Abstract
Methylation of cytosine in CpG dinucleotides promotes transcriptional repression in mammals by blocking transcription factor binding and recruiting methyl-binding proteins that initiate chromatin remodeling. Here, we use a novel cell-based system to show that retrovirally expressed Pax-5 protein activates endogenous early B-cell-specific mb-1 genes in plasmacytoma cells, but only when the promoter is hypomethylated. CpG methylation does not directly affect binding of the promoter by Pax-5. Instead, methylation of an adjacent CpG interferes with assembly of ternary complexes comprising Pax-5 and Ets proteins. In electrophoretic mobility shift assays, recruitment of Ets-1 is blocked by methylation of the Ets site (5′CCGGAG) on the antisense strand. In transfection assays, selective methylation of a single CpG within the Pax-5-dependent Ets site greatly reduces mb-1 promoter activity. Prior demethylation of the endogenous mb-1 promoter is required for its activation by Pax-5 in transduced cells. Although B-lineage cells have only unmethylated mb-1 genes and do not modulate methylation of the mb-1 promoter during development, other tissues feature high percentages of methylated alleles. Together, these studies demonstrate a novel DNA methylation-dependent mechanism for regulating transcriptional activity through the inhibition of DNA-dependent protein-protein interactions.
The development of specialized cells from multipotent progenitors is the result of a complicated array of events involving the initiation of new gene expression and silencing of unnecessary or inappropriate genes. Transcriptional regulation of genes occurs at several levels. Tissue-specific and temporally regulated transcription factors activate and/or repress genes that define lineage- and differentiation stage-specific characteristics. However, accessibility of genes to these factors is imparted by chromatin architecture, which is itself controlled through two known mechanisms: cytosine methylation within CpG dinucleotides, and chromatin remodeling initiated by histone acetyltransferases and histone deacetylases.
DNA methylation has long been recognized as a contributor to epigenetic regulation of transcription (reviewed in reference 2). In higher eukaryotes, transcriptionally active genes and their promoters are generally hypomethylated, while silenced genes are hypermethylated. Two models have been proposed to account for the effects of cytosine methylation on transcription. The first suggests that methylation at CpG dinucleotides blocks binding of transcription factors to their sites through steric hindrance (7). However, this model is limited to a select set of protein-DNA interactions, as many regulatory target sites of DNA-binding proteins do not contain CpG dinucleotides and are not affected by DNA methylation. In addition, some DNA-binding proteins, such as Sp1, are able to bind DNA regardless of its methylation status (23). The second model suggests that specialized methyl-binding proteins bind methylated DNA and recruit chromatin remodeling factors, leading to the formation of a repressive chromatin structure, thus making DNA inaccessible to transcription factors (reviewed in reference 39). While the second model may be more useful for explaining repression of a larger number of genes, it should be noted that these models are not mutually exclusive.
The complex processes driving B-lymphocyte development require a high level of coordination between B-cell-specific factors and other nuclear proteins to activate distinct sets of genes at the appropriate times. One of the key transcription factors responsible for B-cell-specific transcription is Pax-5 (also known as B-cell-specific activator protein, or BSAP; reviewed in reference 35). Pax-5 is a member of the paired-domain family of transcription factors that bind DNA through a highly conserved bipartite DNA-binding domain (DBD). Pax-5 is expressed early in B-cell development but is shut off in terminally differentiated plasma cells. In addition to B cells, Pax-5 is expressed in the developing midbrain, but it is expressed only at low levels in the adult brain (1). Binding sites for Pax-5 in the promoters of the B-cell-specific genes CD19 and mb-1 have been identified and confirmed in functional assays (12, 31). Pax-5 is also implicated in the positive regulation of N-myc, LEF-1, and BLNK and the negative regulation of PD-1 and XBP-1 (38, 40, 41).
We have previously shown that Pax-5 recruits proteins of the Ets family of transcription factors (Ets-1 and GABPα) to bind an adjacent suboptimal site within the promoter of the mb-1 gene (12), which encodes the essential B-cell-specific signaling protein Ig-α. Importantly, Ets proteins do not bind this suboptimal sequence significantly in the absence of Pax-5, due to a single base difference within the consensus Ets binding site (46). Pax-5 enhances binding of Ets proteins (Ets-1) to this site by >1,000-fold (D. Fitzsimmons and J. Hagman, unpublished data). Mutation of either the Pax-5 or Ets binding sites results in similarly decreased transcriptional activity, indicating synergistic interactions between these proteins. Indeed, recent analysis of factor binding to the promoter in intact cells by in vivo footprinting demonstrated coordinate occupancy of both Pax-5 and Ets binding sites in all mb-1-expressing cells (43). The recently determined crystal structure of the Pax-5-Ets-1 complex on DNA confirmed the suspected interactions between the DBDs of these two proteins (14).
To further examine requirements for functional Pax-5-Ets ternary complex formation on the mb-1 promoter, we developed a novel cell-based system for studying transcriptional activation of endogenous mb-1 genes within the context of DNA methylation and chromatin architecture. We present evidence that the methylation status of a single, specific cytosine within the mb-1 promoter dictates the ability of Pax-5 to activate transcription of the endogenous gene in B cells. This is a novel mechanism in that the cytosine is not within the Pax-5 binding site, but is instead within the binding site of its Ets partner. This is the first report of a methylated cytosine inhibiting transcriptional activation driven by a factor whose binding site does not contain a CpG dinucleotide sequence. These data lead us to hypothesize that demethylation of the Ets site is a prerequisite for activation of the mb-1 promoter by Pax-5 during lymphopoiesis.
MATERIALS AND METHODS
Mice.
Six- to 12-week-old BALB/cJ and C57BL/6 mice were obtained from the Jackson Laboratory, Bar Harbor, Maine. Pax-5−/− mice (kindly provided by M. Busslinger, IMP, Vienna, Austria) were maintained in a pathogen-free animal facility at the National Jewish Medical and Research Center (NJMRC).
Cell lines and cell culture.
558LμM cells (25) were kindly provided by M. Reth (University of Freiburg, Freiburg, Germany) and were cultured in RPMI medium containing 10% fetal bovine serum (FBS; Gemini Bioproducts, Woodland, Calif.), 2 mM l-glutamine, 50 μg of gentamicin/ml, 1× HT Media Supplement (50×), 0.3 μg of xanthine/ml, and 1 μg of mycophenolic acid/ml. For 5-azacytidine (5-azaC) treatment, 558LμM DBD clones were treated with 2 μM 5-azaC (Sigma, St. Louis, Mo.) for 48 h and analyzed by flow cytometry. For trichostatin A (TSA; Sigma) treatment, 558LμM DBD clones were treated with 5 to 50 nM TSA for 48 h and analyzed by flow cytometry. Dead cells were excluded from the analysis by staining with propidium iodide. The pre-B-cell lines 40E1 and 70Z/3 were cultured in Iscove’s modified Dulbecco’s medium (IMDM) containing 10% FBS, 2 mM l-glutamine, 50 μg of gentamicin/ml, and 50 μM β-mercaptoethanol. M12.4.1, A20, S194, and the ΦNX retroviral packaging cell line (kindly provided by P. Marrack [44]) were cultured in IMDM medium containing 10% FBS, 2 mM l-glutamine, and 50 μg of gentamicin/ml. All cell lines were grown at 37°C in 6% CO2.
Thymocytes were purified using nylon wool columns as previously described (30). Pax-5-deficient pre-BI cells were isolated from the bone marrow of 2- to 3-week-old Pax-5-deficient mice by sorting for B220+ cells on a MoFlo cell sorter (Cytomation, Inc., Ft. Collins, Colo.). Wild-type pre-B cells (CD19+ membrane immunoglobulin M-negative [mIgM−]) were sorted from the bone marrow of BALB/cJ mice at 6 to 8 weeks of age. Primary and secondary immune response NP-specific B-cell subsets were stained and sorted as previously described (36). Primary immune response bone marrow plasma cells (B220-CD138+) were stained and sorted 3 weeks postimmunization with 100 μg of ovalbumin (Sigma) in complete Freund's adjuvant (Sigma). Secondary immune response bone marrow plasma cells (B220-CD138+) were stained and sorted 3 weeks postboost from mice boosted on day 31 with 50 μg of ovalbumin in incomplete Freund's adjuvant (Sigma).
Plasmid constructs.
All retroviral constructs were derived from the MSCV2.2-IRES-GFPα retroviral vector (provided by P. Marrack [24]). To express FLAG-tagged human Pax-5, we first ligated oligonucleotides encoding a Kozak translation initiation sequence and FLAG tag coding region, 5′CTCACCATGGATTACAAGGATGACGATGACAAAGGGGCT, at the Ecl136II site of plasmid Pax-5.S1 (12). The insert was excised using Ecl136II and HindIII, blunted with Klenow, and ligated into MSCV2.2-IRES-GFPαStu (at a StuI site inserted in place of XhoI). Plasmids pCMV-Tag2B-mEts-1 and pCMV-Tag2B-mEts-2 were kindly provided by J. Sevinsky and N. Ahn. Retroviral constructs for expression of murine FLAG-tagged Ets-1 or Ets-2 were made by cutting these plasmids with NotI and SpeI, or NotI and partially with BamHI, respectively, filling in ends with Klenow, and ligating the fragments at the filled-in XhoI site of MSCV2.2-IRES-GFPα. The construct for expression of FLAG-early B-cell factor (EBF) was prepared in three steps. First, we added the FLAG tag (described above) to EBF that had been previously modified with a Kozak consensus sequence (21). Second, FLAG-EBF was released by partial digestion with NcoI and complete digestion with SalI, filled in, and used to replace the Ets-2 fragment of MSCV-FLAG-Ets-2 (cut with SalI and BamHI and filled in with Klenow).
MSCV2.2-Pax-5-IRES-GFPα (for expression of full-length Pax-5 without the FLAG-tag) was created by excising Pax-5 from plasmid Pax-5S.1 (12) with Ecl136II and SalI, blunting the fragment with Klenow, and inserting it into the StuI site of MSCV2.2-IRES-GFPαStu. For expression of the Pax-5 DBD (amino acids 1 to 149), we added the simian virus 40 (SV40) nuclear localization signal amino acid sequence GALTGALTPKKKRKVED to its carboxyl terminus in two steps. First, we PCR amplified a synthetic oligonucleotide, 5′CTAATCCTCGACTTTTCGTTTCTTCTTAGGAGTAAGAGCACCTGTCAGAGCCCCTTGGTTGGGTGGCTGCTGTAC, with primers 5′ATCATCCGGACAAAAGTACAGCAGCCACCCAACCAA and 5′CTAATCCTCGACTTTTCGTTTCTTC (3′ primer). Next, addition of sequences to the 3′ end of the Pax-5 paired domain was accomplished by amplification of ΔPax-5.4 (46) together with the amplified fragment, primer 5′CTCATCATGGATTTAGAGAAAAATTATCC, and the 3′ primer. The resulting fragment was ligated into the Ecl136II site of Bluescript KS(+), excised using Ecl136II and HindIII, blunted with Klenow, and ligated into the StuI site of MSCV2.2-IRES-GFPαStu. The plasmid pCL-Eco (37) was a kind gift of I. Verma.
Luciferase promoter plasmids were created by blunt ligation of mb-1 promoter fragments into the SmaI site of pGL3-Basic (Promega, Madison, Wis.). Promoter fragments were PCR amplified from TMβPy, TMβPy-mut1, TMβPy-mut2, and TMβPy-mut1/2 plasmids (12) using 5′CTAGAGAGAGACTCAAGGGAATTG and 5′TCTCCCAGTGAGTCGGTTAGTTTG. pRL-TK and pGL3-Promoter plasmids were purchased from Promega.
The plasmid for synthesis of the glutathione S-transferase (GST)-murine Ets-1 (amino acids 4 to 123) fusion protein was made by ligation of the filled-in (Klenow fragment) EagI-EcoRV fragment of cloned murine Ets-1 cDNA (12) into the filled-in BamHI site of pGEX-4T-3 (Amersham) to make pGEX-Ets-1(4-123).
Plasmid standards for methylation-sensitive-single-nucleotide primer extension (MS-SNuPE) were prepared by modification and amplification of the plasmid pM9-4.6, which includes a 4.6-kb BamHI fragment of murine genomic DNA including the mb-1 promoter and downstream sequences (provided by R. Grosschedl). pM9-4.6 DNA was treated with HpaII methylase or was untreated prior to bisulfite conversion (see below). Converted plasmid sequences were PCR amplified using primers 5′CTCAAAAATCAATAATAATAAACCAA and 5′TAGGGTTTTGAGGGTTTT, and amplified products were ligated into the EcoRV site of pBluescript KS(+). Plasmids (pMb1AS-methyl and pMb1AS-unmethyl) were sequenced to confirm complete conversion by bisulfite and integration of a single insert.
Retrovirus production and transduction.
ΦNX packaging cells were plated on poly-d-lysine-coated 100-mm dishes and cultured overnight to reach 60 to 80% confluency. Cells were cotransfected with 8.4 μg of pCL-Eco plasmid DNA and 31.6 μg of MSCV plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Four hours following transfection, FBS was added to a concentration of 5%, and cells were cultured overnight at 37°C. One day posttransfection, the medium was discarded and fresh medium (IMDM, 20 mM l-glutamine, 10% FBS) was added. Two days posttransfection, the medium was collected and centrifuged at 800 × g for 15 min, and the virus-containing supernatant was used to transduce cells. Transductions were performed in six-well dishes using 3 ml of retroviral supernatant, 1 ml of cells (5 × 105 cells/ml), and 6.25 μg of Polybrene (Sigma)/ml. One day posttransduction (1 dpt), cells were transferred to fresh medium. Cells were analyzed for green fluorescent protein (GFP) and mIgM expression 3 dpt. Unless stated otherwise, all experiments were performed with Pax-5 or Pax-5 DBD vectors lacking a FLAG tag.
Production of murine Ets-1(4-123)-specific antiserum.
Recombinant murine Ets-1(4-123) protein was prepared in Escherichia coli. Single colonies were picked from plates of freshly transformed bacteria and inoculated into Luria broth supplemented with carbenicillin (500 μg/ml) and chloramphenicol (34 μg/ml). Cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 and induced with isopropyl-β-d-thiogalactopyranoside (1 mM) for 3 h prior to harvest by centrifugation. Bacterial pellets were resuspended in ice-cold phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4; pH 7.3), lysed by sonication, and centrifuged for 60 min in a Sorvall SS34 rotor at 15,000 rpm to remove bacterial debris. Soluble GST-Ets-1(4-123) protein was purified from bacterial lysates using glutathione-Sepharose 4B (Amersham Biosciences, Inc.) beads according to the manufacturer's instructions for batch purification. Ets-1(4-123) was enzymatically cleaved from bound GST by using recombinant thrombin (Amersham Biosciences, Inc.). Following dialysis against PBS, the concentration and purity of Ets-1(4-123) was assessed by the Bradford assay (Bio-Rad) and sodium dodecyl sulfate (SDS)-4-to-20% polyacrylamide gel electrophoresis and Coomassie brilliant blue staining, respectively.
For immunization, a New Zealand White rabbit was injected intradermally with 500 μg of Ets-1(4-123) emulsified with Freund's complete adjuvant in a total volume of 500 μl, followed by boosting with 500 μg of antigen in Freund's incomplete adjuvant at 4-week intervals. The rabbit was bled via ear vein at 6 weeks postsecondary immunization and at 4-week intervals.
Flow cytometry and Western blotting.
For flow cytometry, biotin-conjugated anti-IgM and phycoerythrin-conjugated anti-CD19 antibodies were purchased from Caltag Laboratories (Burlingame, Calif.). Anti-B220-phycoerythrin, biotin-conjugated CD138 (Syndecan-1), and streptavidin-conjugated allophycocyantin were purchased from BD Pharmingen (San Diego, Calif.). Dead cells were excluded by staining with propidium iodide. Flow cytometry was performed on a FACScalibur machine (Becton Dickinson, San Diego, Calif.). For Western blotting, rabbit anti-human Pax-5 antibody was purchased from Geneka (Montreal, Canada), and horseradish peroxidase (HRP)-conjugated anti-FLAG tag antibody (M2) was purchased from Sigma. Rabbit anti-Ig-α antibody was kindly provided by J. C. Cambier (NJMRC). Rabbit anti-Sp1 antibody was purchased from Santa Cruz Biotech (Santa Cruz, Calif.). HRP-conjugated F(ab)′2 donkey anti-rabbit IgG was purchased from Amersham Biosciences Corp. (Piscataway, N.J.). Whole-cell lysates prepared from similar numbers of cells with 2× SDS sample buffer (10% glycerol, 2% SDS, 62.5 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 0.02% bromophenol blue) or nuclear extracts (25 μg) were separated on 4-to-20% Tris-HCl Ready gels using the Mini Trans-Blot cell system (Bio-Rad, Richmond, Calif.). Separated proteins were transferred to a Hybond ECL nitrocellulose membrane (Amersham) according to the manufacturer's instructions. Western blotting was performed according to the manufacturer's instructions, and HRP-conjugated antibodies were detected using ECL Western blotting detection reagents (Amersham).
Real-time RT-PCR.
Extraction of RNA, cDNA synthesis, primer sequences, and real-time reverse transcription-PCR (RT-PCR) for detection of β-actin and mb-1 transcripts were described previously (43).
Nuclear extracts and electrophoretic mobility shift assays (EMSAs).
Nuclear extracts were prepared using a modification of the method of Schrieber et al. (42). Cell pellets were resuspended in 4 pellet volumes of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of leupeptin/ml, and 2 μg of aprotinin/ml) and incubated for 15 min on ice. A 1/16 volume (of buffer A used) of 10% NP-40 was added, vortexed for 10 s, and then centrifuged at 18,000 × g for 30 s at 4°C. The supernatant (cytoplasmic fraction) was discarded, and 1 pellet volume of buffer C (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, plus protease inhibitors) was added. Pellets were vortexed at 4°C for 2 min and then rotated for 15 min at 4°C. Extracts were centrifuged for 15 min at 15,000 rpm and the supernatant was aliquoted, frozen in a dry ice-ethanol bath, and stored at −80°C. A 0.5-μg aliquot of nuclear extract was used in each binding reaction. Purified anti-Pax-5 antibody and anti-Ets-1 antisera for supershifts are described in the Western blotting section above.
Annealing of oligonucleotides, labeling of DNA probes, and EMSAs were performed as previously described (46). The wild-type mb-1 promoter probe sequences were as follows: sense, 5′TCGAAGGGCCACTGGAGCCCATCTCCGGCACGGC; antisense, 5′TCGAGCCGTGCCGGAGATGGGCTCCAGTGGCCCT (cytosine bases in CpG dinucleotides are underlined). Methylated probes were synthesized by Integrated DNA Technologies. Oligonucleotides with methylated bases were annealed with unmethylated or methylated strands to make hemimethylated or bimethylated probes, respectively. Sequences were the same as above, except 5-methyl cytosine was incorporated at the underlined position in either the Ets-binding site or control site. Highly purified recombinant Pax-5 (1-149) and Ets-1 (280-440) were kindly provided by C. Garvie and C. Wolberger (14).
Transient transfections and luciferase assays.
Methylated plasmids were prepared by methylating the internal cytosine of 5′-CCGG sequences using HpaII methylase (New England Biolabs, Beverly, Mass.) according to the manufacturer's instructions. Efficiency of the methylation reaction was checked by digestion with HpaII and MspI restriction enzymes. Both enzymes digest 5′-CCGG sequences, but HpaII is sensitive to methylation of the internal cytosine, while MspI is methylation insensitive. Unmethylated plasmids were mock methylated by exclusion of S-adenosylmethionine from the reaction. Transfection of M12.4.1 cells was performed by incubating 2.5 × 106 cells in 0.65 ml of a 0.7-mg/ml solution of DEAE-dextran (Pharmacia, Uppsala, Sweden) in 1× TS (140 mM NaCl, 5 mM KCl, 25 mM Tris HCl [pH 7.4], 0.7 mM Na2HPO4, 1 mM MgCl2, 1 mM CaCl2) containing 5 μg of reporter plasmid and 0.1 μg of pRL-TK (Promega) at 20°C for 30 min. Transfected cells were incubated in medium containing 10% serum and 100 μg of chloroquine diphosphate (Sigma)/ml for 30 min at 37°C. Cells were pelleted, resuspended in 5 ml of medium, and cultured in six-well plates for 48 h. Protein extracts were prepared and luciferase assays were performed using the Dual-Luciferase reporter assay system (Promega). Luciferase activities were generated using 20% of total extracts and were normalized to pRL-TK activity. In each experiment, each determination was performed in triplicate. The data shown in Fig. 3 are the combined results of three independent experiments.
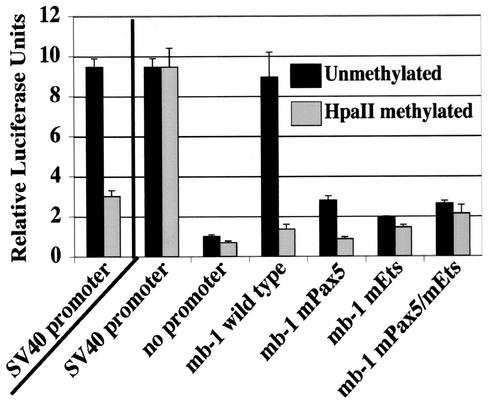
FIG. 3.
Methylation of the Pax-5-dependent Ets site in the minimal mb-1 promoter inhibits transcriptional activation. M12.4.1 B cells were transiently transfected with the indicated pGL3 luciferase reporter constructs. Left panel: HpaII methylation of the SV40 promoter construct results in a 67.3% decrease in luciferase activity. Right panel: Luciferase activities of HpaII methylated constructs were corrected for the effects of methylation of the luciferase gene as determined at left. Black bars, unmethylated reporter constructs; gray bars, HpaII methylase-treated reporter constructs.
MS-SNuPE.
Genomic DNA was treated with bisulfite as follows: ≤1 μg of DNA in 20 of μl sterile water was sheared by pipetting and denatured in freshly prepared 0.3 M NaOH for 30 min at 75°C. Next, 250 μl of freshly prepared 4.8 M sodium bisulfite (Sigma), pH 5.0, and 14 μl of 10 mM hydroquinone (Sigma) were added, and the reaction mixture was overlayed with mineral oil and incubated in the dark at 55°C for 18 h. Modified DNA was purified using the QiaQuick PCR Purification kit (Qiagen, Inc., Valencia, Calif.). DNA was eluted in 40 μl of TE and desulfonated in 0.3 M NaOH at 37°C for 20 min. DNA was ethanol precipitated overnight, dissolved in 20 μl of H2O, and stored at −80°C. Two to 4 μl of modified DNA was used for nested PCR amplification. Primary PCRs comprised 2 to 4 μl of bisulfite-modified DNA, 1× AmpliTaq buffer, 1.5 mM MgCl2, 100 ng of “outside” forward and reverse primers, a 0.2 mM concentration of deoxynucleoside triphosphates (dNTPs), and 2 U of AmpliTaq DNA polymerase (Roche, Indianapolis, Ind.) in a 50-μl volume. Primary reaction conditions were 95°C for 5 min, followed by 30 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. Five microliters of the primary reaction mixture was transferred to a secondary reaction mix containing 1× AmpliTaq buffer, 1.5 mM MgCl2, 200 ng of “inside” forward and reverse primers, 0.2 mM concentration of dNTPs, and 2 U of AmpliTaq DNA polymerase in a 50-μl volume. Secondary reaction conditions were the same as the primary reaction conditions. Following amplification, PCR products were gel isolated using the Qiaex II gel purification kit (Qiagen). Primer sequences for amplification of the bisulfite-treated antisense strand of the mb-1 promoter were as follows: outside forward, 5′CTCAAAAATCAATAATAATAAACCAA; outside reverse, 5′TAGGGTTTTGAGGGTTTT; inside forward, 5′TAAACCACCCTCTCCC; and inside reverse, 5′GGGTTTTTAGATTTTTTGGTAT.
MS-SNuPE was performed as previously described (16) with the following modifications. Due to the presence of a CpG site within the primer annealing site for MS-SNuPE, a mixture of two primers (1 μM each) differing at one base (underlined) was used to compensate for heterogeneity of the template following bisulfite conversion. The primer sequences were 5′GTTTTATTTTTTGTTTAGTTGTGT and 5′GTTTTATTTTTTGTTTAGTCGTGT. Conditions for primer extension reactions were 95°C for 1 min, 49°C for 1 min, 72°C for 2 min. A 25-μl aliquot of 1/5× TE was added to each reaction mixture, and reaction mixtures were purified using a CentriSep Spin column (Princeton Separations, Adelphia, N.J.) equilibrated with 1/5× TE containing 10 μg of yeast tRNA/ml. Purified reaction mixtures were transferred to scintillation vials, and 32P activity was counted. The assay was validated by measuring cytosine versus thymidine incorporation into titrated mixtures of plasmid templates (pMb1AS-methyl and pMb1AS-unmethyl) that represent promoters with fully methylated or unmethylated Ets ternary complex binding sites. Curves for control methylated and unmethylated bisulfite-converted products were generated, and R values were routinely greater than 0.99 (data not shown). The following equation was used to determine the percent methylation of each sample: % methylation = (dCTP cpm)/(dCTP cpm + dTTP cpm) × 100%. Assays on all samples were performed in duplicate. Data presented are the combined results of two to three independent experiments.
RESULTS
Pax-5 induces mb-1 expression in the 558Lμm plasmacytoma cell line.
Previous studies investigating transcriptional regulation of B-cell-specific genes have relied upon EMSAs and plasmid-based reporter systems to determine factors necessary for transcriptional activation. While these systems certainly help define potential binding sites for regulatory factors, they do not accurately reproduce conditions necessary for activation of an endogenous gene within the context of its natural state of DNA methylation and chromatin architecture. Thus, a transcription factor may appear to activate transcription of a target gene in vitro, while it may not have access to this gene in vivo due to DNA methylation and/or a repressive chromatin structure.
To determine requirements for activation of the early B-cell-specific mb-1 gene in its natural chromatin context, we took advantage of the characteristics of the terminally differentiated B-cell line 558Lμm (25). 558Lμm is a derivative of the J558L plasmacytoma line that has been stably transfected with a construct expressing the membrane form of IgM (mIgM). Thus, 558Lμm expresses IgM heavy chains, λ light chains, and Ig-β, three of the four components necessary for display of mIgM on the cell surface. However, 558Lμm cells have transcriptionally inactive mb-1 genes and therefore lack Ig-α, the fourth component necessary for mIgM display. Due to the absence of Ig-α, mIgM remains in the cytoplasm. By showing that forced expression of transfected mb-1 genes activated cell surface expression of the BCR, 558LμM cells were used to identify Ig-α as a necessary component of the B-cell receptor on the plasma membrane (26). Thus, because mb-1 expression is the limiting factor for display of mIg on the cell surface, 558LμM cells provide a useful system for testing requirements for the activation of endogenous mb-1 gene transcription.
To express potential activators of mb-1 gene expression in 558LμM cells, we generated recombinant mouse stem cell viruses (MSCV) that transcriptionally link expression of enhanced GFP via an internal ribosomal entry sequence (IRES) to expression of the test protein (24). Therefore, flow cytometry enables the levels of mIgM expression to be analyzed with respect to the relative expression of test genes in individual cells as indicated by GFP fluorescence intensity. Moreover, expression of GFP allows for cell sorting and recovery of relatively homogenous populations of transduced cells for biochemical analysis. Flow cytometry thus provides a more informative assay than is normally the case with standard reporter gene assays, which, in general, provide only a single number as a readout of reporter gene activity.
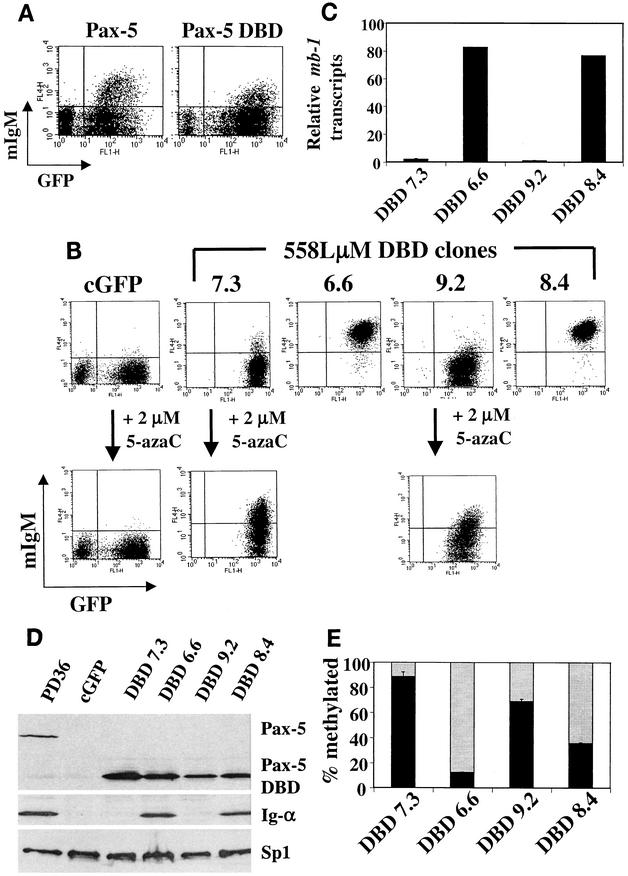
Functionally important binding sites for the transcription factors Pax-5, early B-cell factor (EBF), Ets-1, and Ets-2 were identified previously within the mb-1 promoter (11, 12, 19, 20, 22). Importantly, 558LμM cells do not express endogenous Pax-5 or EBF. Therefore, we transduced 558Lμm cells with retroviruses encoding these proteins with FLAG epitope tags for detection using anti-FLAG antibodies. Three days following infection, the cells were stained with an anti-IgM antibody and analyzed by flow cytometry (Fig. 1A). Transduction with a control GFP retrovirus (hereafter referred to as cGFP) did not induce mIgM expression in 558Lμm cells. However, retroviral expression of FLAG-tagged Pax-5 induced cell surface expression of mIgM on approximately 14% of GFP-positive (transduced) cells. In contrast, retroviruses expressing the FLAG-tagged transcription factors EBF, Ets-1, or Ets-2 had no effect on mIgM expression. Western blotting of whole-cell lysates probed with anti-FLAG antibodies confirmed expression of each of the FLAG-tagged proteins (Fig. 1B).
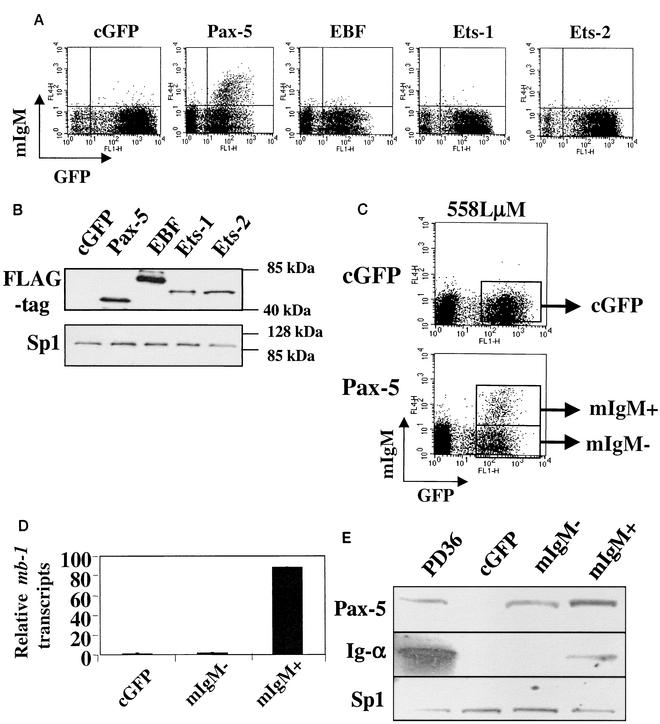
FIG. 1.
Pax-5 activates transcription of endogenous mb-1 genes in the 558LμM cell line. (A) 558LμM cells were transduced with retroviral vectors expressing GFP alone (cGFP) or the indicated FLAG-tagged factors and analyzed 3 dpt for cell surface expression of mIgM. mIgM expression is indicative of mb-1 transcription (D) and Ig-α production (E). (B) Western blotting of whole-cell lysates for expression of retrovirally expressed FLAG-tagged proteins (indicated above). Top: Detection of retrovirally expressed proteins using anti-FLAG antibodies. Bottom: Detection of endogenous Sp1 as a loading control. (C) Flow cytometric analysis of sorted 558LμM populations analyzed by MS-SNuPE. Boxes indicate gates used during cell sorting. (D) Real-time RT-PCR quantitation of mb-1 transcripts in sorted cGFP and Pax-5-expressing mIgM− and mIgM+ 558LμM cells. (E) Western blotting of sorted cGFP and Pax-5(GFP)-expressing mIgM− and mIgM+ 558LμM whole-cell lysates using Pax-5-, Ig-α-, or Sp1-specific antibodies. The pre-B-cell line PD36 served as a positive control for all three antibodies.
To confirm that expression of mIgM on Pax-5-transduced 558Lμm cells was due to transcriptional activation of the mb-1 gene and translation of its product Ig-α, Pax-5-transduced (no FLAG tag; see Materials and Methods) 558Lμm cells were sorted into mIgM+ and mIgM− populations by fluorescence activated cell sorting (FACS) for subsequent analysis (Fig. 1C). Real-time PCR of cDNA synthesized from sorted cell RNA indicates that Pax-5-transduced mIgM+ 558Lμm cells express significant mb-1 transcript levels, while Pax-5-transduced mIgM− 558Lμm cells express nearly undetectable levels. mb-1 transcripts were not detected in cGFP-transduced cells (Fig. 1D). Western blot analysis of whole-cell lysates of sorted cGFP, mIgM−, mIgM+, and control PD36 pre-B cells showed that, while Pax-5-transduced mIgM+ and mIgM− cells express Pax-5, mIgM+ cells contain detectable levels of Ig-α protein (Fig. 1E). Therefore, we conclude that Pax-5 activates transcription of previously silent mb-1 genes in the mIgM+ cells.
mb-1 promoter methylation prevents formation of Pax-5-Ets-DNA ternary complexes.
The lack of mIgM expression in a large percentage of Pax-5-transduced cells was a surprising result. We noted that cells expressing high levels of GFP tended to express higher levels of mIgM, indicating a concentration-dependent effect of Pax-5 on mb-1 transcription. Similar Pax-5 concentration-dependent effects were also noted for the CD19 gene (38). However, the lack of mIgM expression on a significant proportion of cells expressing high levels of GFP (and therefore Pax-5) suggested that a more complicated mechanism is responsible for the lack of mb-1 transcription. To determine the molecular basis for the heterogeneous expression of Ig-α, we examined the mb-1 promoter for regulatory motifs that might explain this phenomenon.
Among many possible mechanisms that could govern the ability of Pax-5 to activate the mb-1 gene in 558LμM cells, we considered epigenetic mechanisms, including inhibition of DNA binding by CpG methylation. Inspection of the minimal functional mb-1 promoter sequence (−186 to +24) identified five CpG dinucleotides. Surprisingly, the Pax-5 recognition sequence (−85 to −71) does not contain CpG dinucleotides, indicating that DNA methylation does not directly modulate Pax-5 DNA binding. Instead, the mb-1 promoter region includes a CpG dinucleotide within the adjacent Pax-5-dependent Ets binding site identified in our laboratory. Previously, we showed that Pax-5 recruits Ets family partner proteins, including Ets-1 and GABPα (together with GABPβ), to bind an adjacent, suboptimal Ets site within the mb-1 promoter (12, 13, 46). Binding of this site by Ets proteins is nearly undetectable in the absence of Pax-5, which facilitates assembly of ternary complexes required for transcriptional activity.
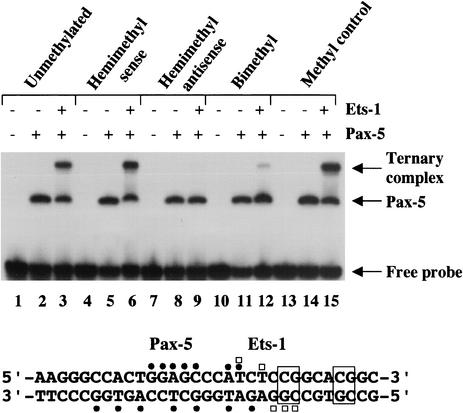
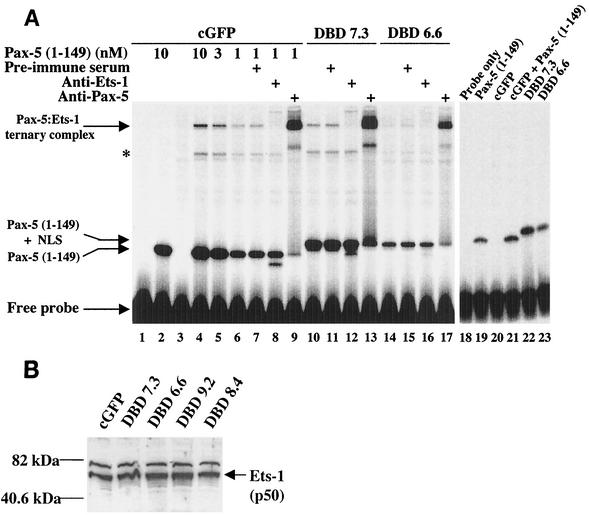
Because it has previously been shown that CpG methylation prevents binding of Ets proteins to promoters of the Ets-regulated genes Surf-1 and Surf-2 (15), we asked whether methylation of the Ets binding site within the mb-1 promoter prevents the recruitment of Ets-1 by Pax-5. EMSA was performed using unmethylated 32P-labeled double-stranded oligonucleotide probes or probes methylated at CpG dinucleotides on the sense strand, antisense strand, or both strands. Recombinant Pax-5 (1-149) paired domain bound each of the probes equivalently by itself (Fig. 2, lanes 2, 5, 8, 11, and 14). In contrast, Pax-5 recruited recombinant Ets-1 (280-440) to bind only the unmethylated and sense-strand methylated probes (lanes 3 and 6), but Pax-5 did not recruit Ets-1 to bind probe DNA methylated on the antisense strand (lane 9). This result is consistent with steric hindrance by 5-methyl cytosine of contacts made by the Ets-1 protein in the ternary complex (14). Although ternary complex assembly was largely inhibited, a small amount was detected when both strands of the probe were methylated (lane 12); however, other data (Fig. 3) suggest that bimethylation of the Pax-5-dependent Ets site blocks functional formation of ternary complexes. Methylation of a second CpG immediately downstream of the Ets site had no effect on ternary complex formation (lane 15). These data indicate that methylation of the promoter on the antisense strand of the Pax-5-dependent Ets site blocks recruitment of Ets-1 by Pax-5 to bind this site.
FIG. 2.
Methylation of the Pax-5-dependent Ets binding site prevents ternary complex assembly. EMSA was performed with recombinant Pax-5(1-149) and Ets-1(280-440) proteins as previously described (46). CpG methylation status of probe DNAs is indicated above as hemimethylated on sense or antisense strands, or bimethylated on both strands. Pax-5 and Pax-5-Ets-1 ternary complexes are indicated at right. The mb-1 probe sequence is shown below. Protein-DNA contacts made by Pax-5 (closed circles) or Ets-1 (open squares) in the crystal structure are indicated (14). The two CpG dinucleotides in the probe are boxed. Effects of modifying the downstream CpG were only tested with methylation of both strands (Methyl control).
Methylation of the Pax-5-dependent Ets binding site inhibits mb-1 transcription.
We have shown previously that mb-1 promoter activity is dependent on functional binding sites for Pax-5 and Ets proteins (12, 43). Cooperative DNA binding increases the apparent affinity of Pax-5 and Ets-1 for sites in the mb-1 promoter by 40- and >1,000-fold, respectively (D. Fitzsimmons and J. Hagman, unpublished). Therefore, we predicted that methylation of the ternary complex Ets site, which blocks DNA binding by Ets-1 in vitro, should greatly reduce functional activity of the transfected mb-1 promoter. To test this hypothesis, transfection assays were conducted to determine whether methylation of the Pax-5-dependent Ets binding site inhibits transcription of a luciferase reporter gene driven by a minimal mb-1 promoter. The Ets binding site CpG resides within the only 5′-CCGG motif present within the promoter. The sequence 5′-CCGG is methylated on both strands in vitro by the HpaII methylase enzyme, resulting in selective methylation of a single CpG within the 210-bp promoter, while the four other CpG dinucleotides present in the promoter remain unmethylated. As an important concern, the luciferase gene itself contains 10 5′-CCGG motifs. Therefore, it was necessary to control for other effects (e.g., reduced transcriptional elongation) on the luciferase reporter gene. To quantitatively determine effects of methylation of the luciferase reporter, we utilized a reporter plasmid driven by the SV40 early region promoter, which does not contain 5′-CCGG motifs, as a control. The mb-1 promoter (−185 to +24) was inserted into the luciferase reporter plasmid pGL3-Basic. Each plasmid was methylated with HpaII methylase, and methylation was confirmed by complete digestion with the methylation-insensitive restriction endonuclease MspI, but not with the methylation-sensitive HpaII endonuclease (data not shown).
To quantitatively measure effects of methylation of the Ets binding site on promoter function, we transfected the B-cell line M12.4.1, which expresses abundant mb-1 transcripts (43). Cells were transfected with methylated or unmethylated plasmids and analyzed for luciferase activity 48 h later. Methylation of the luciferase gene adjacent to the SV40 promoter (Fig. 3, SV40 promoter) resulted in a 67.3% decrease in luciferase activity relative to the unmethylated plasmid. Therefore, all data obtained with methylated plasmids were corrected for this effect (Fig. 3, right box). The unmethylated wild-type mb-1 promoter produced luciferase activity approximately ninefold (P = 0.0001) over that of the promoter-less pGL3-Basic construct (no promoter). Similar to our previous studies (12, 43), mutation of the Pax-5 (mPax-5), Ets (mEts), or both ternary complex sites (mPax-5/mEts) resulted in decreases in promoter function of 68.7% (P = 0.0007), 77.5% (P = 0.0004), and 70.6% (P = 0.0008), respectively. With correction for effects of luciferase gene methylation, HpaII methylation of the wild-type mb-1 promoter resulted in an 84.7% (P = 0.0002) decrease in transcriptional activity, indicating a very strong effect of methylation on transcriptional activity mediated by Pax-5-Ets ternary complexes. Notably, levels obtained with the methylated wild-type promoter are similar to levels obtained with the Ets site mutation in the unmethylated promoter. These data, together with our EMSA data, suggest that transcriptional activity of the mb-1 gene is greatly decreased upon methylation of the Pax-5-dependent Ets site due to the blockade of ternary complex formation.
mb-1 expression correlates with reduced promoter methylation in 558LμM cells.
To determine whether the methylation status of the 558LμM mb-1 promoter correlates with the ability to express surface mIgM, we performed MS-SNuPE (16), which quantitatively determines the methylation status of a single CpG site on a single DNA strand. In MS-SNuPE, genomic DNA is modified with sodium bisulfite, resulting in deamination of unmethylated cytosine to produce uracil. 5-Methyl cytosine is protected from deamination. Modified DNA is PCR amplified using primers specific for converted DNA. Importantly, because the bisulfite reaction changes DNA sequences via cytosine conversion, sense and antisense strands of the template DNA are no longer complementary. Therefore, sense and antisense strands can be amplified separately with strand-specific primers. After amplification, PCR products are subjected to single nucleotide primer extension using a primer ending one base 5′ of the cytosine of interest. α32P-labeled dCTP or dTTP are included in a standard PCR in the absence of other nucleotides, and a single round of extension is performed. If the cytosine of interest was originally methylated, radiolabeled cytosine is incorporated in the MS-SNuPE reaction. If the cytosine was originally unmethylated, the base is converted and a thymine is incorporated. A comparison of dCTP and dTTP incorporation is made to determine the methylation status of the cytosine of interest.
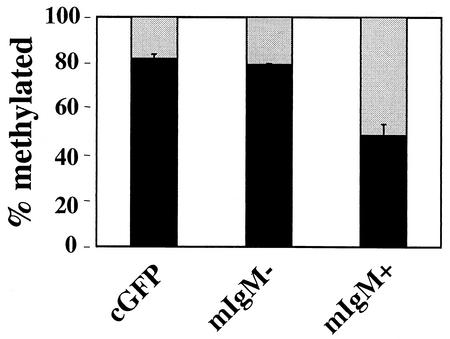
MS-SNuPE was used to determine the methylation status of the Pax-5-dependent Ets binding site on the antisense strand, because Ets-1 binding is blocked by its methylation (Fig. 2). cGFP or Pax-5-transduced 558LμM cells were sorted based on GFP expression and, in the case of the Pax-5-transduced cells, mIgM expression (Fig. 1C). The cGFP and mIgM− populations were 82 and 79% methylated, respectively, while the mIgM+ population was only 49% methylated (Fig. 4). Although it is not possible to determine allelic differences using MS-SNuPE, the data suggest that the majority of 558LμM cells in the starting population had two methylated mb-1 alleles, while the sorted mIgM+ cells had on average one methylated allele and one unmethylated allele.
FIG. 4.
Methylation of the Ets-binding site CpG correlates with an inability to activate endogenous mb-1 gene transcription. Shown are the results of MS-SNuPE (see Materials and Methods) analysis of the antisense strand Pax-5-dependent Ets site in DNA isolated from sorted cGFP and Pax-5-expressing mIgM− and mIgM+ 558LμM cells (see Fig. 1C). Percent methylation was determined as follows: dCTP cpm/(dCTP cpm + dTTP cpm) · 100%. Black bars indicate percentages of alleles methylated at the Pax-5-dependent Ets site.
The differing levels of methylation between mIgM− and mIgM+ cells could be explained by either of two mechanisms: (i) Pax-5 directly or indirectly induces demethylation of the promoter in a subpopulation of 558LμM cells, or (ii) expression of Pax-5 activates mb-1 transcription in a population of cells that have reduced methylation of the promoter relative to the total population. In the latter model, mb-1 genes in 558LμM cells may be heterogeneously methylated. In select clones, lower levels of methylation are predicted to predispose these cells to activation of their mb-1 genes by Pax-5.
To address these two possibilities, two cloning strategies were employed to determine whether the ability to activate the mb-1 gene is a clonal property and whether the methylation status of the mb-1 gene dictates or is a result of mb-1 transcriptional activation. First, 558LμM cells were transduced with a Pax-5 DBD retrovirus. Previously, carboxyl-terminal sequences of Pax-5 were shown to be dispensable for activation of the mb-1 gene in Pax-5-deficient pre-B cells (38), which is likely due to the ability of the DBD to recruit Ets proteins as well as the full-length protein. The Pax-5 DBD is sufficient for activation of mb-1 transcription in 558LμM cells, although at somewhat lower levels than that obtained with full-length Pax-5 (Fig. 5A). We chose to use the Pax-5 DBD instead of full-length Pax-5 in this cloning strategy because the DBD is expressed more stably in 558LμM cells than full-length Pax-5, perhaps due to toxicity of the full-length protein. Although full-length-Pax-5-expressing cultures did not display increased numbers of dead or dying cells compared to untransduced or cGFP-transduced cultures, downregulation of GFP and full-length Pax-5 expression was detected within 1 week following transduction (data not shown). Use of the Pax-5 DBD in place of full-length Pax-5 enabled long-term cloning strategies to be performed without a loss of Pax-5 DBD expression. 558LμM cells were cloned by limiting dilution following transduction with Pax-5 DBD retroviruses and analyzed for surface mIgM expression by flow cytometry. Eighteen clones were examined, of which 12 expressed surface mIgM and 6 did not. Four clones were subjected to an additional round of subcloning to ensure the isolation of single cell clones, and two mIgM− clones (DBD 7.3 and DBD 9.2) and two mIgM+ clones (DBD 6.6 and DBD 8.4) were obtained (Fig. 5B, upper panel). Three and 2% of DBD 7.3 and DBD 9.2 cells expressed surface mIgM, respectively, while 99 and 100% of DBD 6.6 and DBD 8.4 cells expressed surface mIgM, respectively. The clones maintained their phenotypes stably for more than 2 months in culture. Despite high expression levels of DBD protein in all four clones, only the mIgM+ clones DBD 6.6 and DBD 8.4 expressed detectable levels of mb-1 transcripts (Fig. 5C) and Ig-α protein (Fig. 5D). Notably, the highest levels of Pax-5 DBD were expressed in DBD 7.3 cells, which did not express mIgM on the cell surface, indicating that Pax-5 DBD protein levels are not alone responsible for these phenotypes. MS-SNuPE analysis of DBD 7.3 and DBD 9.2 cells detected 88.5 and 67% methylation of mb-1 genes, respectively, while DBD 6.6 and DBD 8.4 were 12.6 and 35% methylated, respectively (Fig. 5E). Thus, levels of methylation in these cells compare favorably with their mIgM expression profiles.
FIG.5.
Pax-5 DBD-expressing clones exhibit differential transcription of the mb-1 gene. (A) Retrovirally expressed Pax-5 DBD activates mb-1 transcription in 558LμM cells similar to full-length Pax-5, as determined by surface mIgM expression. (B) DBD-expressing cells were cloned by limiting dilution, expanded, and analyzed for mIgM expression by flow cytometry (upper panels). Expression of cell surface mIgM in two clones, DBD 6.6 and DBD 8.4, was efficiently activated by Pax-5 DBD. In contrast, expression of mIgM was only weakly detected on the surface of DBD 7.3 and DBD 9.2. Treatment of these cells with 2 μM 5-azaC for 48 h resulted in upregulation of surface mIgM expression (lower panels). (C) Real-time RT-PCR quantitation of mb-1 transcripts in the DBD-expressing clones. (D) Western blot analysis of the indicated whole-cell lysates for Pax-5, Ig-α, and Sp1 expression. PD36 pre-B cells served as a positive control, and detection of Sp1 served as a loading control. (E) MS-SNuPE analysis of the antisense strand Pax-5-dependent Ets site was performed using DNA isolated from the DBD-expressing clones. Black bars show the percentage of alleles methylated at the Pax-5-dependent Ets site in the indicated population.
To further confirm that methylation of the mb-1 gene correlates with the lack of mIgM expression in clones DBD 7.3 and DBD 9.2, these mIgM− clones were treated with 5-azacytidine (5-azaC), an inhibitor of methyltransferases that causes progressive demethylation of the genome as cells divide. A total of 45% of DBD 7.3 and 23% of DBD 9.2 cells expressed surface mIgM following 48 h of treatment with 2 μM 5-azacytidine, while cGFP-transduced cells still did not express detectable levels of surface mIgM (Fig. 5B, lower panel). Therefore, 5-azacytidine facilitates a significant Pax-5-dependent increase of mIgM+ cells. In contrast, treatment of clones with the histone deacetylase inhibitor TSA had no effect on surface mIgM expression (data not shown).
Another possible explanation for the inability of Pax-5 to activate mb-1 transcription in these clones is the lack of Pax-5 partner proteins. To address this issue, nuclear extracts were prepared from cGFP-transduced 558LμM cells, as well as the Pax-5 DBD-transduced DBD 7.3 (mIgM−) and DBD 6.6 (mIgM+) clones. The extracts were used in EMSA to detect Pax-5 ternary complex assembly with endogenous proteins (Fig. 6A). Probes included the unmethylated and antisense hemimethylated probes used in Fig. 2. Complexes were not detected using cGFP nuclear extract in the absence of Pax-5 (lane 3). However, with the addition of 10 nM recombinant Pax-5 (1-149) DBD protein, Pax-5-Ets ternary complexes were readily detected (lane 4). Importantly, ternary complex formation was dependent on the concentration of Pax-5, because reducing recombinant Pax-5 decreased ternary complex assembly accordingly (lanes 5 and 6). To confirm the identities of proteins in the detected complexes, we tested factor binding in the presence of specific antibodies recognizing Pax-5 or Ets-1, because recent studies suggest that Ets-1 is the most likely functional partner of Pax-5 in B cells (D. Fitzsimmons and J. Hagman, unpublished). Supershifting by Ets-1- (lane 8) or Pax-5-specific antibodies (lane 9) confirmed the presence of these proteins in the ternary complex bands. Faster-migrating complexes did not react with the Ets-1 antibody, but contained Pax-5. It is likely that these complexes contain partially degraded Ets-1 that has lost sequences recognized by the antiserum. Similar analysis of DBD 7.3 and DBD 6.6 extracts suggests that differential assembly of ternary complexes is not responsible for differences between these clones. In these clones, the Pax-5 DBD migrates more slowly due to the presence of the SV40 nuclear localization sequence at the carboxyl terminus. Similar to results with cGFP-transduced cells, ternary complex assembly is dependent on the concentration of the retrovirally expressed Pax-5 DBD (lanes 10 and 14, respectively), and complexes show similar patterns of immunoreactivity with Pax-5- and Ets-1-specific antibodies. Interestingly, higher levels of Pax-5 DBD and ternary complexes are detected in the mIgM− DBD 7.3 clone relative to the mIgM+ DBD 6.6 clone. As a control for specificity of these complexes, ternary complexes do not assemble on the antisense hemimethylated probe (lanes 18 to 23), further confirming that methylation of the Ets binding site blocks ternary complex assembly.
FIG. 6.
Expression of Ets partner proteins in mb-1-expressing and -nonexpressing DBD clones. (A) EMSA detection of Pax-5-Ets ternary complexes. Nuclear extracts (0.5 μg) prepared from cGFP-transduced 558LμM, DBD 7.3, and DBD 6.6 cells were used in EMSAs to detect binding to the mb-1 ternary complex probe. cGFP extracts were tested alone or with increasing concentrations of recombinant Pax-5(1-149). Supershifting with anti-Ets-1 antisera and anti-Pax-5 antibodies detected these proteins in ternary complexes. Bands indicated by an asterisk are faster-migrating complexes that did not react with the Ets-1 antibody but contained Pax-5. The presence of a fast-migrating band below Pax-5 in lanes 8, 12 and 16 is an artifact generated by the anti-Ets-1 antiserum that was not consistently observed. In lanes 18 to 23, nuclear extracts and recombinant Pax-5(1-149) were tested with the antisense hemimethylated ternary complex probe (see Fig. 2). (B) Western blotting of cGFP-transduced 558LμM cells, DBD 7.3, DBD 6.6, DBD 9.2, and DBD 8.4 nuclear extracts (25 μg) probed with anti-Ets-1 antisera. The faster-migrating band is Ets-1(p50). The slower-migrating band is likely an alternatively spliced form of Ets-1, or a highly homologous protein that cross-reacts with the anti-Ets-1 antisera.
To assess levels of total Ets-1 protein in the clones, we performed Western blotting on nuclear extracts prepared from cGFP-transduced 558LμM cells, or Pax-5 DBD-transduced DBD 7.3, DBD 6.6, DBD 9.2, or DBD 8.4 cells with the anti-Ets-1 antibody. The data indicate that Ets-1 is expressed in equivalent amounts in each of the five cell populations (Fig. 6B). These experiments show that proteins necessary for Pax-5-Ets ternary complex assembly are present in each of these cell populations. Together, results in Fig. 6A and B suggest that regulatory mechanisms (e.g., DNA methylation) other than levels of ternary complex proteins are more likely to be responsible for the different phenotypes of these cells.
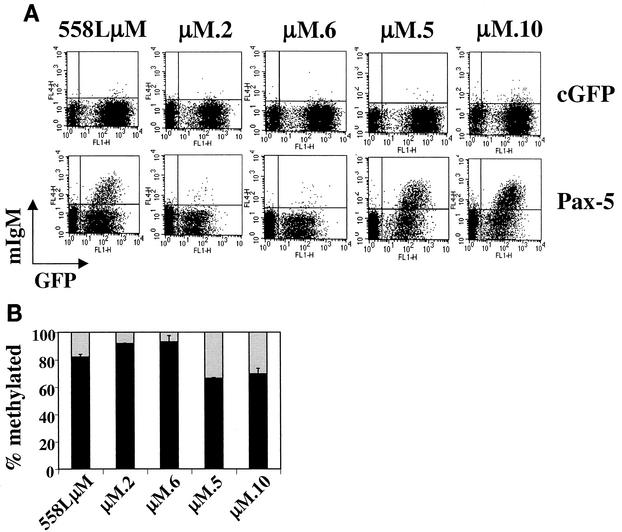
To address whether the methylation status of the Pax-5-dependent Ets site predetermines whether Pax-5 is able to activate mb-1 transcription, cells from the 558LμM cell line were cloned by limiting dilution, and 10 resulting clones were transduced with cGFP or Pax-5-expressing retroviruses and analyzed for their ability to display surface mIgM. Three out of 10 clones yielded high percentages of mIgM expressing cells (27.8, 34.3, and 41.4%) when transduced with the Pax-5 retrovirus. The remaining seven clones yielded only very low percentages of mIgM-expressing cells (less than 4% mIgM+). The parent population and four representative clones are shown in Fig. 7A. The low-frequency clones μM.2 and μM.6 produced 3 and 4% surface mIgM-expressing cells, respectively, while the high-frequency clones μM.5 and μM.10 produced 34.3 and 41.1% surface mIgM-expressing cells, respectively. To determine whether the low-frequency and high-frequency clones had intrinsic differences in the methylation status of their mb-1 promoters, MS-SNuPE was performed on the Pax-5-dependent Ets site in the expanded clones prior to viral transduction (Fig. 7B). Low-frequency clones μM.2 and μM.6 were 92 and 93% methylated, respectively. These levels were approximately 10% higher than the level of methylation in the 558LμM parent population. Strikingly, the high-frequency clones μM.5 and μM.10 were 66 and 69% methylated, respectively, which is a 13 to 16% decrease compared to the parent population and a 23 to 26% decrease compared to the low-frequency clones. These data strongly suggest that the Pax-5-dependent Ets site CpG must be unmethylated in order for Pax-5 to recruit its Ets partner protein, form a stable ternary complex, and activate transcription.
FIG. 7.
Differential activation of mb-1 transcription by Pax-5 in 558LμM clones. (A) 558LμM cells were cloned by limiting dilution prior to transduction with retroviruses for expression of full-length Pax-5. 558LμM is the parent population. μM.2, μM.6, μM.5, and μM.10 are subclones derived from 558LμM. After cloning by limiting dilution, cells were transduced either with cGFP or Pax-5-expressing retroviruses and analyzed for mIgM expression at 3 dpt. (B) MS-SNuPE analysis of the antisense strand Pax-5-dependent Ets site was performed using DNA isolated from the 558LμM parent population and clones. Black bars show the percentage of alleles methylated at the Pax-5-dependent Ets site in the indicated population.
Methylation status of the mb-1 promoter in B-cell lines.
Having established that methylation of the Pax-5-dependent Ets binding site in the mb-1 promoter inhibits transcription, we asked whether methylation of this site might be a regulatory mechanism responsible for modulating mb-1 promoter activity during B-cell development. We compared transcript levels and Ets site methylation levels in five B-cell lines representing various stages of B-cell development (Table 1). 40E1 pre-BI cells expressed high levels of mb-1 transcripts as determined by a quantitative real time RT-PCR assay and displayed only 1.4% methylation of the Ets binding site CpG. 70Z/3 pre-BII-like cells expressed mb-1 transcript levels that were approximately 46% of 40E1 and were 1.7% methylated. The A20 cell line, which has undergone Ig heavy chain class switching and thus represents a more mature B-cell stage, expressed mb-1 transcript levels approximately 21-fold below those for 40E1 and was 5.3% methylated. The plasmacytoma cell lines S194 and J558L contained nearly undetectable levels of mb-1 transcripts (471- and 112-fold below 40E1 levels, respectively) and had much higher levels of Ets site methylation, 57.1 and 82.1%, respectively. In summary, terminally differentiated cell lines exhibit high levels of methylation of the Pax-5-dependent Ets site, together with a lack of mb-1 transcripts. However, methylation of the mb-1 promoter does not directly correlate with levels of transcripts, per se. Instead, other studies in our laboratory suggest that levels of mb-1 transcripts correlate best with other parameters, including levels of EBF. Together, these data suggest that an unmethylated Pax-5-dependent Ets site is necessary but not sufficient for activation of mb-1 transcription.
TABLE 1.
Comparison of mb-1 transcript levels with methylation levels at the mb-1 promoter Pax-5-dependent Ets site
| Cell line | Relative concna | % DNA methylationb |
|---|---|---|
| 40E1 | 471.1 ± 0.1 | 1.4 |
| 70Z/3 | 215.3 ± 0.2 | 1.7 |
| A20 | 22.5 ± 0.2 | 5.3 |
| S194 | 1.0 ± 0.3 | 57.1 |
| J558L | 4.2 ± 0.2 | 82.1 |
Fold increase of mb-1 transcripts versus S194 cells as determined by real-time PCR quantitation. Values were previously reported in Sigvardsson et al. (43).
MS-SNuPE quantitation of CpG methylation levels at the mb-1 promoter Pax-5-dependent Ets site.
The Pax-5-dependent Ets site is demethylated at all stages of B-cell development.
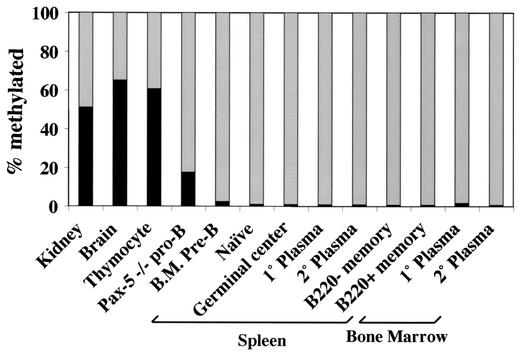
To determine whether methylation of the Pax-5-dependent Ets site is tissue-specifically and developmentally regulated, MS-SNuPE was performed on non-B cells and on primary B cells representing various stages of differentiation (Fig. 8). Whole kidney and whole brain exhibited 51 and 65% methylated alleles, respectively. Thymocytes showed similar levels, with 61% methylation. This is in contrast to previously published results, which indicated that human pre-T-cell lines are completely methylated (18). However, the method for analyzing methylation was not as sensitive as the MS-SNuPE assay, and pre-T-cell lines may not accurately reflect the phenotype of primary thymocytes. In contrast, CD19+ IgM− pre-B cells isolated from bone marrow were 2.4% methylated. Interestingly, B220+ pre-BI cells isolated from bone marrow of Pax-5−/− mice were 17.5% methylated, suggesting that Pax-5 plays a role in maintaining the unmethylated state of the mb-1 promoter during early stages of B-cell development. B cells from later stages of differentiation were all unmethylated (>98%), including B220+ IgM+ IgD+ naïve B cells, day 7 primary immune response (NP-KLH) germinal center B cells (IgD− NP+ B220+), day 7 primary splenic plasma cells (IgD− NP+ CD138+), day 5 secondary response splenic plasma cells (IgD− NP+ CD138+), day 5 secondary response splenic memory B cells (IgD− NP+ B220− and IgD− NP+ B220+) and primary and secondary response B220-Syndecan+ plasma cells sorted from bone marrow. Notably, bone marrow-derived plasma cells, which represent normal counterparts of plasmacytoma cell lines, did not reproduce the high levels of mb-1 promoter methylation observed in the tumor cells. The data indicate that methylation of the Pax-5-dependent Ets binding site is not modulated during normal B-cell development, but other tissues feature high percentages of methylated mb-1 alleles.
FIG. 8.
Methylation of the Pax-5-dependent Ets site in non-B-cell tissues and sorted ex vivo B cells at different stages of development. MS-SNuPE analysis of the antisense strand Pax-5-dependent Ets site was performed using DNA isolated from the indicated tissues and ex vivo-sorted cell populations. Black bars indicate the percentage of alleles methylated at the Pax-5-dependent Ets site. B.M., bone marrow-derived. Pax-5−/− pre-B cells were derived from bone marrow of Pax-5 knockout mice (38). The phenotype of the J558L plamacytoma cells (the parent cell line of 558LμM) most resembles bone marrow-derived plasma cells.
DISCUSSION
Methylation of cytosine within CpG dinucleotides is the major modification of DNA in mammalian genomes. With few exceptions, DNA methylation represses transcription either by blocking binding of transcription factors to their CpG-containing binding sites or by recruiting methyl-binding proteins, which in turn recruit chromatin-remodeling machinery to induce the formation of a repressive chromatin structure (2, 39, 45). Most DNA methylation occurs symmetrically. During the cell cycle, newly replicated DNA incorporates unmethylated cytosine. The DNA methyltransferase 1 protein (Dnmt1) recognizes hemimethylated DNA and methylates the daughter strand, reestablishing a fully methylated state. Another form of DNA methylation is mediated by the de novo methyltransferases Dnmt3a and Dnmt3b. Unlike Dnmt1, these two proteins show no preference for hemimethylated substrates, and their expression patterns correlate with de novo methylation during development (39). In addition to their DNA-methylating capabilities, Dnmt proteins recruit histone deacetylases to DNA, which in turn repress transcription (reviewed in reference 4). Thus, DNA methylation and histone deacetylation appear to be intimately associated processes leading to transcriptional repression.
In contrast to the detailed understanding of the DNA methylation process, the mechanisms promoting demethylation are unclear. As no DNA demethylase has yet been identified, the mechanism of demethylation likely involves remodeling of the chromatin architecture, allowing sequence-specific transcription factors to access their binding sites, activate transcription, and prevent Dnmt1 from methylating newly synthesized strands during DNA replication. Thus, over several rounds of division, daughter cells effectively become demethylated at recently activated genes.
In the immune system, DNA methylation plays an important role in the transcriptional regulation of several lineage-specific genes. Rearrangement of genes to assemble the mature BCR or T cell receptor (TCR) genes in B or T cells, respectively, generally proceeds in multiple, developmentally regulated steps. The methylation status of gene segments encoding BCR or TCR chains has been correlated with V(D)J rearrangement (3, 10, 17, 28), although demethylation alone is not sufficient to activate recombination (6). In B cells, a rare recessive mutation in the Dnmt3b gene causes an immunodeficiency disease called ICF (immunodeficiency, centromeric region instability, and facial anomalies). Individuals with ICF have normal numbers of B cells, no or reduced levels of serum immunoglobulin, and altered expression levels of many B-cell-specific genes (8), suggesting that the methylation status of these genes contributes to their regulation. More recently, the Dnmt1 gene was conditionally deleted in T cells, resulting in reduced cell viability and increased expression of cytokines (33). Reduced methylation of cytokine genes directly correlated with increased levels of cytokine expression compared to wild-type T cells. In addition, CD8αβ, which is normally restricted to T cells expressing TCRαβ, was expressed inappropriately by TCRγδ cells in Dnmt1 knockout mice. This aberrant expression correlated strongly with demethylation of CD8α and CD8β genes in Dnmt1-deficient TCRγδ cells, suggesting that demethylation promotes expression of these genes. Methylation also controls transcription of the interleukin-4 (IL-4) cytokine gene during Th1/Th2 differentiation of T cells (29, 32).
Here we report a novel DNA methylation-dependent mechanism for regulating transcriptional activity through the inhibition of DNA-dependent protein-protein interactions. The data indicate that methylation of a single specific cytosine within the B-cell-specific mb-1 promoter blocks the formation of functional Pax-5-Ets ternary complexes. Intriguingly, CpG methylation does not directly affect binding to the promoter by Pax-5. Instead, methylation of a CpG residing within the Pax-5-dependent Ets binding site interferes with assembly of ternary complexes comprising Pax-5 and Ets proteins. Three pieces of evidence support this conclusion. First, although binding of the promoter by Pax-5 is not affected by DNA methylation, Pax-5-Ets-1 ternary complexes do not assemble when the promoter is methylated at a CpG within the Ets site on the antisense strand. Second, methylation of the mb-1 promoter Ets binding site greatly reduces promoter activity in transfection assays. Finally, activation of endogenous mb-1 gene transcription in 558LμM cells requires prior demethylation of the promoter Ets site. Thus, we conclude that methylation of the Pax-5-dependent Ets site CpG results in the inability of Pax-5 to activate mb-1 transcription.
Analysis of the effects of methylation upon activation of the mb-1 gene in 558LμM cells using retrovirally expressed Pax-5 is complicated. Pax-5-transduced mIgM− cells did not exhibit methylation levels higher than the control population (cGFP), as might be expected due to separation from the less-methylated mIgM+ cells. This observation can be explained by the downregulation of mIgM and GFP expression over time, which we often observe in cells transduced with full-length Pax-5. In multiple experiments, mIgM and GFP expression peaked on day 3 posttransduction and declined by approximately 50% over an 8-day time course (data not shown). Thus, it is possible that some cells initially expressing mb-1 transcripts (and possessing a lower level of methylation) did not maintain surface Ig levels effectively, and as a result they were excluded from the mIgM+ population. In addition, comparison of the GFP intensity of mIgM+ cells versus mIgM− cells suggests dose-dependent effects of Pax-5. A similar dose dependency was observed for Pax-5-dependent activation of the target gene CD19 (38). It is possible that many of the cells with lower methylation levels express insufficient Pax-5 for induction of detectable mIgM surface expression.
Another level of complexity becomes apparent when examining the data obtained for clones μM.5 and μM.10 (Fig. 7). Although these clones have a reduced level of Ets site methylation compared to the parent population, they maintain a higher level of methylation than one might expect from a clonal population. One possible interpretation of these results is that in the absence of Pax-5, the heterogeneous methylation status of mb-1 genes in these populations is maintained. The mechanism controlling methylation in these cells may be stochastic, where the methylation status of mb-1 promoters is a function of the concentration of one or more factors involved in regulating methylation. Future studies will attempt to address the identity(ies) of this factor(s).
Pax-5 itself is unlikely to facilitate demethylation of the mb-1 promoter. Instead, activation of transcription by Pax-5 is dependent on the prior demethylation of the Pax-5-dependent Ets site. Experiments (Fig. 5) indicate that even very high Pax-5 protein levels (as in clone DBD 7.3) are not sufficient for activation of Ig-α expression in the presence of high levels of DNA methylation. However, we cannot rule out the possibility that another factor(s) contributes to the ability of Pax-5 to activate mb-1 transcription in these clones. This may account for the observation that DBD 9.2 cells, which have 33% unmethylated mb-1 alleles, do not activate mb-1 transcription in response to Pax-5 DBD expression (Fig. 5C and E). Ets partners of Pax-5 are obvious candidates for factors that augment Pax-5 function, and recent analyses of requirements for activation of mb-1 transcription by Pax-5 suggest that Ets-1, and not GABPα/β, is the most likely partner of Pax-5 in B cells. However, differences in Ets-1 expression were not observed in our cloned lines (Fig. 6). EBF, which was not detected significantly in any of the clones, is also unlikely to account for the observed differences. As an alternative hypothesis, chromatin architecture and sublocalization of mb-1 genes within heterochromatic regions of the nucleus could potentially block transcriptional activity by preventing access to the transcriptional machinery (9), but additional studies are necessary to explore these possibilities.
The simplest explanation for differences between Pax-5-responsive and nonresponsive clones is the relative status of mb-1 promoter DNA methylation. Activation of the mb-1 gene in Pax-5 DBD-expressing mIgM− clones is induced by treatment with 5-azacytidine, a drug that inhibits methyltransferase activity and therefore indirectly demethylates genes in dividing cells (Fig. 5B). As one caveat, we cannot rule out that this treatment regimen does not affect the expression of other factors necessary for mb-1 transcription in these cells. However, the ability to form ternary complexes (including Ets-1) is already present in the expressing and nonexpressing clones used in our studies. In addition, EMSA and luciferase assay data strongly support the conclusion that demethylation of the Ets site is necessary for transcriptional activation of the mb-1 gene. Therefore, even if 5-azacytidine has other effects, demethylation of the mb-1 promoter must occur prior to transcriptional activation.
Transcription of the mb-1 gene is differentially regulated during B-cell development, but mb-1 genes are completely demethylated in early pre-B cells through terminally differentiated plasma cells. B-lineage cell lines possess higher frequencies of methylated alleles. Analysis of these lines detected high levels of expression in cells representing early B-cell progenitors, lower levels in mature and germinal center B cells, and a lack of expression in terminally differentiated plasma cells (H. Maier, unpublished data, and reference 43). Lower levels of transcripts correlate with reduced occupancy of factors on promoter regions in vivo in cell lines representing these stages of development. The data presented in Table 1 suggest that DNA methylation is not the cause of reduced mb-1 transcription, since the A20 cell line, which expresses low levels of mb-1 transcripts, maintains highly unmethylated mb-1 alleles (although levels of methylation are greater than in cell lines expressing higher levels of mb-1 transcripts). In addition, primary germinal center B cells, which express 5- to 10-fold less mb-1 transcripts than primary resting B cells (data not shown), and splenic and bone marrow plasma cells, which express little if any mb-1 transcripts, all have completely unmethylated mb-1 alleles (Fig. 8). Instead of increased mb-1 promoter methylation, reduced mb-1 expression at later stages of B-cell development is more likely due to reduced levels of key tissue-specific transcription factors necessary for activation, such as EBF. EBF transcript levels correlate exceptionally well with mb-1 transcripts, and footprint analysis of the mb-1 promoter indicates that levels of transcripts correlate well with occupancy of the promoter EBF site in vivo (43).
Methylation of the mb-1 promoter may not be important for transcriptional regulation within the B lineage but may aid in preventing inappropriate transcription in other tissues. As suggested by our analysis of plasmacytoma cell lines, DNA methylation is a potential mechanism for controlling the ability of Pax-5 and related proteins to form ternary complexes on gene targets. Differential methylation in non-B cells versus B cells suggests that this mechanism may be important for limiting access of factors to mb-1 alleles in non-B-cell lineages. In this regard, various Pax and Ets proteins are widely expressed in humans and other mammals, and protein-protein interactions are predicted to enhance their DNA binding in vivo (5, 13). Pax-5 and its close relatives Pax-2 and Pax-8 have degenerate (and overlapping) DNA recognition properties that are insufficient for restricting their DNA-binding specificity in vivo. These proteins share the highly conserved Pax β-hairpin motif, which participates in interactions with Ets proteins (13, 46). Indeed, we have shown that Pax family members including Pax-2 and more distantly related family members can assemble ternary complexes with many different Ets proteins on the mb-1 promoter in vitro (13, 46), suggesting an evolutionarily conserved mechanism for regulating transcription. Taken together, these observations suggest that, in other tissues, methylation of the mb-1 promoter Pax-5-Ets ternary complex site may be necessary to prevent binding by related proteins and inappropriate activation of mb-1 transcription during development.
Mechanisms that initiate demethylation of inactive genes are poorly understood. Our data suggest that Pax-5 is an unlikely candidate for an initiator of mb-1 gene demethylation during lymphopoiesis, because its prolonged expression is unable to decrease methylation in 558LμM cells, even after >16 rounds of DNA replication. Therefore, in contrast with reports of viral proteins that promote demethylation by binding directly to transiently hemimethylated sites during DNA replication (27), other mechanisms must be invoked to establish the hypomethylated state of the mb-1 promoter in B cells. A role for Pax-5 is not completely ruled out, because B-cell precursors in Pax-5-deficient mouse bone marrow exhibit much higher frequencies of methylated alleles relative to their normal counterparts. We propose that, prior to the onset of Pax-5 function, other factors assemble complexes that stimulate chromatin remodeling and reduce methylation. Candidates include the multiprotein complex we have detected upstream of the Pax-5 binding sites, including EBF, E2A basic helix-loop-helix proteins, and/or related proteins (43). Notably, EBF has now been shown to mediate subnuclear localization of one target gene, λ5, within active regions of chromatin (34). Although we currently do not know the stage at which the mb-1 gene becomes hypomethylated during development, our results suggest that it may be coincident with, or before the onset of definitive B-lineage-specific factors. Clearly, this is an important mechanism with implications for the general control of tissue-specific transcription, cell differentiation, and hematopoiesis.
Acknowledgments
We thank James DeGregori and Arthur Gutierrez-Hartmann for their critical reading of the manuscript and Robert Scheinman, John Kappler, V. Michael Holers, Raul Torres, Randy Noelle, and Michael McHeyzer-Williams for helpful discussions. We thank Roberta Pelanda and Philippa Marrack for providing mice for these studies, Michael Weaver for his bisulfite treatment protocol, Meinrad Busslinger for his gift of Pax-5-deficient mice, and Michael McHeyzer-Williams and Louise McHeyzer-Williams for providing NP-specific sorted cells for MS-SNuPE analysis.
H.M. is supported by National Institutes of Health training grant T32 AI 07405. D.F. was supported by funds from NJMRC. D.R.C. and J. H. are recipients of grants from the Cancer League of Colorado. J.H. is supported by grants from the National Institutes of Health (R01 AI37574 and P01 AI22295), from the Rocky Mountain Chapter of the Arthritis Foundation, and by a generous gift from the Milheim Foundation.
REFERENCES
- 1.Adams, B., P. Dorfler, A. Aguzzi, Z. Kozmik, P. Urbanek, I. Maurer-Fogy, and M. Busslinger. 1992. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 6:1589-1607. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression—belts, braces and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 3.Burger, C., and A. Radbruch. 1990. Protective methylation of immunoglobulin and T cell receptor (TcR) gene loci prior to induction of class switch and TcR recombination. Eur. J. Immunol. 20:2285-2291. [DOI] [PubMed] [Google Scholar]
- 4.Burgers, W. A., F. Fuks, and T. Kouzarides. 2002. DNA methyltransferases get connected to chromatin. Trends Genet. 18:275-277. [DOI] [PubMed] [Google Scholar]
- 5.Busslinger, M., and P. Urbanek. 1995. The role of BSAP (Pax-5) in B-cell development. Curr. Opin. Genet. Dev. 5:595-601. [DOI] [PubMed] [Google Scholar]
- 6.Cherry, S. R., C. Beard, R. Jaenisch, and D. Baltimore. 2000. V(D)J recombination is not activated by demethylation of the kappa locus. Proc. Natl. Acad. Sci. USA 97:8467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eden, S., and H. Cedar. 1994. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 4:255-259. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich, M., K. L. Buchanan, F. Tsien, G. Jiang, B. Sun, W. Uicker, C. M. Weemaes, D. Smeets, K. Sperling, B. H. Belohradsky, N. Tommerup, D. E. Misek, J. M. Rouillard, R. Kuick, and S. M. Hanash. 2001. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum. Mol. Genet. 10:2917-2931. [DOI] [PubMed] [Google Scholar]
- 9.Emerson, B. A. 2002. Specificity of gene regulation. Cell 109:267-270. [DOI] [PubMed] [Google Scholar]
- 10.Engler, P., A. Weng, and U. Storb. 1993. Influence of CpG methylation and target spacing on V(D)J recombination in a transgenic substrate. Mol. Cell. Biol. 13:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldhaus, A., D. Mbangkollo, K. Arvin, C. Klug, and H. Singh. 1992. BlyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol. Cell. Biol. 12:1126-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzsimmons, D., W. Hodsdon, W. Wheat, S. M. Maira, B. Wasylyk, and J. Hagman. 1996. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10:2198-2211. [DOI] [PubMed] [Google Scholar]
- 13.Fitzsimmons, D., R. Lutz, W. Wheat, H. M. Chamberlin, and J. Hagman. 2001. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 29:4154-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvie, C. W., J. Hagman, and C. Wolberger. 2001. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell 8:1267-1276. [DOI] [PubMed] [Google Scholar]
- 15.Gaston, K., and M. Fried. 1995. CpG methylation has differential effects on the binding of YY-1 and ETS proteins to the bi-directional promoter of the Surf-1 and Surf-2 genes. Nucleic Acids Res. 23:901-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalgo, M. L., and P. A. Jones. 1997. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (MS-SNuPE). Nucleic Acids Res. 25:2529-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodhardt, M., P. Cavelier, N. Doyen, S. Kallenbach, C. Babinet, and F. Rougeon. 1993. Methylation status of immunoglobulin kappa gene segments correlates with their recombination potential. Eur. J. Immunol. 23:1789-1795. [DOI] [PubMed] [Google Scholar]
- 18.Ha, H., B. L. Barnoski, L. Sun, B. S. Emanuel, and P. D. Burrows. 1994. Structure, chromosomal localization, and methylation pattern of the human mb-1 gene. J. Immunol. 152:5749-5757. [PubMed] [Google Scholar]
- 19.Hagman, J., C. Belanger, A. Travis, C. W. Turck, and R. Grosschedl. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760-773. [DOI] [PubMed] [Google Scholar]
- 20.Hagman, J., and R. Grosschedl. 1992. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc. Natl. Acad. Sci. USA 89:8889-8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagman, J., M. J. Gutch, H. Lin, and R. Grosschedl. 1995. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 14:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagman, J., A. Travis, and R. Grosschedl. 1991. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 10:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington, M. A., P. A. Jones, M. Imagawa, and M. Karin. 1988. Cytosine methylation does not affect binding of transcription factor Sp1. Proc. Natl. Acad. Sci. USA 85:2066-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136-138. [PubMed] [Google Scholar]
- 25.Hombach, J., L. Leclercq, A. Radbruch, K. Rajewsky, and M. Reth. 1988. A novel 34-kd protein co-isolated with the IgM molecule in surface IgM-expressing cells. EMBO J. 7:3451-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hombach, J., T. Tsubata, L. Leclerq, H. Stappert, and M. Reth. 1990. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature 343:760-762. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh, C.-L. 1999. Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol. 19:46-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh, C. L., G. Gauss, and M. R. Lieber. 1992. Replication, transcription, CpG methylation and DNA topology in V(D)J recombination. Curr. Top. Microbiol. Immunol. 182:125-135. [DOI] [PubMed] [Google Scholar]
- 29.Hutchins, A. S., A. C. Mullen, H. W. Lee, K. J. Sykes, F. A. High, B. D. Hendrich, A. P. Bird, and S. L. Reiner. 2002. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell 10:81-91. [DOI] [PubMed] [Google Scholar]
- 30.Julius, M. H., M. Janusz, and J. Lisowski. 1988. A colostral protein that induces the growth and differentiation of resting B lymphocytes. J. Immunol. 140:1366-1371. [PubMed] [Google Scholar]
- 31.Kozmik, Z., S. Wang, P. Dorfler, B. Adams, and M. Busslinger. 1992. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol. 12:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, D. U., S. Agarwal, and A. Rao. 2002. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16:649-660. [DOI] [PubMed] [Google Scholar]
- 33.Lee, P. P., D. R. Fitzpatrick, C. Beard, H. K. Jessup, S. Lehar, K. W. Makar, M. Perez-Melgosa, M. T. Sweetser, M. Schlissel, S. Nguyen, S. R. Cherry, J. H. Tsai, S. M. Tucker, W. M. Weaver, A. Kelso, R. Jaenisch, and C. B. Wilson. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function and survival. Immunity 15:763-774. [DOI] [PubMed] [Google Scholar]
- 34.Lundgren, M., C. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 35.Maier, H., and J. Hagman. 2002. Roles of EBF and Pax-5 in B lineage commitment and development. Semin. Immunol. 14:415-422. [DOI] [PubMed] [Google Scholar]
- 36.McHeyzer-Williams, L. J., M. Cool, and M. G. McHeyzer-Williams. 2000. Antigen-specific B cell memory: expression and replenishment of a novel B220-memory B cell compartment. J. Exp. Med. 191:1149-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naviaux, R. K., E. Constanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nutt, S. L., A. M. Morrison, P. Dorfler, A. Rolink, and M. Busslinger. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ordway, J. M., and T. Curran. 2002. Methylation matters: modeling a manageable genome. Cell Growth Differ. 13:149-162. [PubMed] [Google Scholar]
- 40.Reimold, A. M., P. D. Ponath, Y. S. Li, R. R. Hardy, C. S. David, J. L. Strominger, and L. H. Glimcher. 1996. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schebesta, M., P. Pfeffer, and M. Busslinger. 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity 17:781-793. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigvardsson, M., D. R. Clark, D. Fitzsimmons, M. Doyle, P. Åkerblad, T. Breslin, S. Bilke, Y.-F. Liu, C. Yeamans, G. Zhang, and J. Hagman. 2002. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B-cell-specific mb-1 promoter. Mol. Cell. Biol. 22:8539-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swift, S., J. B. Lorens, P. Achacoso, and G. P. Nolan. 2002. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems, unit 10.17C. In J. E. Coligan et al. (ed.), Current protocols in immunology. John Wiley and Sons, Inc., Hoboken, N.J. [DOI] [PubMed]
- 45.Wade, P. A. 2001. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene 20:3166-3173. [DOI] [PubMed] [Google Scholar]
- 46.Wheat, W., D. Fitzsimmons, H. Lennox, S. R. Krautkramer, L. N. Gentile, L. P. McIntosh, and J. Hagman. 1999. The highly conserved beta-hairpin of the paired DNA-binding domain is required for assembly of Pax-Ets ternary complexes. Mol. Cell. Biol. 19:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]