quink, and F. Wang. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: Genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, J. D., D. Nguyen, and T. A. Kunkel. 1993. Frameshift fidelity during replication of double-stranded DNA in HeLa cell extracts. Biochemistry 32:4083-4089. [DOI] [PubMed] [Google Scholar]
- 36.Silverman, R. H. D., D. Watling, F. R. Balkwill, J. Trowsdale, and I. M. Kerr. 1982. The ppp(AA2′p)nA and protein kinase systems in wild-type and resistant Daudi cells. Eur. J. Biochem. 126:333-341. [DOI] [PubMed] [Google Scholar]
- 37.Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296:1260-1262. [DOI] [PubMed] [Google Scholar]
- 38.Streisinger, G., and J. E. Owen. 1985. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109:633-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vidal, S., J. Curran, and D. Kolakofsky. 1990. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 9:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viguera, E., D. Canceill, and S. D. Ehrlich. 2001. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 20:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, et al. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue, S.-A., Q.-L. Lu, R. Poulsom, L. Karran, M. D. Jones, and B. E. Griffin. 2000. Expression of two related viral early genes in Epstein-Barr virus-associated tumors. J. Virol. 74:2793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue, S.-A. 1999. Expression of two related viral early genes in EBV associated tumours. Ph.D. thesis, University of London, London, United Kingdom.
- 45.Xue, S.-A., L. G. Labrecque, Q.-L. Lu, S. K. Ong, I. A. Lampert, et al. 2002. Promiscuous expression of Epstein-Barr virus genes in Burkitt's lymphomas from the central African country, Malawi. Int. J. Cancer 99:635-643. [DOI] [PubMed] [Google Scholar]
- 46.Yurieva, O., M. Skangalis, J. Kuriyan, and M. O'Donnell. 1997. Thermus thermophilis [sic] dnaX homolog encoding gamma- and tau-like proteins of the chromosomal replicase. J. Biol. Chem. 272:27131-27139. [DOI] [PubMed] [Google Scholar]