Abstract
The process of cellular morphogenesis is highly conserved in eukaryotes and is dependent upon the function of proteins that are centrally involved in specification of the cell cycle. The human enhancer of invasion clone 10 (HEI10) protein was identified from a HeLa cell library based on its ability to promote yeast agar invasion and filamentation. Through two-hybrid screening, the mitotic cyclin B1 and an E2 ubiquitin-conjugating enzyme were isolated as HEI10-interacting proteins. Mutation of the HEI10 divergent RING finger motif (characteristic of E3 ubiquitin ligases) and Cdc2/cyclin binding and phosphorylation sites alter HEI10-dependent yeast phenotypes, including delay in G2/M transition. In vertebrates, the addition of HEI10 inhibits nuclear envelope breakdown and mitotic entry in Xenopus egg extracts. Mechanistically, HEI10 expression reduces cyclin B levels in cycling Xenopus eggs and reduces levels of the cyclin B ortholog Clb2p in yeast. HEI10 is itself a specific in vitro substrate of purified cyclin B/cdc2, with a TPVR motif as primary phosphorylation site. Finally, HEI10 is itself ubiquitinated in egg extracts and is also autoubiquitinated in vitro. These and other points lead to a model in which HEI10 defines a divergent class of E3 ubiquitin ligase, functioning in progression through G2/M.
The spatial and temporal regulation of cell division is essential for development and normal cellular replacement in both unicellular and multicellular organisms. Cell cycle control and cell morphology are coordinated by complex signaling networks, the inappropriate perturbation of which can lead to either programmed cell death (apoptosis) or indefinite cell division cycles (cancer). The fact that many protein constituents of such pathways are conserved over great phylogenetic distances among eukaryotes emphasizes their importance. Since the 1980s, complementation studies in which human genes were identified based on the specific rescue of, or genetic interaction with, orthologous pathways in yeast have proven to be a fruitful means of gaining insight into mammalian growth controls. For example, the central cell cycle kinases CDC2 (32) and CDK2 (11) and the C, D, and E cyclins (15, 24, 53, 57) were identified by complementation of yeast mutants.
Analysis of yeast budding controls has been of particular interest as a model of cellular morphogenesis and has provided an interpretive framework for studies of signal transduction and morphological control processes in less experimentally amenable multicellular organisms (see, for example, reference 26). Bud emergence in vegetatively growing yeast is a complex process that depends on the rearrangement of the cytoskeleton throughout the different phases of the cell cycle (8, 56). The site of bud emergence is specified by signals that mark the location of a previous bud. In addition to the spatial regulation of budding, bud emergence is temporally synchronized with the different phases of the cell cycle and is sensitive to the environment. A coordinated response to changes in these diverse signals allows yeast the flexibility to pursue additional developmental fates beyond vegetative budding. These include the shmooing of haploids in response to mating factors, the formation of spores during nutrient deprivation and, notably, the hyperpolarization and polarity changes characteristic of pseudohyphal or filamentous growth.
The pseudohyphal phenotype was first described in modern laboratory yeast cells during growth under conditions of significant nitrogen limitation (16). It was subsequently found that pseudohyphal growth is stimulated by RAS overexpression (16) and protein kinase A (PKA) activation (39, 45) and, in addition, involves signaling through components of the mitogen-activated protein (MAPK) cascade (35, 39, 44, 45, 48). Compared to vegetatively growing yeast, the cell cycle in filamentously growing yeast is characterized by a prolonged G2/M phase and an accompanying loss of cell growth in G1, a hyperelongated shape, a switch in the yeast budding pattern from bipolar to unipolar, and the continued attachment of daughter cells to mother cells after septation (16, 26, 27, 38). The transition to pseudohyphal growth mode, therefore, occurs through coordinated reprogramming of nutrient response, morphoregulatory signaling cascades, and the cell cycle.
We have previously used the pseudohyphal phenotype in yeast to screen for human cDNAs which when overexpressed cause diploid yeast growing on low-nitrogen medium to develop into hyperpolarized stellate colonies. This screen led to the isolation of human enhancer of filamentation 1 (HEF1) (29), a protein that we have subsequently shown to be involved in the control of human mitotic cell cycle regulation (31), morphogenesis (13), and apoptosis (30, 42, 43). Toward the goal of performing a higher-throughput screen for similar genes, we have exploited the observation of Roberts and Fink (49) that pseudohyphally growing diploid yeast invade agar when grown on low-nitrogen media but not on rich media (49). In screening for human genes that cause yeast to invade the agar on rich medium, we have identified a novel gene, human enhancer of invasion 10 (HEI10), and other genes which will be described elsewhere. As described here, HEI10 encodes a novel protein that induces multiple canonical changes associated with a pseudohyphal transition when expressed in diploid yeast. We find that HEI10 interacts physically and functionally with a mitotic cyclin and the ubiquitin ligation machinery in yeast cells and in higher eukaryotes. Based on structure-function studies exploiting HEI10-induced yeast phenotypes, we identified here an amino-terminal RING-like finger motif, characteristic of E3 ubiquitin ligases, and additional sequences including consensus cyclin and cyclin-dependent kinase (cdk) phosphorylation sites. These and other supporting data suggest the involvement of HEI10 in the regulation of cell cycle controls governing progression through G2.
MATERIALS AND METHODS
Cell lines, yeast and bacterial strains, and plasmids.
MCF-7 human breast adenocarcinoma, HeLa human cervical carcinoma, and U2OS human osteosarcoma cell lines were maintained in a solution containing Dulbecco modified Eagle medium,10% fetal bovine serum, penicillin, and streptomycin (and glutamine, for maintaining U2OS cells). The Saccharomyces cerevisiae strain EGY48 (MATα ura3 trp1 his3 6lexAop-LEU2) was used for yeast two-hybrid screening. CGX74 (MATa/α trp1/trp1) and CGX75 (MATa/α trp1/trp1 ura3/ura3) (gifts of C. Gimeno) were used in the agar invasion screen as described in Results and in a functional assessment of HEI10. The bacterial strain BL21(DE3) (Novagen) was used for expression and purification of glutathione S-transferase (GST)-fused proteins. The Clb2p expression plasmids were obtained from S. Holloway and are described elsewhere (20). The FLO11-LacZ plasmid was a kind gift of Anne Dranginis. The HEI10 cDNA (834 bp; 277 amino acids [aa] plus the stop codon) used as the basis of the constructs described here was PCR amplified from the original clone obtained in the agar invasion screen with EcoRI and XhoI sites added to the 5′ and 3′ ends, respectively. The HEI10 endpoints of all constructs used are provided in Results. Chromosomal assignment of HEI10 was based on comparison of the cDNA to genomic clones in the human genome sequencing database.
Antibodies.
The anti-HEI10 (α-HEI10) antibody was raised in rabbits immunized with a HEI10 carboxy terminally derived peptide (aa 259 to 277; SRELEQQQVSSRAFKVKRI, showing no similarity to other sequences in GenBank; Research Genetics). HEI10-specific antibody was affinity purified against the synthesized peptide as described previously (31). Other antibodies used in the present study included α-LexA rabbit polyclonal antibody (12), α-phosphothreonine-specific polyclonal antibody from Zymed (South San Franscisco, Calif.), α-GST, and α-hemagglutinin (HA) mouse monoclonal antibodies from Santa Cruz (Santa Cruz, Calif.), α-ubiquitin rabbit polyclonal antibody from Sigma, α-alpha tubulin monoclonal antibody from Sigma, α-TUB2 polyclonal antibody (a gift from V. Guacci), α-cyclin B rabbit polyclonal (46), and secondary α-rabbit and α-mouse antibodies conjugated to horseradish peroxidase for chemiluminescence detection from Amersham Life Sciences (Arlington Heights, Ill.) and rhodamine-conjugated α-rabbit antibodies from Molecular Probes (Eugene, Oreg.).
Northern blot analysis.
The full-length HEI10 cDNA (834 bp) was labeled with [α-32P]dCTP by random priming and used to probe a multiple-tissue Northern blot containing 2 μg of mRNA per lane prepared from different human tissues (Clontech, Palo Alto, Calif.). An equivalent mRNA load in each lane was confirmed through probing with similarly labeled actin-cDNA, and this procedure has been described elsewhere (29).
Preparation of cell lysates and Western blot analysis.
Cell lysates were prepared as previously described (29, 31) by lysis in PTY buffer supplemented with protease inhibitors. Western blot analysis was performed and proteins were visualized by using goat α-rabbit or α-mouse antibodies (1:5,000) in conjunction with enhanced chemiluminescence (NEN Life Science, Boston, Mass.). The signal intensity was quantitated by using scanning blots and by analyzing the data with NIH Image software.
Yeast microscopy.
CGX74 yeast containing pJG4-4 plasmids as described in Results were streaked from a W-Glu master plate to single cells on synthetic low-nitrogen (SLAHGR [2% galactose, 1% raffinose] or SLAHD [2% dextrose]) plates and allowed to grow at 30°C for 15 to 18 h. The colonies were visualized under an inverted microscope (Nikon TE300) with a ×20 objective lens. For time-lapse viewing, a ×40 lens was used to visualize the budding cells for 18 to 20 h after plating. Images were captured by using a Quantix-cooled charge-coupled device camera (Photometrics). For bud scar staining, cell cultures were grown in W-Glu overnight at 30°C and then transferred to W-Gal/Raff and grown to an optical density at 600 nm (OD600) of 0.4. The cultures were fixed in formaldehyde, harvested by centrifugation, washed with 1× phosphate-buffered saline, resuspended in Calcofluor stain (Sigma; prepared in distilled water at 1 mg/ml), and incubated at room temperature in the dark for 5 min. Cells were then washed twice with distilled water and mounted by using 1% low-melting-point agarose. Bud scars were visualized under a Nikon E800 microscope, with counts made of yeast with at least four detectable bud scars.
Fluorescence-activated cell sorting (FACS) analysis.
CGX75 yeast containing the constructs to be assessed (see Results) were grown in UW-Glu liquid medium overnight at 30°C. Cells were diluted in UW-Gal medium to an OD600 of 0.15 and grown for ∼6 h (two to three divisions) up to an OD600 of 0.4 to 0.6. Yeast cells were fixed in 70% ethanol, pelleted, and sonicated in Tris-HCl (pH 7.5). Cells were incubated with 50 μl of RNase A (10 mg/ml) for 1 h at 37°C. Cells were then mixed with 5 μl of proteinase K (20 mg/ml) and incubated at 50°C for 1 h. Each sample was supplemented with 5 μl of propidium iodide (2.5 mg/ml; Sigma) and incubated at room temperature in the dark for 1 h. The samples were analyzed for propidium iodide content on a Becton Dickinson FACScan.
Interaction trap/two-hybrid analysis.
The reagents and technical details are described at length by Golemis and Khazak (17). LexA fused to the amino terminus of HEI10 (aa 1 to 143; note that full-length HEI10 strongly activates transcription) was used as bait in a yeast two-hybrid screen of a human fetal brain library by using standard approaches (17). Over 600,000 primary transformants were screened, and positive results were confirmed by retransformation into naive EGY48/pJK103/LexA-HEI10N yeast. As controls for specificity, HEI10 interactions were separately evaluated with cyclins C, D2, and E; cyclin B and UbcH7 interactions were evaluated with negative control baits, including LexA-bicoid and LexA-HEF1. All were negative.
Immunofluorescence.
Cells were seeded on coverslips at a level of 25% confluence at 24 to 36 h prior to processing. Cells were fixed with 3.5% formaldehyde, permeabilized with buffer KB (10 mM Tris [pH 7.5], 150 mM NaCl, 0.1% bovine serum albumin) with 0.2% Triton X-100 for 5 min, washed in buffer KB, and then treated for immunofluorescence by standard procedures (43) with antibody to HEI10 and rhodamine-conjugated anti-rabbit antibody for the secondary antibody, with DAPI (4′,6′-diamidino-2-phenylindole) added in some experiments to visualize the nuclei. In peptide block experiments, 200 μg of the HEI10 C-terminal peptide was included in incubation with primary antibody. Coverslips were then washed six times (90 s each) with buffer KB and mounted on a slide by using Vectashield antifade mounting medium (Vector Laboratories).
Subcellular fractionation.
MCF7 and U2OS cells expressing endogenous HEI10 were grown to 80 to 90% confluence. The plates were washed once with phosphate-buffered saline and then incubated for 10 min on ice in a hypotonic buffer (10 mM Tris [pH 7.4], 0.2 mM MgCl2, and 5 mM KCl, supplemented with 1 mM NaVO4, 10 μg of leupeptin and aprotinin/ml, and 20 μg of phenylmethylsulfonyl fluoride/ml). Cells were scraped off the plate and Dounce homogenized (Wheaton 7-ml type A pestle, 25 strokes). The homogenate was supplemented with sucrose (to 250 mM) and EDTA (pH 8.0; to 1 mM) and was centrifuged at 1,000 × g for 5 min at 4°C. The pellet fraction (P1 fraction, nuclei) was resuspended in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4; 1% NP-40; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA). The supernatant was centrifuged in a Beckman swinging-bucket rotor (SW 55 Ti) at 100,000 × g for 1 h at 4°C. The resulting supernatant (S100) represents the cytosolic fraction.
Production and purification of GST fusions.
GST fusions were produced from the vector pGEX in the bacterial strain BL21 under standard conditions (1). After lysis of bacteria, GST fusions were purified by using glutathione-Sepharose beads (Pharmacia), eluted from beads on ice, and either used fresh or stored at −80°C.
Kinase assay.
For each assay, an active cyclin-cdk complex was first generated by mixing 125 μl of Sf9 cell lysates overexpressing cdk2 or cdc2 and 125 μl of His-tagged cyclin E or B bound to nickel beads in the presence of ATP (to 1 mM) and MgCl2 (to 10 mM) and then rocked at room temperature for 30 min, as described earlier (28). Activation of the complexes was determined by assessing kinase activity on histone H1 substrate (Sigma) as described below. For the kinase assay, 1 μl of the complexed and activated cyclin-cdk was mixed with 1 μg of substrate (HEI10-GST, HEI10-GST mutant, or GST alone), 10 μCi of [γ-32P]ATP (for autoradiography), or 1 mM ATP (for antibody visualization of phosphothreonine and mobility shift), and the volumes of the mixes were brought to 30 μl by using a kinase buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, and freshly added 1 mM dithiothreitol). The kinase reaction was allowed to proceed for 30 min at 30°C. The reactions were stopped by adding sodium dodecyl sulfate (SDS)-containing sample buffer to the tubes, and the mixtures were resolved by SDS-polyacrylamide gel electrophoresis (PAGE).
Cell cycle progression.
Cycling Xenopus egg extract was prepared and used to monitor cell cycle progression as described earlier (40). Sperm nuclei were added to the extract at 500 nuclei/μl, together with either buffer or 1 μg of GST or GST-HEI10 fusion proteins. Aliquots were taken at the times indicated in Results and either fixed for immunofluorescence or used for Western analysis to detect cyclin B or tubulin expression.
Ubiquitin ladder formation in Xenopus eggs.
Cytostatic factor-arrested Xenopus egg extract was prepared as described by Murray (40). The extract (45 μl) was mixed with 1 μg of GST (5-μl volume) or GST-HEI10 and incubated at room temperature for 20 min. GST beads were then added to the reaction mix to pull down the recombinant proteins. The beads were washed five times with NETN (20 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.5% NP-40; 100 mM NaCl) and processed subsequently by standard SDS-PAGE and immunoblotting.
Ubiquitination assay.
E1, E2, and E3 proteins were separately expressed in bacteria and prepared as crude cell lysates. A mixture was prepared by combining 75 μl of ubiquitination buffer (20 mM Tris-HCl, pH 7.4; 50 mM NaCl; 4 mM ATP; 0.2 mM dithiothreitol; 10 mM MgCl2) with 5 to 10 μl of crude E2, 2 μl of ubiquitin (1 μg/μl; Sigma), 5 μl of crude GST-E3 (1 μg of GST-HEI10), and 5 μl of crude E1, followed by incubation at 30°C for 25 min. GST-Sepharose beads (40 μl/reaction) were added, and incubation continued at 4°C for 45 min on a nutator. Beads were spun down at 1,000 rpm at 4°C for 1 min, washed three times with 500 μl of EBC buffer (50 mM Tris HCl, 120 mM NaCl, 0.5 mM EDTA, 0.5% NP-40), and samples were processed subsequently by standard SDS-PAGE and immunoblotting. Ubiquitination was visualized by using rabbit α-ubiquitin antibody (Sigma) at a 1:2,000 dilution.
RESULTS
HEI10 isolation based on regulation of the yeast pseudohyphal program.
To screen for candidate human growth regulatory genes, a HeLa cDNA library cloned in the vector pJG4-4 and expressed under the control of the GAL1 promoter was transformed into the CGX74 diploid strain of S. cerevisiae. About 500,000 primary transformed colonies were obtained, scraped, pooled, and screened for invasion promoting genes by plating library-containing CGX74 on inducing (galactose) rich medium for 60 h; plates were then washed with vigorous blasts of distilled water. Microcolonies growing beneath the agar surface were isolated and retested for agar invasion on galactose- but not glucose-containing rich medium. Plasmids were recovered from colonies with this phenotype, transformed versus vector pJG4-4 into naive CGX74, and reassayed for agar invasion (Fig. 1A). Clones recapitulating the initial phenotype were designated HEI genes (for human enhancers of invasion) and sequenced. We focus here on the characterization of the novel HEI10 gene; other genes arising from the screen are described elsewhere.
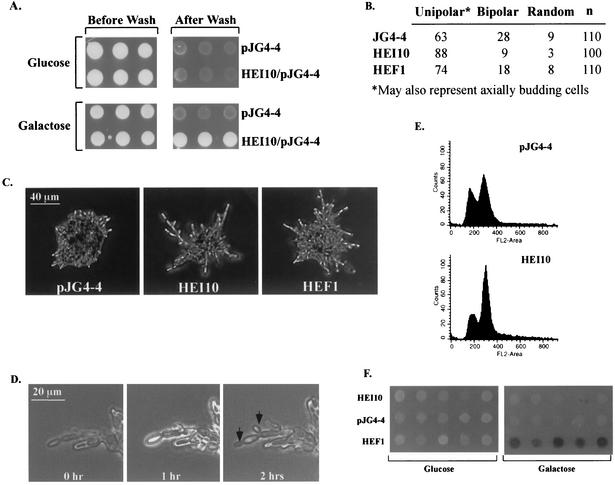
FIG. 1.
HEI10 profile in yeast. (A) Expression of HEI10 in yeast induces agar invasion. HEI10 was expressed in yeast under the control of the GAL1 promoter from the vector pJG4-4. Yeast expressing HEI10 remain attached to the agar after exposure to vigorous blasts of water. (B) Budding patterns of yeast expressing HEI10. Yeast were stained with Calcofluor to visualize their previous budding sites. Yeast were grouped according to budding pattern under unipolar (bud scars clustered at one pole), bipolar (bud scars at opposite poles), and random budding. (C) Filamentation phenotype of yeast expressing pJG4-4, HEI10/pJG4-4, or HEF1/pJG4-4 after culture on SLAHGR medium for 24 h. (D) Bud emergence in HEI10-expressing yeast occurs simultaneously in mother and daughter, a characteristic of pseudohyphal growth. HEI10-expressing yeast cells were streaked onto single cells on SLAHGR plates, grown for 18 h, and imaged at 1-h time intervals (magnification, ×28). Arrows indicate bud emergence sites. (E) HEI10 expression leads to accumulation in G2. A FACS profile of yeast expressing pJG4-4 versus HEI10 is shown. (F) HEI10 fails to activate the FLO11-LacZ reporter. Yeast cells expressing pJG4-4, HEI10, or HEF1, together with a FLO11pro-LacZ reporter, were plated on uninducing (glucose) or inducing (galactose) media containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to assess reporter activation.
The program of pseudohyphal growth induction in yeast is becoming well documented and is characterized by a set of stereotypic changes in cell morphology, cell signaling, and cell cycle. Some key hallmarks of pseudohyphal growth include (i) filamentation of diploid yeast propagated on low nitrogen, characterized by cellular hyperelongation; (ii) conversion of the budding pattern from bipolar to unipolar; (iii) alteration in the kinetics of bud emergence such that mother and daughter cells bud simultaneously; and (iv), correspondingly, enlargement of the G2 compartment of the cell cycle (2, 10, 25). We assessed multiple parameters to confirm that HEI10 specifically regulated pseudohyphal conversion, as opposed to inducing nonspecific cellular aggregation, to allow us to consider HEI10 function in the context of known yeast pseudohyphal regulatory pathways.
First, the ability of HEI10 to enhance the filamentous growth of yeast on low-nitrogen medium was studied. Diploid strains expressing HEI10, HEF1 (as positive phenotypic control), or empty vector were streaked onto single cells on low-nitrogen plates and grown at 30°C for 18 h. Pseudohyphal yeast form a chain of attached cells with a hyperelongated shape marked by an axial ratio (AR) of ∼3.0, whereas vegetatively growing yeast have an AR of 1.5, reflecting an average of 6 μm in length and 4 μm in width (16). These values correspond well to the ratios observed in yeast expressing HEI10 (9.2-μm length, 3.0-μm width, AR 3.1) and the previously identified HEF1 (10.6-μm length, 3.0 μm-width, AR 3.6) versus cells expressing empty vector (6.3-μm length, 4.1-μm width, AR 1.5). Second, the percentage of cells showing buds concentrated at one end of the cell (unipolar or axial budding) or at both ends of the cell (bipolar budding) was determined (Fig. 1B). Under the conditions of the assay (49), the strong prediction is that enhancement of the unipolar-axial compartment reflects increased unipolar budding. Diploid strains overexpressing either HEI10 or HEF1 have a significantly higher ratio of unipolar-axial to bipolar budding cells compared to yeast transformed with pJG4-4, similar to previously reported values for enhanced filamentation. Consistent with the patterns of AR and unipolar budding increase, HEI10 microcolonies on low-nitrogen medium have the stellate appearance associated with pseudohyphal growth (Fig. 1C). Third, the kinetics of bud emergence were examined in yeast expressing HEI10 or empty pJG4-4 vector. Bud emergence in HEI10-expressing yeast occurs simultaneously in the mother and daughter cells, conforming to the pattern expected in pseudohyphal growth (Fig. 1D). Finally, induction of HEI10 in yeast leads to a significant accumulation of cells in G2 (Fig. 1E).
Pseudohyphal growth is frequently, although not inevitably, characterized by an increase in signaling through a STE20/STE11/STE7/STE12 MAPK cascade or, alternatively, through the activation of PKA signaling. A consequence of either MAPK or PKA pathway activation is induction of transcription through the STE12/Tec1 responsive FLO11 promoter (34). The ability of HEI10 to activate the FLO11 promoter was studied (Fig. 1F). The diploid strain CGX75 was transformed with HEI10, HEF1, or pJG4-4, along with FLO11-LacZ (34). HEI10 and pJG4-4 both failed to activate the transcription of LacZ from the FLO11 promoter, whereas HEF1 strongly induced the expression of FLO11-LacZ (Fig. 1F).
These results supported the idea of a HEI10 function specific to pseudohyphal signaling but also suggested that HEI10 impact on yeast growth does not involve canonical activation of the MAPK or the PKA pathways.
Definition of the HEI10 gene and protein.
The sequence of the HEI10 clone obtained from the screen contains an open reading frame of 834 bp encoding a 277-aa protein with a predicted molecular mass of 32 kDa (Fig. 2A). The sequence was used as the basis of a BLAST probe of the human expressed sequence tag and The Institute for Genomic Research databases, and a longer clone of 1,503 bp was assembled that includes ∼469 bp of the 5′-untranslated region, the 834-bp coding region, and ∼200 bp of the 3′-untranslated region. The assembled HEI10 sequence has an upstream stop codon (at position −177 bp) and no intervening in-frame methionine codons, indicating that the clone identified by invasion screening encodes the full-length protein. HEI10 is expressed as a single transcript of ∼1.6 kb in a number of human tissues (Fig. 2B), a finding compatible with the estimation described above. The expression level varied among tissues, from lower levels in the placenta, brain, and lung to moderate levels in the kidney and liver to the highest levels in the heart. The HEI10 gene resides on human chromosome 14q11.1 and encompasses HEI10 within three exons. Of potential interest, one recent report has demonstrated that HEI10 is a component of a translocation fusion to the HMGIC gene in a uterine leiomyoma (37), whereas some studies have suggested involvement of this locus in the development of additional cancers. Two pseudogenes for HEI10 (based on 85 to 90% overall sequence homology, coupled with the lack of internal splice sites) exist: one at 20q11.2-13.2 and one at 1p34.
FIG. 2.
Structure of the HEI10 protein and HEI10 RNA expression. (A) HEI10 encodes an 277-aa open reading frame. A diagram of HEI10 domains is presented, with structural motifs as described in Results. (B) HEI10 transcript is expressed to different levels in a number of human tissues. (C) Sequence alignment of HEI10 candidate RING domain versus other RING fingers. The c-Cbl RING finger is presented as a reference C3HC4 RING consensus sequence, with core cysteines indicated in green. Deviation of HEI10 orthologs from five different vertebrate species from the c-Cbl RING consensus residues is indicated in red. Other RING fingers from proteins with confirmed E3 ligase activity are presented as context; these include the proteins HHAR1, Doa10, TEB4, HRD1, and Parkin.
Analysis of both the primary amino acid sequence of HEI10 and secondary structure predictions revealed several notable features. The first (aa 4 to 50) is closely related to a C3HC4 RING finger motif (7), although sufficiently divergent not to be predicted by domain prediction computational algorithms (Fig. 2A and C). RING fingers have been specifically implicated in functions, including cell cycle progression signal transduction and other processes (14), and are found in E3 ubiquitin ligases. Second, aa 108 to 196 are predicted to form a coiled coil (using the structure prediction engine at www.expasy.ch). Other candidate motifs of interest to HEI10 function are discussed below. Closely related protein sequences can be identified in numerous vertebrate species, although not in lower eukaryotes. The 87% amino acid identity from human to mouse orthologs of HEI10 is slightly higher than the average (36), whereas the 100% identity between aa 1 to 83, encompassing the RING motif, is indicative of an important function.
HEI10 interaction with cyclin B and UbcH7 and delineation of HEI10 functional motifs.
To gain insight into the mechanism of HEI10 action both in yeast and in human cells, the two-hybrid system was used to identify potential HEI10 interactive partners by using a bait representing the amino-terminal 143 aa of HEI10. Clones encoding cyclin B1 (aa 315 to end [5]) and the E2 ubiquitin-conjugating enzyme UbcH7 (aa 29 to end [41]) were isolated as candidate interacting partners of HEI10. Subsequent specificity analysis indicated that neither interacted with a set of negative control baits (see Materials and Methods). These two potential interactors were of considerable interest, based on their defined functions in regulating cell cycle progression and cellular signaling. Notably, cyclin B1 is homologous to the S. cerevisiae mitotic cyclin Clb2p. Reduction of Clb2p levels or activity promotes filamentous growth via extension of G2 (2, 10), whereas overexpression of Clb2p can reverse cell cycle mutations promoting pseudohyphal growth (3, 10, 50). Moreover, a single point mutation of Cdc28p (C127Y), which causes defective interaction between Cdc28p and Clb2p, is also sufficient to induce filamentous growth (10). Furthermore, the carboxy-terminal end of HEI10 encompasses two S/TPX (basic residue) cell cycle kinase consensus sequences (NTPVR at aa 220 to 224 and VSPSR at aa 256 to 260) (Fig. 2A). Based on a detailed analysis of the recognition preference of cdk-cyclin partners, the NTPVR site has been reported as a favored substrate of cyclin B/cdc2 (21). The presence of a RING finger-like domain at the amino terminus of HEI10 was also of immediate relevance to further analysis of a HEI10-UbcH7 interaction, given the known action of RING fingers in E3 proteins in conferring association with E2 ubiquitin-conjugating enzymes (23, 47).
We next utilized the HEI10-dependent yeast phenotypes to establish the importance of the RING finger and the NTPVR and VSPSR motifs to HEI10 functionality. The results, summarized in Fig. 3A, show that the deletion (HEI10-ΔRING) or point mutation (HEI10-CHC-AAA) of the RING finger sequence abrogated the ability of HEI10 to induce either agar invasion or filamentation (also data not shown). In contrast, deletion of the carboxy-terminal 78 aa of HEI10 (HEI10-Δ200-277, eliminating both potential cdk-cyclin phosphorylation motifs) did not result in a reduction of these phenotypes (Fig. 3A), although further truncation of HEI10 to a fragment corresponding to the two-hybrid bait (HEI10-N, aa 1 to 143) eliminated HEI10 invasiveness (not shown). FACS analysis of yeast expressing HEI10 and derivatives mirrors the agar invasion results. Cells expressing HEI10-ΔRING had a G1 population similar to that observed with vector only, but those expressing HEI10-Δ200-277 had a reduction in G1 compartmentation that was comparable to or slightly greater than that obtained with full-length HEI10 (Fig. 3B). Finally, Western analysis of yeast lysates expressing HEI10 and derivatives excludes reduction in HEI10 protein levels associated with particular mutants as an explanation for these results (data not shown).
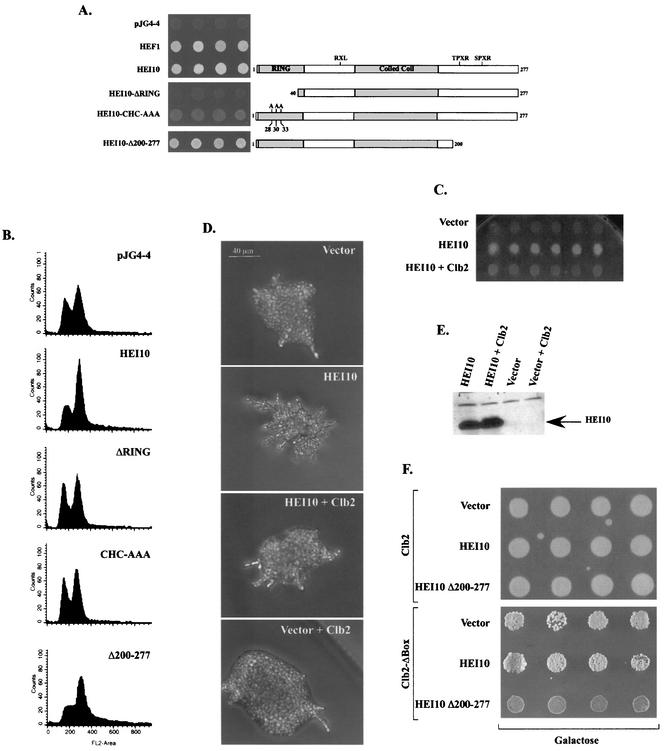
FIG. 3.
Structure-function relationship of HEI10 domains and budding phenotypes. (A) HEI10 and HEI10 mutants assessed are shown (diagram on right). The ability of HEI10 and each derivative to induce agar invasion and aggregation is shown; each patch represents an independent transformant. For this assay, HEI10 or HEI10 mutants were expressed under the control of the GAL1 promoter and were grown on glucose or galactose and then washed vigorously with water. (B) FACS profiles of the yeast strains shown in panel A. (C) HEI10's ability to induce invasion on rich media is impaired upon coexpression of Clb2p. Yeast express proteins are as indicated; the wash assay was as described for panel A, with each patch representing an individual transformant. (D) HEI10 induction of filamentation on low-nitrogen media is impaired by coexpression with Clb2p. Filamentation assay was as shown in Fig. 1C; a representative colony is shown. (E) Western blot analysis of HEI10 expression in yeast cells in the presence or absence of Clb2p. (F) Growth on rich medium with galactose for yeast-expressing vector, HEI10, or HEI10-Δ200--277, together with Clb2p or a Clb2p-Δbox mutant. Each patch represents an individual transformant.
Clb2p overexpression inhibits yeast filamentation in HEI10 expressing yeast.
In order to further characterize the functional relationship between HEI10 and Clb2p, we studied the ability of Clb2p overexpression to inhibit HEI10 function in yeast. HEI10 and vector, or HEI10 and Clb2p, were coexpressed in CGX75, and yeast cells were grown on rich media to observe their ability to invade the agar substratum. Whereas HEI10-expressing cells efficiently invaded the agar, cells coexpressing Clb2p with HEI10 failed to invade the substratum (Fig. 3C). In parallel, the ability of Clb2p to inhibit HEI10 induction of filamentation on low-nitrogen media was studied (Fig. 3D). Similar to the effect observed on rich media, the coexpression of Clb2p interfered with HEI10 ability to induce filamentation on SLAHGR plates (Fig. 3D). Expression of HEI10 was monitored in both cases in order to confirm that failure to induce filamentation is not due to loss of HEI10 expression in the presence of excess Clb2p (Fig. 3E). These results are in accord with the hypothesis that HEI10 and Clb2p function in the same pathway in an antagonistic manner.
Yeast cells unable to degrade Clb2p undergo arrest at the G2/M boundary. The Clb2p-Δbox mutant (20), lacking both a cyclin destruction box and a KEN box, shows a slow-growth phenotype due to the cell's very limited ability to degrade this mutant form of Clb2p. To further probe genetic interactions of HEI10 with Clb2p, we examined the effect of HEI10 or HEI10 derivatives on the growth of yeast coexpressing the Clb2p-Δbox mutant (Fig. 3F and data not shown). The coexpression of HEI10 and nonfunctional derivatives (e.g., HEI10-ΔRING) did not significantly affect the Clb2p-Δbox reduced growth phenotype or the growth of yeast overexpressing wild-type Clb2p. In contrast, cells coexpressing HEI10-Δ200-277 and Clb2p-Δbox completely failed to grow, although cells coexpressing this HEI10 derivative with wild-type Clb2p were unaffected. This allele-specific interaction further supported the idea of specific functional interactions between HEI10 and Clb2p, as discussed further below.
HEI10 expression in human cell cycle.
Although pseudohyphal and two-hybrid results predicted that HEI10 functions in cell cycle control, direct demonstration of this point was necessary in higher eukaryotes. Western analysis with an antibody specific for the carboxy-terminal 19 aa of HEI10 (HEI10-19C, Fig. 4) applied to whole-cell lysates prepared from a number of different cell lines recognized 32- and 19-kDa species; in one cell line, MCF-7, an additional ∼29-kDa species was recognized (Fig. 4A, left two lanes, and data not shown). The 32-kDa species shows a gel mobility identical to that of exogenously expressed HEI10 (data not shown), and the reactivity of the anti-HEI10 antibody with this and the 29-kDa species is lost if the antibody is preincubated with HEI10-19C peptide (Fig. 4A, right two lanes), suggesting the 32-kDa species represents endogenous HEI10. In contrast, the ∼19-kDa species is still recognized by anti-HEI10 antibdoy preincubated with HEI10-19C (Fig. 4A), suggesting a nonspecific cross-reaction. Immunofluorescence analysis of HEI10 in U20S, HeLa, or MCF-7 cells indicates that the predominant intracellular localization of the protein is nuclear (Fig. 4B and results not shown), whereas minor signal is also detected at the cell periphery. This staining pattern is not observed when cells are incubated with peptide-blocked antibody or secondary antibody alone (Fig. 4B and results not shown).
FIG. 4.
Expression of endogenous HEI10. (A) Western visualization of HEI10 in MCF-7 or U20S cells with anti-HEI10 antibodies (left) or with antibodies plus blocking peptide (right panel). (B) Immunofluorescence analysis of MCF-7 cells show that HEI10 with the antibody is predominantly a nuclear protein (left panel); in contrast, staining with the peptide-blocked anti-HEI10 shows only weak cytoplasmic staining (center panel). In cells undergoing mitosis (right panels), HEI10 signal colocalizes with DAPI-stained DNA (brightness adjusted versus other images). (C) Cell fractionation of MCF-7 lysates. HEI10 is present in the nuclear fraction of MCF-7 lysates (N) but not in the cytosolic fraction (C). A Western blot of the same lysates with antipaxillin, a cytosolic protein, is shown as a control for the purity of the fractions. The 19-kDa band is predominantly present in the cytosolic fraction.
Although all cells demonstrate similar weak juxtamembrane staining, most cells have bright nuclear staining (between 80 to 90%), whereas some cells have little or no nuclear staining (10 to 20%) (results not shown). To independently confirm the localization patterns predicted by immunofluorescence, MCF7 cells were fractionated (31) into P1 (nuclear) versus S100 (cytoplasmic) fractions and then probed with anti-HEI10 (Fig. 4C). The 32-kDa species and the 29-kDa species were restricted exclusively to the P1 nuclear fraction, whereas the 19-kDa nonspecific band was almost exclusively restricted to the S100 cytoplasmic fraction, supporting the immunofluorescence results. It was of clear interest to determine whether HEI10 localization was subject to cell cycle control. The frequency of HEI10 nuclear staining was compared in asynchronous exponentially growing cells versus cells in stationary phase (predominantly G0/G1) and hydroxyurea-arrested cells (predominantly in S phase). Strong nuclear staining for HEI10 is observed in 66% of exponentially growing cells and in 41% of cells in stationary phase but only in 16.5% of cells arrested in S phase. Intriguingly, upon careful examination of cells progressing through mitosis, we observed that HEI10 was associated with segregating chromosomes during metaphase and anaphase (Fig. 4B, right panel, and results not shown), suggesting that HEI10 may associate with DNA or chromatin-binding proteins under at least some cell cycle conditions.
HEI10-dependent inhibition of mitotic entry in higher eukaryotes: functional interactions with cyclin B1 and Clb2p.
Repeated efforts by multiple approaches to overexpress HEI10 in mammalian cells were unsuccessful in that they resulted in the appearance of aggregated proteins associated with the perinuclear endoplasmic reticulum (results not shown). Although gross protein truncation yielded HEI10 derivatives with nuclear localization, it was undesirable to use such truncations for functional experiments. We therefore utilized a heterologous vertebrate system to analyze HEI10 function in higher eukaryotes. Xenopus oocyte extracts undergo controlled replicative cycles in vitro and have been heavily exploited as a biochemical model for mammalian cell cycle, as discussed earlier (51). After stimulation of cycling, egg extracts undergo a synchronized entry into M phase that is characterized by the breakdown of the nuclear envelope. Extracts were stimulated to cycle in the presence of assay buffer, GST only, or GST-HEI10. As shown (Fig. 5A), the addition of HEI10 resulted in both a significant delay in the timing of nuclear envelope breakdown and a substantial reduction in the absolute number of cells showing such breakdown. These results reinforced the yeast results showing a HEI10-dependent delay of entry into M phase, strongly implying that HEI10 would also function in the control of mitotic entry in human cells.
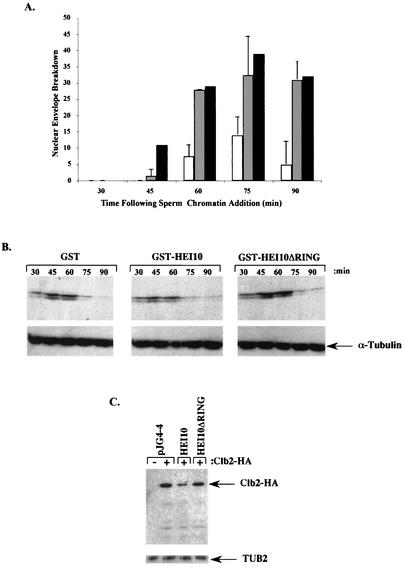
FIG. 5.
HEI10 expression limits mitotic entry and limits cyclin B-Clb2p accumulation. (A) Nuclear envelope breakdown of Xenopus extracts after sperm addition in the presence of GST-HEI10 (open bars), buffer (shaded bars), or GST (solid bars). (B) Egg lysates shown after incubation for the times indicated after sperm addition in the presence of GST, GST-HEI10, or GST-HEI10-ΔRING were used for Western analysis with anti-cyclin B (top) or tubulin (bottom). (C) Western analysis of Clb2p-HA levels in yeast transformed with pJG4-4 vector, HEI10, or HEI10-ΔRING. The anti-tubulin control (TUB2) is shown at the bottom.
Based on the protein interaction and yeast results presented above, the most plausible explanation for the disruption of mitotic entry would be interference with the accumulation of cyclin B through function involving activity as a ubiquitin ligase. We first examined the profile of cyclin B accumulation in Xenopus egg extracts at various time periods after the stimulation of cycling in the presence of GST only, GST-HEI10, or GST-HEI10-ΔRING (Fig. 5B). In the presence of GST-HEI10, the total levels of cyclin B accumulation were depressed (Fig. 5B), and accumulation was delayed versus the other two conditions. Quantitation of results indicated a 50% decrease in cyclin B levels in GST-HEI10 versus the other two conditions at 45 and 60 min after the initiation of cycling, times corresponding to the time of normal nuclear envelope breakdown. As a separate measure of HEI10 involvement in control of mitotic cyclin stability, we transformed yeast with plasmids expressing HEI10, HEI10-ΔRING, or vector, together with plasmids expressing HA epitope-tagged Clb2p or vector only, and then examined the Clb2p levels (Fig. 5C). In findings paralleling the Xenopus results, HEI10 expression caused a significant reduction in levels of HA-Clb2p, whereas deletion of the HEI10 RING motif eliminated this activity. For yeast cells, quantitation of Westerns indicated a reduction in Clb2p levels of >50%. These results, obtained across a considerable evolutionary distance, strongly imply HEI10 would also function in the control of mitotic entry in human cells in a mechanism involving regulation of cyclin B stability.
Definition of HEI10 as an E3 ubiquitin ligase.
To directly examine ubiquitination contingent on HEI10, either GST or GST-HEI10 was added to Xenopus egg extracts arrested either in interphase or in M phase; GST proteins were then collected with glutathione-Sepharose beads, and Western analysis was used to determine whether significant posttranslational modifications of HEI10 itself occurred commensurate with interactions with proposed HEI10 partners. As shown (Fig. 6A), GST-HEI10 incubated with noncycling Xenopus extracts developed an extensive ladder of higher migrating species. The modified forms were most abundant in interphase extracts, although they were observed in mitotic extracts as well. The periodicity of molecular masses observed approximated the ∼9-kDa ladder characteristic of ubiquitin modification; in fact, reprobing of lanes with antibody to ubiquitin (Fig. 6B) showed significant reactivity in lanes containing GST-HEI10 but not in lanes containing GST.
FIG. 6.
HEI10 and ubiquitination. (A) Bacterially purified GST-HEI10 is shown as an input species (IN) or after incubation with synchronized interphase (IP) or M-phase (MP) Xenopus egg extract after visualization with anti-HEI10. (B) GST-HEI10 or GST bacterially purified proteins, after incubation with interphase (IP) or mitotic (MP) extract, visualized with anti-ubiquitin antibody. (C) Purified HEI10 was incubated in an in vitro ubiquitination cocktail containing either UbcH7, UbcH8, or E2D2 as E2 ubiquitin-conjugating enzyme and then visualized with to anti-ubiquitin antibody.
Finally, to probe the functionality of the HEI10 RING finger-like domain, we utilized an in vitro ubiquitin transfer assay. Purified GST-HEI10 was incubated in reactions containing either UbcH7, UbcH8, or E2D2 as E2 ubiquitin ligase. E3 proteins frequently show autoubiquitination as an indicator of activity, as discussed earlier (47). As shown (Fig. 6C), GST-HEI10 is ubiquitinated in the presence of UbcH7 but not in the presence of either of the other E2 proteins. These results indicate that HEI10 is active as an E3 ligase in a pure system and at least moderately specific in its requirement for an E2 activity.
HEI10 is a specific substrate of cyclin B-cdc2 in vitro.
Finally, we sought to determine whether HEI10 is a cyclin B-cdc2 substrate. Purified bacterially expressed GST-HEI10 was incubated with purified active cyclin B-cdc2 complex in the presence of [γ-32P]ATP. The presence of an ∼65-kDa band corresponding to autophosphorylated cyclin B indicates that the cyclin B-cdc2 complex is active (Fig. 7A, lanes 1 to 4). When HEI10-GST was added to the reaction as a substrate, an additional phosphorylated band (∼60 kDa) that comigrates with HEI10-GST was observed (Fig. 7A, lanes 2 and 3). The HEI10-GST band increased in intensity when increased amounts of active cyclin B-cdc2 were present in the reaction (Fig. 7A and B). In contrast, no phosphorylation of GST alone was observed under the same conditions (lane 1). To determine whether the phosphorylation of HEI10 specifically required to cyclin B-cdc2, we performed parallel in vitro kinase reactions to compare the ability of similarly prepared cyclin E-cdk2 versus cyclin B-cdc2 to use HEI10 as a substrate. HEI10-GST was not phosphorylated by an active (based on significant phosphorylation of histone H1) cyclin E-cdk2 complex under the same conditions that led to HEI10-GST phosphorylation by cyclin B-cdc2 (results not shown). We noted that the specificity of the HEI10 interaction with cyclin B was further supported by specificity testing in the yeast two-hybrid system, in which HEI10 failed to interact with a series of DNA binding-domain fused cyclins other than B1 (results not shown).
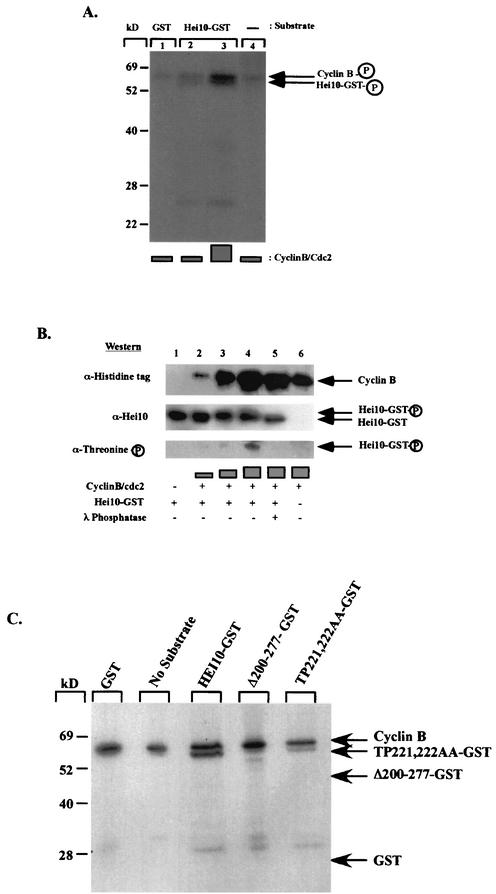
FIG. 7.
HEI10 is phosphorylated in vitro by cyclin B1/Cdc2. (A) Autoradiograph showing increased incorporation of radiolabeled [γ-32P]ATP in HEI10-GST with an increased amount of cyclin B-cdc2 (lanes 2 and 3). (B) Upon incubation with increasing amounts of cyclin B-cdc2, HEI10 mobility was decreased (lanes 2, 3, and 4). The mobility was increased if λ phosphatase was added to the reaction (lane 5). A band with a size similar to that of HEI10-GST was visualized by Western blot by using antiphosphothreonine-specific antibodies when HEI10-GST was incubated with increasing amounts of cyclin B-cdc2 (lanes 2, 3, and 4). (C) Autoradiograph showing in vitro phosphorylation of HEI10 and HEI10 mutants. Phosphorylation was abrogated in HEI10-Δ200-277, whereas it was strikingly reduced in upon mutation of TP221,222. The arrows on the left show the predicted mobilities of the various proteins.
As noted above, plausible sites for HEI10 phosphorylation by cyclin B-cdc2 were the motifs NTPVR at aa 220 to 224 and VSPSR at aa 256 to 260. Phosphorylation of HEI10-GST in a reaction with unlabeled ATP and increasing levels cyclin B-cdc2, followed by Western analysis with antibody to phosphothreonine, or in parallel to HEI10, indicated that appearance of a slower-migrating form of HEI10 corresponded to the appearance of an antiphosphothreonine-reactive species (Fig. 7B, lanes 3 and 4). This species and retarded migration of HEI10 are absent after incubation of reactions with lambda phosphatase (Fig. 7B, lane 5). Finally, full-length HEI10, HEI10-Δ200-277, and HEI10-TP221,222AA were purified from bacteria in parallel, and phosphorylation by cyclin B-cdc2 was analyzed by using radiolabeled [γ-32P]ATP (Fig. 7C). This analysis confirms that the primary site of cyclin B-cdc2 phosphorylation was the NTPVR motif, since mutation of this site eliminated the majority of HEI10 phosphorylation, whereas truncation of the carboxy terminus of HEI10 eliminated all cyclin B-cdc2 phosphorylation, suggesting that the VSPSR motif at aa 256 to 260 was the sole alternative phosphorylation site.
DISCUSSION
Based on the sum of the data presented above, we propose that HEI10 is a novel factor contributing to the control of entry into mitosis via regulation of the accumulation of cyclin B1. The data further suggest that HEI10 regulation of cyclin B may involve direct interaction of HEI10 with cyclin B and the function of HEI10 in the control of protein degradation.
Over the last decade, the pseudohyphal program in S. cerevisiae has been mechanistically dissected as a manipulable and informative model of cellular morphogenesis. Although several early studies of pseudohyphal approach emphasized the contribution of STE pathway MAPK signaling and, more recently, PKA signaling to this developmental switch, the failure of HEI10 to activate the FLO11 reporter excludes this explanation for its mode of function. A recent comprehensive review of the process by Rua et al. has characterized the regulation of filamentation as an excellent model demonstrating different means of signaling to the core cell cycle apparatus (50). Downregulation of Clb2p levels (by direct mutation of CLB2 [2] or by control of CLB2 transcription or Clb2p protein degradation) is a central inducing stimuli for filamentous growth.
We demonstrate here that HEI10 specifically induces G2 delay and associated filamentation processes dependent upon an intact RING finger motif in HEI10. The facts that HEI10 expression results in a decrease of steady-state levels of coexpressed Clb2p and that overexpression of Clb2p is sufficient to block HEI10-dependent phenotypes are compatible with the model that Clb2p is an important direct target in HEI10 induction of pseudohyphal growth. The fact that the RING finger of HEI10 is required for this phenotype is further compatible with the idea that HEI10 downregulates Clb2p through a mechanism involving its proteolysis rather than by affecting its transcription or through other means. However, these results do not exclude the possibility that HEI10 interacts with other yeast proteins involved in filamentation response, that HEI10 may affect Clb2p levels or activity by an indirect means, or that Clb2p in turn phosphorylates and hence regulates HEI10. In the context of the last point, the synthetic lethal expression observed between HEI10-Δ200-277 and Clb2p-Δbox is suggestive. One explanation for this phenotype would be to hypothesize that HEI10 normally interacts with and promotes the degradation of Clb2p and other substrates, whereas Clb2p in conjunction with Cdc28p phosphorylates and reciprocally downregulates the activity, but not the protein levels, of HEI10. With a nondegradable Clb2p and a noninactivatable HEI10, cells may be terminally blocked at G2/M. Further tests of this model are in progress.
Analysis of HEI10 in the context of Clb2p were prompted by the observations that a bait consisting of the amino-terminal 143 aa of HEI10 isolated cyclin B1 from a two-hybrid screen and that HEI10 is a specific target of phosphorylation by cyclin B-cdc2 in vitro. HEI10 addition to cycling Xenopus extracts grossly reduced nuclear envelope breakdown, a process dependent upon cyclin B activity, and resulted in a simultaneous reduction in cyclin B levels. HEI10 also interacts with an E2 ubiquitin-conjugating enzyme, specifically autoubiquitinates in the presence of this E2, UbcH7 in vitro, and is ubiquitinated in a cell cycle-specific manner in Xenopus egg extracts. These results, coupled with the results in yeast, raise the clear possibility that HEI10 acts as an E3 ligase for cyclin B1 in higher eukaryotes and is itself downregulated through action of the cyclin B-Cdc2 mitotic kinase. However, technical limitations in the study of HEI10 have thus far prevented the direct confirmation of this model. HEI10 expressed in bacteria does not promote the ubiquitination of cyclin B in vitro. This may be due to the requirement for additional partners for the reaction, or it may reflect the fact that UbcH7 is not in fact the correct E2-conjugating enzyme for HEI10 (a possibility discussed in greater detail below), or it may suggest that HEI10 must be activated by some eukaryote-specific posttranslational modification to be effective. Although it would be possible in principle to immunoprecipitate and thus purify HEI10 from a eukaryotic source, in practice the antibody available to HEI10 does not immunoprecipitate effectively, in part because endogenous HEI10 tends to be insoluble unless harsh cell lysis are used. These technical problems also render moot the possibility of experiments to coimmunoprecipitate HEI10 with antibodies to cyclin B, whereas expressing HEI10 as a fusion protein to an immunoprecipitable epitope tag results in loss of HEI10 functional activities in yeast (results not shown), and other problems related to HEI10 overexpression in mammalian cells are discussed further below.
In further comment upon the HEI10 ubiquitination data presented here, it is also not clear whether HEI10 autoubiquitinates in vivo or whether UbcH7 is involved in this process. We note that, whereas the yeast ortholog of UbcH7 protein is restricted to a localization in the endoplasmic reticulum and involved in the unfolded protein response, the mammalian protein is more broadly distributed throughout the cell (54) and has been shown to function in conjunction with diverse other classes of protein (4, 52, 58), making an interaction plausible. However, reviews of E3 activity (47) have noted the possibility of mechanistically informative artifacts in which a proubiquitination activity can be revealed in vitro but differs from the observed specificity of a protein in vivo. In the context of this discussion, we note that the HEI10 RING finger sequence, although highly conserved among HEI10 orthologs, is divergent from other previously described RINGs. An alignment of the HEI10 RING sequence versus other previously described E3 proteins (Fig. 2C) reveals two points of divergence from the established RING consensus. RING finger proteins usually have a C3HC4 sequence of cysteines and histidines: HEI10 instead shows a C3HCHC2 sequence. Further, the spacing between the first two cysteines in the consensus is described as CxxC; the first Zn-binding moiety of HEI10 is Cx4C. Nevertheless, scrutiny of a number of recently defined RING finger proteins identifies several, including Parkin and HHAR1, that differ significantly from the consensus but nevertheless retain E3 activity (see reference 4). HEI10 further extends the allowable sequence variation in this functional group.
It is additionally of interest that the Northern data for HEI10 distribution raise the possibility that HEI10 is differentially expressed in different cell types, given recent proposals that different anaphase-promoting complex (APC) activators may contribute to the temporal and spatial control of APC outside the cell cycle (see, for example, reference 55). Certainly, many researchers have investigated the regulation of cyclin B proteolysis by the anaphase-promoting complex in detail without having previously identifyied HEI10. However, recent studies demonstrating combined action of multiple different destruction motifs for APC/C-mediated degradation in vivo have reopened the question of proteolytic targeting controls (18, 20, 33, 59). It is becoming clear that important cell cycle regulatory proteins are degraded through the use of different targeting proteins at different points throughout the cell cycle. This fine control is known to involve a switch between Cdh1p-targeted degradation and Cdc20-targeted degradation: the destruction box, the KEN box, and the recently defined A box are some motifs that have been shown to be differentially required for these targeting events. It is likely that further specificity determinants, which may include HEI10, remain to be assigned. Given the interest of this finding, we continue to investigate this point, as well as the still-extent possibility that HEI10 control of cyclin B and Clb2p levels is indirect.
An important question is the degree to which phenotypes associated with HEI10 expression in yeast and Xenopus reflect the activities of HEI10 in human cells. Given that parallel phenotypes are observed in these evolutionarily diverged species, it is likely that HEI10 functions in cell cycle control in humans, but the chief current impediment to studies of HEI10 is the inability to overexpress HEI10 in mammalian cells in a manner reflecting the endogenous localization of the protein. Other reports (6) have noted that some cellular proteins are mislocalized when overexpressed in the absence of a binding partner, offering one possible explanation for this difficulty. Alternatively, one hypothetical possibility is that overexpressed HEI10 is interacting with cyclin B2, which is similar to cyclin B1 in sequence but is constitutively associated with the Golgi apparatus (19, 22). Nevertheless, investigations to date of the endogenous HEI10 are compatible with an interaction with cyclin B1, which migrates constantly between the nucleus and the cytoplasm at prophase and then translocates entirely into the nucleus at the end of prophase and binds the mitotic apparatus, as discussed earlier (9). The observation that HEI10 nuclear levels vary throughout the cell cycle but colocalize with segregating chromosomes during mitosis is of considerable interest.
Finally, the recent identification of HEI10 as a fusion breakpoint in a uterine leiomyoma (37) is of considerable interest. The reported breakpoint produces a chimeric protein expressing HMG1C sequences translated in frame with aa 99 to 277 of HEI10. This fusion excises the HEI10 RING and RXL sequences while maintaining the remainder of the protein. Such a truncation suggests that the chimera may act as dominant negative for HEI10 activities by, for example, competing with endogenous HEI10 for interactions with additional complexed proteins. Conversely, HMG1C has been reported frequently as a breakpoint in uterine leiomyomas and other tumors, as discussed by Mine et al. (37), whereas ours is the first report of HEI10 involvement in such an event: it may be that the sole tumor-promoting activity derives from truncation of HMG1C. Although it is important not to overinterpret this result at this stage, further study of the HEI10 locus, in conjunction with the functional data presented here, is certainly warranted.
Acknowledgments
We are grateful to Margret Einarson, Randy Strich, and Peter Adams for helpful discussion of the manuscript. Olli Carpen and Mikaela Gronholm made many useful suggestions. We thank Natasha Frolova for superb help with Xenopus experiments, and we thank Thanos Halazonetis and members of his laboratory for help in the performance of in vitro ubiquitination assays and for many useful suggestions. We thank Sandra Holloway for reagents to study Clb2p and Anne Dranginis for the FLO11-LacZ plasmid. Jonathon Boyd provided expert assistance in microscopy.
This work was supported by a grant from the National Institutes of Health to E.A.G. (CA80991) and by NIH core grant CA-06927 to the Fox Chase Cancer Center.
REFERENCES
- 1.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. J. Kaelin. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. H., A. Acurio, and S. J. Kron. 1999. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell 10:3301-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, S. H., B. T. Tobe, J. N. Fitz Gerald, S. L. Anderson, A. Acurio, and S. J. Kron. 2001. Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol. Biol. Cell 12:3589-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardley, H. C., N. G. Tan, S. A. Rose, A. F. Markham, and P. A. Robinson. 2001. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. J. Biol. Chem. 276:19640-19647. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, J. R., P. N. Friedman, D. R. Marshak, C. Prives, and D. Beach. 1990. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc. Natl. Acad. Sci. USA 87:4766-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S., C. K. Suzuki, J. Lippincott-Schwartz, A. M. Weissman, and R. D. Klausner. 1989. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J. Cell Biol. 109:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden, K. L., and P. S. Freemont. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6:395-401. [DOI] [PubMed] [Google Scholar]
- 8.Casamayor, A., and M. Snyder. 2002. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5:179-186. [DOI] [PubMed] [Google Scholar]
- 9.Draviam, V. M., S. Orrechia, M. Lowe, R. Pardi, and J. Pines. 2001. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 152:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgington, N. P., M. J. Blacketer, T. A. Bierwagen, and A. M. Myers. 1999. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elledge, S. J., and M. R. Spotswood. 1991. A new human p34 protein kinase, CDK2, identified by complementation of a cdc28 mutation in Saccharomyces cerevisiae, is a homolog of Xenopus Eg1. EMBO J. 10:2653-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fashena, S. J., M. B. Einarson, G. M. O'Neill, C. P. Patriotis, and E. A. Golemis. 2002. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J. Cell Sci. 115:99-111. [DOI] [PubMed] [Google Scholar]
- 14.Freemont, P. S. 2000. RING for destruction? Curr. Biol. 10:R84-R87. [DOI] [PubMed] [Google Scholar]
- 15.Ghiara, J. B., H. E. Richardson, K. Sugimoto, M. Henze, D. J. Lew, C. Wittenberg, and S. I. Reed. 1991. A cyclin B homolog in Saccharomyces cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell 65:163-174. [DOI] [PubMed] [Google Scholar]
- 16.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 17.Golemis, E. A., and V. Khazak. 1997. The interaction trap and interaction mating, p. 197-218. In R. Tuan (ed.), Expression and detection of recombinant genes, vol. 63. Humana Press, Clifton, N.J. [DOI] [PubMed]
- 18.Hagting, A., N. Den Elzen, H. C. Vodermaier, I. C. Waizenegger, J. M. Peters, and J. Pines. 2002. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 157:1125-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagting, A., M. Jackman, K. Simpson, and J. Pines. 1999. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 9:680-689. [DOI] [PubMed] [Google Scholar]
- 20.Hendrickson, C., M. A. Meyn III, L. Morabito, and S. L. Holloway. 2001. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase-to-anaphase transition. Curr. Biol. 11:1781-1787. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, J. K., and M. J. Solomon. 1996. A predictive scale for evaluating cyclin-dependent kinase substrates: a comparison of p34cdc2 and p33cdk2. J. Biol. Chem. 271:25240-25246. [DOI] [PubMed] [Google Scholar]
- 22.Jackman, M., M. Firth, and J. Pines. 1995. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 14:1646-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. R. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 24.Koff, A., F. Cross, A. Fisher, J. Schumacher, K. Leguellec, M. Philippe, and J. M. Roberts. 1991. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66:1217-1228. [DOI] [PubMed] [Google Scholar]
- 25.Kron, S. J., and N. A. R. Gow. 1995. Budding yeast morphogenesis: signalling, cytoskeleton, and cell cycle. Curr. Opin. Cell Biol. 7:845-855. [DOI] [PubMed] [Google Scholar]
- 26.Kron, S. J., C. A. Styles, and G. R. Fink. 1994. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5:1003-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubler, E., H.-U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 28.Kumagai, A., and W. G. Dunphy. 1995. Control of the Cdc2/cyclin B complex in Xenopus egg extracts arrested at a G2/M checkpoint with DNA synthesis inhibitors. Mol. Biol. Cell 6:199-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law, S. F., J. Estojak, B. Wang, T. Mysliwiec, G. D. Kruh, and E. A. Golemis. 1996. Human enhancer of filamentation 1 (HEF1), a novel p130Cas-like docking protein, associates with FAK, and induces pseudohyphal growth in yeast. Mol. Cell. Biol. 16:3327-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law, S. F., G. M. O'Neill, S. J. Fashena, M. B. Einarson, and E. A. Golemis. 2000. The docking protein HEF1 is an apoptotic mediator at focal adhesion sites. Mol. Cell. Biol. 20:5184-5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law, S. F., Y.-Z. Zhang, A. Klein-Szanto, and E. A. Golemis. 1998. Cell-cycle regulated processing of HEF1 to multiple protein forms differentially targeted to multiple compartments. Mol. Cell. Biol. 18:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. G., and P. Nurse. 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327:31-35. [DOI] [PubMed] [Google Scholar]
- 33.Littlepage, L. E., and J. V. Ruderman. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, W.-S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 36.Makalowski, W., J. Zhang, and M. S. Boguski. 1996. Comparative analysis of 1196 orthologous mouse and human full-length mRNA and protein sequences. Genome Res. 6:846-857. [DOI] [PubMed] [Google Scholar]
- 37.Mine, N., K. Kurose, H. Konishi, T. Araki, H. Nagai, and M. Emi. 2001. Fusion of a sequence from HEI10 (14q11) to the HMGIC gene at 12q15 in a uterine leiomyoma. Jpn. J. Cancer Res. 92:135-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosch, H.-U., and G. R. Fink. 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145:671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosch, H. U., E. Kubler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, A. W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581-605. [PubMed] [Google Scholar]
- 41.Nuber, U., S. Schwarz, P. Kaiser, R. Schneider, and M. Scheffner. 1996. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J. Biol. Chem. 271:2795-2800. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill, G. M., S. J. Fashena, and E. A. Golemis. 2000. Integrin signaling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 10:111-119. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill, G. M., and E. A. Golemis. 2001. Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics. Mol. Cell. Biol. 21:5094-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567-572. [DOI] [PubMed] [Google Scholar]
- 45.Pan, X., and J. Heitman. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patra, D., and W. G. Dunphy. 1996. Xe-p9, a Xenopus Suc1/Cks homolog, has multiple essential roles in cell cycle control. Genes Dev. 10:1503-1515. [DOI] [PubMed] [Google Scholar]
- 47.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 50.Rua, D., B. T. Tobe, and S. J. Kron. 2001. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4:720-727. [DOI] [PubMed] [Google Scholar]
- 51.Spodik, B., S. H. Seeholzer, and T. R. Coleman. 2002. Using egg extracts to modify recombinant proteins, p. 355-374. In E. Golemis (ed.), Protein interactions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Tanaka, K., T. Suzuki, T. Chiba, H. Shimura, N. Hattori, and Y. Mizuno. 2001. Parkin is linked to the ubiquitin pathway. J. Mol. Med. 79:482-494. [DOI] [PubMed] [Google Scholar]
- 53.Tassan, J. P., M. Jaquenoud, P. Leopold, S. J. Schultz, and E. A. Nigg. 1995. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc. Natl. Acad. Sci. USA 92:8871-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiwari, S., and A. M. Weissman. 2001. Endoplasmic reticulum (ER)-associated degradation of T-cell receptor subunits: involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem. 276:16193-16200. [DOI] [PubMed] [Google Scholar]
- 55.Wan, Y., and M. W. Kirschner. 2001. Identification of multiple CDH1 homologues in vertebrates conferring different substrate specificities. Proc. Natl. Acad. Sci. USA 98:13066-13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wendland, J. 2001. Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 34:63-82. [DOI] [PubMed] [Google Scholar]
- 57.Xiong, Y., T. Connolly, B. Futcher, and D. Beach. 1991. Human D-type cyclin. Cell 65:691-699. [DOI] [PubMed] [Google Scholar]
- 58.Yokouchi, M., T. Kondo, A. Sanjay, A. Houghton, A. Yoshimura, S. Komiya, H. Zhang, and R. Baron. 2001. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 276:35185-35193. [DOI] [PubMed] [Google Scholar]
- 59.Zur, A., and M. Brandeis. 2002. Timing of APC/C substrate degradation is determined by fzy/fzr specificity of destruction boxes. EMBO J. 21:4500-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]