Abstract
The folate receptor type α (FR-α) is a promising tumor marker and target. Here, we investigate the mechanistic basis for the tumor specificity and vast overexpression of FR-α. Among representative FR-α-positive (HeLa and JAR) and FR-α-negative (MG63, Caki1, and HT3) cell lines, the transcription rates of the endogenous FR-α gene, as well as the FR-α promoter activity, were relatively weak and comparable, but the FR-α transcript was abundant only in total RNA and nuclear RNA from the FR-α-positive cells. Rous sarcoma virus (RSV) promoter-driven expression of the FR-α gene was 7 to 30 times greater in the FR-α-positive than in FR-α-negative cells, both at the protein and mRNA levels, independently of intron sequences. Through the use of chimeric FR-α/FR-β cDNAs, the above pattern of FR-α expression was attributed to a 60-bp sequence in the FR-α open reading frame. This sequence element, when placed in the 5′ untranslated region of RSV promoter-luciferase, decreased the reporter expression approximately 7- to 20-fold in FR-α-negative cells (MG63, Caki1, HT3, BG1, and MCF7) relative to FR-α-positive cells (HeLa, JAR, and JEG3). Substitution of this FR-α element in FR-β increased the in vivo degradation rate of the transcript in the nuclei of MG63 cells but not in the nuclei of HeLa cells or in the cytosol of MG63 or HeLa cells. The results reveal an efficient mechanism by which a novel sequence element causes differential transcript degradation in the nucleus to ensure narrow tissue specificity for a gene (e.g., that for FR-α) whose transcription is weak and relatively nonselective. FR-α exhibited constitutive mRNA and protein synthesis during the cell cycle and a slow protein turnover, presumably ensuring a high steady-state level of the receptor in cells that could override the nuclear mRNA instability determinant.
The folate receptor (FR) types α and β are glycosyl-phosphatidylinositol-anchored cell surface glycoproteins (11, 40). The expression of FR-α among normal tissues is restricted to the luminal surface of certain epithelial cells (37, 38, 39). FR-α is consistently and vastly overexpressed in major subtypes of ovarian and uterine carcinoma and testicular choriocarcinoma and less frequently in carcinomas of the breast, colon, and kidney (5, 27, 32, 39). Owing to its inaccessibility in normal tissues via the bloodstream and its high expression in malignant cells, FR-α offers a means of obtaining targeted delivery of diagnostic and therapeutic agents to the receptor-rich tumors. Accordingly, a wide variety of FR-α-mediated diagnostic and therapeutic approaches are currently in various stages of preclinical and clinical testing (8, 9a, 12, 15, 17, 21, 25, 30, 36). FR-β bears approximately 70% amino acid sequence identity to FR-α (23, 40) and is limited in its expression to cells in the myelomonocytic lineage (19, 28, 35).
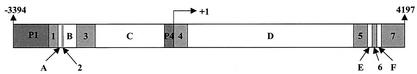
The entire FR-α gene (Fig. 1) consists of seven exons and six introns and two TATA-less promoters, namely, the P1 promoter, located upstream of the first exon, and the P4 promoter, located in the fourth exon (6). The P1 promoter gives rise to multiple, alternatively spliced transcripts, which appear to show some degree of selectivity among different tissues, whereas the P4 promoter generates a single mRNA species (6). P1 promoter transcripts, however, are translated much less efficiently than the P4 promoter transcript (24); moreover, the major FR-α mRNA in malignant cells appears to be P4 promoter driven (6).
FIG. 1.
FR-α gene organization. The 7.5-kb FR-α gene structure is shown with seven exons (labeled 1 to 7) and six introns (labeled A to F). The two promoters, P1 and P4, are shown in shaded boxes. The P1 promoter occurs upstream of exon 1, and the P4 promoter occurs upstream of exon 4. The arrows at the left and right sides indicate nucleotide positions; the transcription start site for the P4 promoter is labeled +1.
Here, we report a systematic investigation of the molecular basis for the narrow tissue and tumor specificity of FR-α. For the detailed studies, we chose two model cell lines (HeLa cervical carcinoma and JAR choriocarcinoma) that are known to overexpress FR-α and three (HT3 cervical carcinoma, MG63 osteosarcoma, and Caki1 kidney carcinoma) that are FR negative, and we subsequently tested additional FR-α-positive and -negative cell lines. Our findings indicate that there is not a tight tissue-specific control of FR-α transcription, since a relatively low level of transcription occurs in FR-α-negative as well as FR-α-positive cells. The results point to differential transcript stability in the nucleus, directed by a novel instability determinant in the open reading frame (ORF) as the principal mechanism governing the tissue specificity of FR-α in the model cell lines. Constitutive gene transcription and protein synthesis throughout the cell cycle, combined with a slow turnover of the protein, appear to ensure a high level of FR-α in HeLa cells despite the low transcription rate.
Cytosolic mRNA stability has emerged as a dominant principle in gene regulation (26), but although the metabolism of snRNAs and snoRNAs has been well studied, gene regulation at the level of mRNA stability in the nucleus has received relatively little attention. The singular role of nuclear mRNA stability in determining the tissue specificity of expression of the FR-α gene is discussed in this context.
MATERIALS AND METHODS
Cell culture and reagents.
All cell culture media were supplemented with 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine. Cells were purchased from the American Type Culture Collection (Manassas, Va.). Tissue culture media were purchased from Irvine Scientific (Santa Ana, Calif.) unless otherwise indicated. HeLa cells were grown in Eagle's minimal essential medium (MEM) with nonessential amino acids and 10% fetal bovine serum (FBS) (Gibco-BRL, Grand Island, N.Y.). JAR cells were grown in RPMI 1640 and 10% FBS. Caki1 and HT3 cells were grown in McCoy's 5A medium and 15% FBS. MG63 cells were grown in Dulbecco's modified Eagle solution with 10% FBS. Restriction enzymes, oligonucleotides, Trizol reagent, and Taq DNA polymerase were purchased from Gibco-BRL. T4 polynucleotide kinase, Klenow DNA polymerase, DNA ligase, and the luciferase assay system were purchased from Promega (Madison, Wis.). [α-32P]dATP, [α-32P]UTP, and [γ-32P]ATP were purchased from DuPont-New England Nuclear (Boston, Mass.). [3H]folic acid was purchased from Moravek Biochemicals (Brea, Calif.). Taq Plus precision DNA polymerase was purchased from Stratagene (La Jolla, Calif.). Fugene 6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, Ind.).
DNA constructs and transfection.
DNA constructs were made either by using natural restriction sites or by creating restriction sites by PCR by using either Taq DNA polymerase or Taq Plus precision DNA polymerase. For cDNA constructs, the templates were either FR-α or FR-β cDNA. For genomic constructs, the FR-α gene cloned from normal uterus was used as the template. To insert short fragments (<80 bp) of FR-α or FR-β into a plasmid, synthetic oligonucleotides corresponding to the complementary strands were purchased, annealed, and phosphorylated before being ligated into the plasmid. Transfection was performed by using Fugene 6 transfection reagent in accordance with the vendor's protocol. Cells were transfected at a confluence of 50 to 70%, and further analyses were performed 2 days after transfection. The transfection efficiency was normalized either by using luciferase values derived from a cotransfected Rous sarcoma virus (RSV) promoter-luciferase reporter plasmid or by Southern blotting to compare the amounts of plasmid present in the cells at the time of harvest.
DNA isolation and Southern blot analysis.
Genomic DNA and plasmid DNA were isolated by following standard protocols (29). In the genomic Southern blot, the isolated DNA was first digested with the restriction enzyme BamHI. The DNA (30 μg) was then electrophoresed on a 1% agarose gel. Blotting and hybridization were performed by using Hybond-N nylon membranes (Amersham, Arlington Heights, Ill.) in accordance with the manufacturer's protocol. Essentially, the DNA was transferred to the nylon membrane by a standard capillary blotting method by using 20× SSC (3 M sodium chloride, 0.3 M sodium citrate) blotting buffer. After the transfer (16 to 18 h), the DNA was immobilized on the membrane by UV cross-linking. FR-α cDNA was labeled by the method of random labeling and used as a probe. The blot was prehybridized for 1 h at 65°C with denatured and sheared salmon sperm DNA (0.5 mg/ml) in 5× SSPE (0.9 M sodium chloride, 0.05 M sodium phosphate, 5.0 mM EDTA [pH 7.7]), 5× Denhardt's solution, and 0.5% sodium dodecyl sulfate (SDS). The labeled probe was added to 106 cpm/ml, and hybridization was allowed to proceed for about 16 h at 65°C. After hybridization, the blot was washed twice in 2× SSPE and 0.1% SDS at room temperature and twice in 1× SSPE and 0.1% SDS at 65°C. The blot was exposed to X-ray film at −80°C. The band intensities were determined by exposing the blots to a phosphor screen (Molecular Dynamics, Sunnyvale, Calif.) and subsequently scanning the screen.
Nuclear run-on.
The procedure is a modification of that described by Garber et al. (7). Briefly, cells (5 × 107) were lysed in 5 ml of lysis buffer A (15 mM HEPES, 60 mM KCl, 15 mM NaCl, 0.15 mM spermin, 0.5 mM spermidine, 0.5 mM egtazic acid, 2 mM EDTA, 0.3 M sucrose, and 14 mM β-mercaptoethanol) containing 0.1% Nonidet P-40. The nuclei were separated by centrifugation, resuspended gently in nuclear storage buffer (50 mM Tris [pH 8.3] containing 40% glycerol, 5 mM MgCl2, and 0.1 mM EDTA), snap-frozen on dry ice, and stored at −80°C for up to 4 weeks. For each reaction, 107 nuclei were thawed on ice and resuspended in 300 μl of reaction buffer (50 mM Tris [pH 8.0], 150 mM KCl, 5 mM MgCl2, 0.5 mM MnCl2, 2 mM dithiothreitol (DTT), 0.1 mM EDTA, and 10% glycerol) and 60 U of RNase inhibitor from Roche Diagnostic Corp. The reaction was initiated by the addition of ribonucleotides to a final concentration of 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, and 100 μCi of [32P]UTP (800 Ci/mmol) (DuPont-New England Nuclear). The reaction mixture was first incubated in a water bath at 30°C followed by digestion with 100 U of DNase I (Roche Diagnostics Corp.) and 33 μg of proteinase K in 33 μl of 10× second-incubation buffer (100 mM Tris-HCl [pH 7.5] containing 5% SDS and 50 mM EDTA) at 37 and 42°C, respectively. Each incubation step lasted for 30 min. The newly transcribed RNA was extracted with phenol-chloroform, precipitated with sodium acetate and ethanol, and purified on P-30 Tris Micro Bio-Spin columns (Bio-Rad Laboratories, Hercules, Calif.). In 500 μl of hybridization buffer, the purified RNA probes (4 × 106 cpm) were hybridized at 45°C for 48 h to FR-α or γ-actin cDNAs or vector DNA alone that had been previously immobilized on nitrocellulose membranes. The membranes were then washed with 2× SSPE at 45°C and were thereafter washed sequentially with the same buffer containing either RNase A (Roche Diagnostics Corp.) or proteinase K at 37°C. The membranes were then exposed to X-ray film. The quantity of radioactivity of each band was measured by phosphorimage analysis.
RNA purification and Northern blot analysis.
Total RNA was isolated by using a single-step phenol and guanidine isothiocyanate extraction method (Trizol reagent, Gibco-BRL) in accordance with the vendor's protocol. RNA (20 μg) was electrophoresed on a 1% agarose gel containing formaldehyde. Blotting and hybridization were performed by using Hybond-N nylon membranes. Essentially, the RNA was transferred to the membrane by a standard capillary blotting method. The RNA was then fixed to the membrane by UV cross-linking. Prehybridization and hybridization were carried out in a solution containing 50% formamide, 5 × SSPE, 2 × Denhardt's reagent, and 0.1% SDS at 42°C. The blot was then washed twice with 2× SSC and 0.1% SDS at room temperature and two more times with 0.2× SSC and 0.1% SDS at 68°C before being exposed to X-ray film at −80°C. To control for the loading efficiencies and to ensure the integrity of the RNA, the same blot was first stripped by soaking in boiling 0.1% SDS and then reprobed with labeled cDNA fragments for either cyclophilin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The band intensities were measured by a phosphorimage analysis.
[3H]folic acid binding assay.
Cells in six-well tissue culture dishes were washed sequentially at 4°C with phosphate-buffered saline (PBS) (10 mM sodium phosphate buffer [pH 7.5] and 100 mM NaCl), acid buffer (10 mM sodium acetate [pH 3.5] and 150 mM NaCl), and again with PBS. The cells were then incubated in a 1-ml solution of PBS containing 3 pmol of [3H]folic acid and 9 pmol of unlabeled folic acid for 1 h at 4°C. The cells were then washed two times with PBS. Ice-cold acid buffer (0.5 ml) was used to remove bound folate, and the eluted radioactivity was measured by liquid scintillation counting.
Isolation of nuclei.
Isolation was carried out according to a published protocol (9). The entire procedure was carried out on ice or at 4°C. Briefly, cells from three 10-cm dishes were scraped off the plates using a rubber policeman. The cells were then washed with PBS two times before being resuspended in 10 ml of swelling buffer (10 mM Tris-HCl [pH 7.9], 10 mM KCl, and 1 mM DTT). The cells were then allowed to stand on ice for 5 min, sedimented at 500 × g for 5 min, and resuspended in 2 ml of homogenization buffer (10 mM Tris HCl [pH 7.9], 0.3 M sucrose, 1.5 mM MgCl2, 0.3% Triton X-100, and 1 mM DTT). The cells were lysed by using a glass Dounce homogenizer. The resulting nuclei were sedimented by centrifugation at 500 × g for 5 min. The nuclei were then washed three times with 10 ml of homogenization buffer to avoid contamination by cytosol. The absence of significant cytosolic contamination was ensured by electron microscopy. Further, the quality of the nuclei was ensured by the biochemical test of enrichment for U3 small nuclear RNA in the nuclear extracts by procedures described below. The U3 RNA, measured by quantitative reverse transcription-PCR (RT-PCR) as described below, was enriched fivefold in the nuclear extracts, consistent with the value expected from the relative distribution of total RNA between the nucleus and the cytosol (2). The nuclei were then stored at −80°C for use in RNA purification.
Luciferase assay.
At 48 h after transfection, cells in each well of six-well plates were washed with PBS and scraped off in 400 μl of lysis buffer (Promega). The lysates were frozen at −80°C and then brought back to room temperature. After centrifugation at 14,000 × g for 1 min, an aliquot of 20 μl of the lysate was assayed by using a luminometer (Lumat LB9501, Wallac Berthold) by following the protocol provided by the vendor.
Quantitative RT-PCR analysis of nuclear RNA.
RT-PCR analysis was used in one of two ways. Real-time RT-PCR was used to measure endogenous FR-α pre-mRNA. For the real-time analysis, reverse transcription was carried out by following a standard procedure. Essentially, 200 ng of nuclear RNA was mixed with random hexamer primers (5 × 10−4 optical density units/μl), RNasin inhibitor (1 U/μl), Moloney murine leukemia virus reverse transcriptase (5 U/μl), and deoxynucleoside triphosphates (1.0 mM each) in reverse transcriptase buffer (50 mM potassium chloride and 10 mM Tris HCl [pH 8.3]). The 10-μl reaction mixture was first incubated at 25°C for 10 min, then at 42°C for 15 min, and finally at 99°C for 6.5 min. The subsequent PCR was carried out in the presence of 12.5 μl of PCR Mastermix (Applied Biosystems, Branchburg, N.J.), 0.5 μl each of the forward and reverse primers (forward primer sequence, 5′-GGT TGT TGA GTT TCC GCA TTA CA-3′; reverse primer sequence, 5′-GAT GGG TCA CTC CAA CTT CAT TTC-3′), and 0.5 μl of the Taqman probe (sequence, 5′-AAA TAA GGA AGC AGG CCC AAA GGA GAG C-3′). Both the primers and the Taqman probe are specific for the FR-α intron D sequence. The PCR conditions were in a sequence of 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. The fluorescence generated by each cycle of amplification was monitored and recorded by a Gene Amp 5700 sequence detection system (Applied Biosystems). A GAPDH control was included at the beginning of the RT-PCR assay with primers and the Taqman probe provided by Applied Biosystems. Control RNA (80 pg, 400 pg, 2 ng, 10 ng, and 50 ng) was used for amplifying GAPDH.
To measure mRNAs for FR-β and chimeric FR-α/FR-β in the nuclei of transfected cells, a standard template cRNA was generated from an FR-β cDNA construct (FR-βΔcDNA) containing a deletion from nucleotides (nt) 642 to 822. FR-βΔcDNA was transcribed in vitro by a standard procedure (29), and the cRNA was purified. One picogram of the cRNA standard was used in each quantitative RT-PCR together with 0.25 to 0.5 μg of nuclear RNA. Reverse transcription was carried out according to the previous procedure. Five microliters of the reverse transcription products was used in the following PCR. The primers used to amplify the cDNA for either FR-β or the chimeric FR were βF2 (5′-TCTGTGTAGTCACCATGTGCAGTG-3′) and βR1 (5′-GGAGCATCTCACCATTCACATGC-3′), which spanned the internal deletion in the FR-βΔcDNA standard. The PCR primer βR1 was labeled with [γ-32P]ATP by the method of end labeling. Labeled primer (0.3 nmol) was mixed with 0.45 nmol of unlabeled primer to obtain a final concentration of approximately 9.4 μM. Of this solution, 1 μl was used in each PCR at a final concentration of 0.15 μM. The PCR was carried out under conditions of 95°C for 2 min followed by 20 cycles of a sequence of 95°C (1 min), 55°C (1 min), and 72°C (1 min) and a final cycle of 72°C (7 min). For detecting the U3 small nuclear RNA, reverse transcription was carried out as described above except that 1 μg of either total RNA or nuclear RNA was used. For PCR, two primers, namely, hu3F (5′-CGGAACTAGCGAAAGTTTCTCGCC-3′) and hu3R (5′-CGGACTAGTCTCTGTCATTGAGCC-3′), were used to amplify a fragment of 356 bp. The PCR conditions were the same as previously described except that the annealing temperature was 60°C. The PCR product was fractionated by electrophoresis on a 6% nondenaturing polyacrylamide gel. The gel was dried and exposed to an X-ray film. The band intensities were measured by phosphorimage analysis. The intensity of the band from the target substrate was normalized to that of the standard within each lane.
Cell cycle analysis.
HeLa cells plated in 35-mm dishes were synchronized at the G1/S boundary by first culturing them in the presence of thymidine (2 mM) for 14 h and by incubation for a further 14 h under normal growth conditions followed by incubation for 14 h in medium containing 5 μg of aphidicolin/ml (Sigma). For release from the G1/S boundary, the medium was changed to regular folate-free RPMI medium. To monitor [3H]thymidine incorporation, [3H]thymidine (7 μCi per 0.7 ml of medium in each well) was added to the wells at each time point; after incubation for 20 min, the DNA was trichloroacetic acid precipitated and neutralized with NaOH, followed by measurement of the incorporated radioactivity by liquid scintillation counting.
Immunoprecipitation.
Protein A agarose (Pierce) was washed twice with PBS containing 1% Triton X-100, and a 1:1 suspension of the gel was prepared in the same buffer. For each sample, 40 μl of the protein A agarose suspension was mixed with 80 μl of affinity-purified anti-FR antibody and 80 μl of the buffer. The gel suspension was incubated at room temperature on a rotary mixer for 4 h. Unbound antibody was removed by washing twice with 200 μl of the above buffer, and the gel was resuspended in an equal volume of the buffer. To each sample (300 μl) of cell lysates and media, 40 μl of the antibody-bound protein A agarose suspension was added and incubated at 4°C overnight on a rotary mixer. The protein A agarose was then washed three times with 300 μl of the buffer. The proteins bound to the gel were solubilized by incubation in the sample buffer for SDS-polyacrylamide gel electrophoresis at 37°C for 30 min. The samples were then separated by electrophoresis on an SDS-12% polyacrylamide gel. The gels were then treated with Entensify (DuPont) followed by autoradiography.
[35S]cysteine pulse-chase.
The cells were incubated with cysteine-free MEM supplemented with 70 μCi of [35S]cysteine/ml for 30 min at 37°C. The medium was then changed to MEM, and the cells were further incubated for various periods. This was followed by immunoprecipitation of the cell lysates.
5′ RACE analysis.
5′ rapid amplification of cDNA ends (RACE) analysis was carried out by using the 5′ RACE system from Invitrogen (Carlsbad, Calif.) in accordance with the manufacturer's protocol. Briefly, 4 μg of RNA was reverse transcribed by using SuperScript II reverse transcriptase and a gene-specific primer for FR (5′-CATGGCTGCAGCATAGAACCTCGC-3′). The first-strand cDNA was then tailed with dC at the 3′ end by using terminal deoxynucleotidyl transferase. The product was used as template in the subsequent PCR by using a nested gene-specific primer for FR (5′-TGAAGTCCACTGCCAGCCCTT-3′) and the abridged anchor primer (provided by Invitrogen). All other experimental procedures were carried out in accordance with the manufacturer's protocol. The PCR products were analyzed by electrophoresis on a 1% agarose gel and identified by ethidium bromide staining.
Tet-Off expression system for FR.
The cDNA for either wild-type FR-β or the FR chimera was subcloned into the pTRE plasmid (Clontech, Palo Alto, Calif.) between the MluI and SalI restriction sites. The insert-containing pTRE and the Tet-Off plasmids were cotransfected into cells by using Fugene. At 48 h posttransfection, the cells were treated with 1 μg of doxycycline (Dox)/ml to selectively turn off the expression of the transfected FR; the cells were frozen at different time points for subsequent RNA preparation.
RESULTS
The amount of a protein in a cell could, in general, be regulated at one or more of the following progressive levels leading to gene expression: (i) gene amplification or reorganization, (ii) transcription, (iii) hnRNA processing, (iv) hnRNA stability, (v) nuclear mRNA export, (vi) cytosolic mRNA stability, (vii) translation, (viii) posttranslational modification, (ix) protein targeting, and (x) protein turnover. The initial experiments in this study were designed to comprehensively examine the level(s) at which differential FR-α gene expression occurred in the model cell lines.
The model cell lines are not significantly different in FR-α gene copy number, gross gene organization, and transcription rates.
Southern blotting and restriction map analysis of genomic DNA from the FR-α-positive and -negative cell lines showed essentially similar patterns and no significant differences in band intensities (result not shown). This result rules out differential gene amplification as a cause of the differential FR-α expression among the cell lines. It further suggests that gross differences in the FR-α gene organization are also unlikely among these cells.
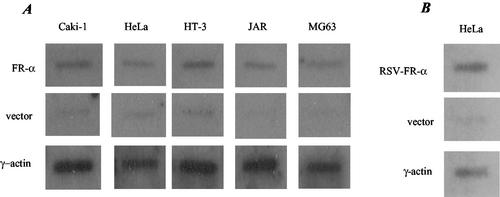
Nuclear run-on experiments gave a relatively strong signal for the γ-actin gene in all of the cell lines tested (Fig. 2). The signals for the FR-α gene were above the background in all cases but did not show differences that could account for overexpression of FR-α in HeLa and JAR cells but not in MG63, Caki1, and HT3 cells (Fig. 2A). In a control experiment, in HeLa cells transfected with the FR-α cDNA under the control of the strong RSV promoter, the intensity of the signal was comparable to that of the endogenous γ-actin gene (Fig. 2B).
FIG. 2.
Nuclear run-on analysis. Newly synthesized and radiolabeled RNA from isolated nuclei were obtained as described in Materials and Methods. (A) RNA (4 × 106 cpm) from each of the indicated cell lines was allowed to hybridize with either FR-α or γ-actin cDNA containing plasmids (10 μg) which were previously immobilized by slot blot on nitrocellulose membranes. Immobilized plasmid vector was used as a negative control for each cell line. (B) As a positive control for FR-α gene transcription, HeLa cells were transfected with the FR-α cDNA under the control of the strong RSV promoter (RSV-FR-α) and subjected to nuclear run-on analysis as described above.
P4 is the major but relatively weak promoter.
From transient transfections with promoter-luciferase reporter constructs (Table 1), a P4 promoter fragment (nt −271 to +33) of the FR-α gene gave ∼25- to ∼80-fold reporter activity relative to that of a P1 promoter fragment (nt −3394 to −2468) in all of the cell lines. Moreover, when the cluster of Sp1 sites in the P4 promoter was deleted from a full-length (P1 plus P4) FR-α promoter-luciferase construct, 90 to 100% of the promoter activity was lost (Table 1). Thus, P4 is the predominant promoter in the model cell lines. The identity of the endogenous FR-α mRNA in the FR-positive cells was also determined by 5′ RACE PCR to be the P4 promoter-driven transcript (results not shown). Compared to the activity of the RSV promoter-luciferase construct, however, the activity of P4 promoter-luciferase was relatively poor (2 to 8% of the RSV promoter activity) and did not show differential activities that correlated with the ability of the cells to express endogenous FR-α (Table 1).
TABLE 1.
Promoter usage in the model cell linesa
| Cell line | Reporter activity
|
||
|---|---|---|---|
| P4 Luc/P1 Luc | Full promoter ΔP4 Luc/ full promoter Luc | P4 Luc/RSV Luc | |
| HeLa | 24.80 ± 2.32 | 0.063 ± 0.003 | 0.080 ± 0.032 |
| Caki1 | 65.24 ± 5.21 | 0.113 ± 0.021 | 0.072 ± 0.025 |
| HT3 | 81.46 ± 7.36 | 0.002 ± 0.001 | 0.021 ± 0.001 |
| JAR | 30.16 ± 4.55 | 0.074 ± 0.022 | 0.042 ± 0.003 |
| MG3 | 53.9 ± 3.12 | 0.001 ± 0.0005 | 0.072 ± 0.002 |
P4 Luc, FR-α P4 promoter fragment (nt −271 to +33) plus luciferase; P1 Luc, FR-α P1 promoter fragment (nt −3394 to −2468) plus luciferase; full promoter Luc, FR-α gene fragment spanning both P1 and P4 (nt −3394 to +33) plus luciferase; full promoter ΔP4 Luc, full promoter Luc containing a deletion of Sp1 sites in P4 (nt −140 to −30); RSV Luc, RSV promoter plus luciferase. The transcription start site in the P4 promoter is considered to be nt +1. The constructs were inserted in the PGL3 basic plasmid. Transfection efficiencies were normalized to that of a cotransfected β-galactosidase control.
Differential FR-α expression reflects relative cytosolic and nuclear transcript levels.
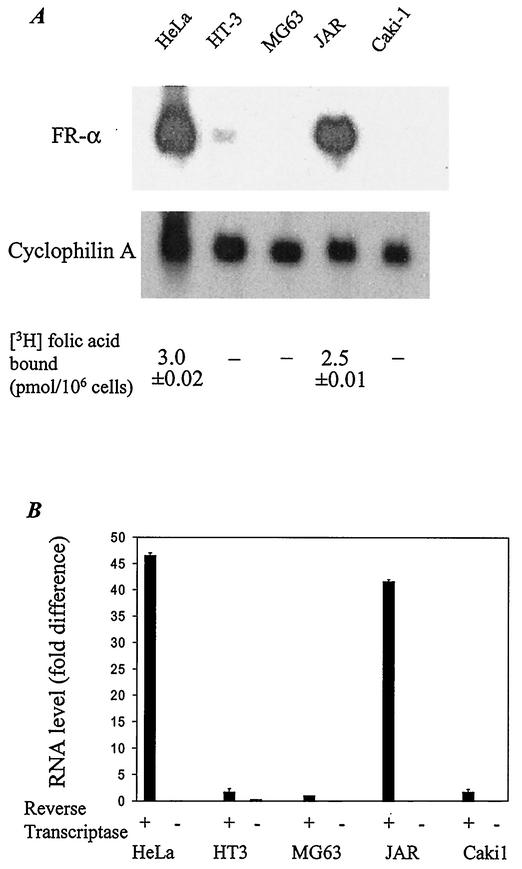
Quantitation of FR expression on the cell surface by a [3H]folic acid binding assay showed virtually insignificant levels in Caki1, MG63, and HT3 cells compared with those in HeLa and JAR cells (Fig. 3A). The relative levels of [3H]folic acid binding protein were reflected in the relative levels of FR-α mRNA from a Northern blot of total RNA from these cells (Fig. 3A). By real-time RT-PCR analysis of nuclear RNA, the FR-α pre-mRNA was observed in HeLa and JAR cells but was relatively insignificant in HT3, Caki1, and MG63 cells (Fig. 3B). These results suggest that the principal regulatory event(s) that leads to differential FR-α expression among the cell lines occurs in the nucleus.
FIG. 3.
FR-α expression at the mRNA and protein levels. (A) Northern blot analysis of 20 μg of total RNA from each cell line. The blot was first probed with 32P-labeled FR-α cDNA and then erased and reprobed with 32P-labeled cDNA for cyclophilin A. Cell surface binding of [3H]folic acid was measured as described in Materials and Methods, and the resulting value is indicated for each cell line. −, specific [3H]folic acid binding cannot be reliably measured above background (0.1 pmol/106 cells). (B) The FR-α pre-mRNA in the nuclei of the cell lines was measured by real-time RT-PCR analysis of nuclear RNA by using an intron D-specific Taqman probe, as described in Materials and Methods. The mRNA levels for GAPDH were similarly determined by using a GAPDH-specific Taqman probe to ensure the integrity of the RNA samples (data not shown). The level of FR-α pre-mRNA in MG63 cells is assigned a value of 1. + and −, presence and absence of reverse transcriptase, respectively.
Differential FR-α expression occurs by a posttranscriptional mechanism and is independent of introns.
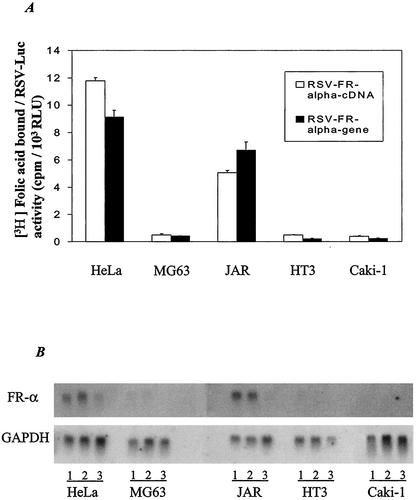
The preceding results, taken together, exclude differential transcription rates as a major mechanistic basis for the tissue specificity of FR-α and further suggest a possible posttranscriptional regulatory event occurring in the nucleus. To establish that the differential FR-α expression among the cell lines can occur independently of a chromosomal or promoter context and, further, to test for a possible influence of intronic sequences, cells were transiently transfected with constructs in which either the entire FR-α gene (including introns) downstream of the P4 promoter or the FR-α cDNA downstream of the P4 promoter was linked to the strong and universally active RSV promoter. The increase in cell surface [3H]folic acid binding due to transfection of the cell lines was measured and normalized in all cases to luciferase activity arising from cotransfection with an RSV promoter-luciferase reporter construct. The normalization to RSV-luciferase activity was carried out to control for variability among the cell lines in transfection efficiency or RSV promoter activity. The results (Fig. 4A) showed relatively high (7- to 30-fold) expression of FR-α in HeLa and JAR cells compared with that in HT3, MG63, or Caki1 cells. Further, the tissue specificity of FR-α expression was retained whether or not intronic sequences were present (Fig. 4A). Northern blot analysis of total RNA from the cells transfected in Fig. 4A showed FR-α mRNA expression patterns similar to those observed for the protein (Fig. 4B). These results strongly support a posttranscriptional regulatory mechanism for the tissue specificity of FR-α at the level of the FR-α transcript but not involving hnRNA splicing.
FIG. 4.
RSV promoter-driven FR-α expression at the protein and mRNA levels. The cells were transected with expression plasmid containing either the FR-α cDNA (RSV FR-α cDNA) or the cDNA plus the intervening introns (RSV FR-α gene). (A) [3H]folic acid binding was measured on the surface of the transfected cells and normalized to a cotransfected RSV promoter-luciferase reporter control. (B) Northern blot of 20 μg of total RNA from cells transfected with RSV FR-α cDNA (lanes numbered 1) or RSV FR-α gene (lanes numbered 2) and also RNA from cells transfected with the vector alone (lanes numbered 3). The blot was first probed with 32P-labeled FR-α cDNA and then erased and reprobed for GAPDH.
Mapping of a regulatory element in FR-α by using chimeric FR-α/FR-β.
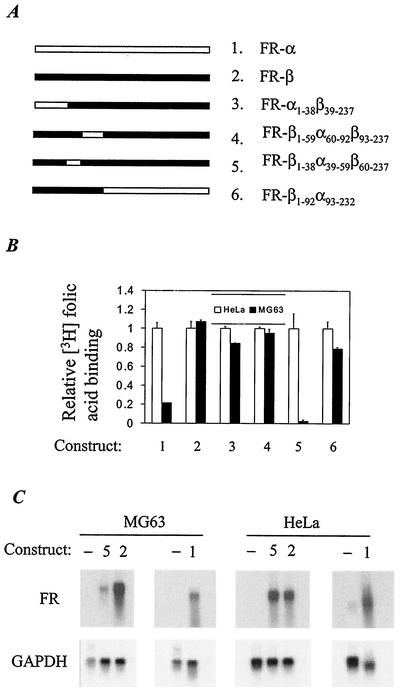
In contrast to results for transfection with FR-α, when HeLa and MG63 cells were transfected with an RSV promoter-FR-β cDNA construct, no significant difference was observed between the two cell lines, either in terms of the increase in cell surface [3H]folic acid binding or FR-β mRNA level (Fig. 5). We took advantage of this observation to map the region of the FR-α cDNA that was responsible for the tissue-specific expression of FR-α. Accordingly, chimeric FR-α/FR-β cDNAs were linked to the RSV promoter (Fig. 5A) and used to transfect HeLa or MG63 cells. Among the chimeras tested, a single construct retained the differential protein (Fig. 5B) and mRNA (Fig. 5C) expression of FR-α. This construct contained 60 bp of an FR-α ORF sequence substituted in FR-β. This element occurs between nt 234 and 293 in the full-length 1,108-bp FR-α cDNA relative to the transcription initiation site of the P4 promoter (nt +1) and the translational start site (nt +45). The tissue specificity of FR-α may thus be attributed to this 60-bp sequence while excluding a similar role for the remainder of the FR-α gene sequence.
FIG. 5.
Mapping the cis element in the FR-α cDNA sequence responsible for its tissue specificity. (A) Schematic representation of the FR-α/FR-β cDNA chimeras used for transfecting HeLa and MG63 cells. The positions of the encoded amino acids are numbered, with the position of the first amino acid in the mature protein numbered +1. The chimeras are FR-α1-38β39-237, FR-β1-59α60-92β93-237, FR-β1-38α39-59β60-237, and FR-β1-92α93-232. (B) Cell surface [3H]folic acid binding. Cells were transfected with 0.5 μg of the indicated cDNA construct (corresponding to the constructs numbered 1 to 6 in panel A) together with 0.5 μg of RSV-Luc. The amount of bound radioactivity was normalized against the corresponding luciferase value. (C) Northern blot analysis of 20 μg of total RNA from HeLa or MG63 cells transfected with either FR-α (construct 1), FR-β (construct 2), FR-β1-38α39-59β60-237 (construct 5), or vector alone (−). The blot was cut into four strips, and each strip was probed separately. FR-α cDNA was used to probe the RNA from cells transfected with FR-α cDNA. A fragment of FR-β cDNA (nt 45 to 206) was used to probe the RNA from cells transfected with cDNA for FR-β or the chimera in order to avoid background due to endogenous FR-α mRNA in HeLa cells. The blots were erased and reprobed for GAPDH.
The 60-bp FR-α element can transfer the tissue specificity of FR-α to luciferase.
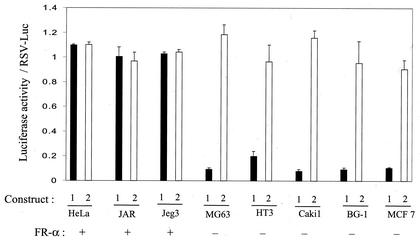
Based on the preceding observation that a 60-bp FR-α ORF sequence is capable of conferring the tissue specificity of FR-α on FR-β, it was of interest to test the ability of this element to transfer the tissue specificity of FR-α to a heterologous gene. The FR-α sequence element was inserted into the 5′ untranslated region (UTR) of the RSV promoter-luciferase construct rather than its ORF in order to retain the ORF sequence of luciferase. Transfection of cells with this construct resulted in a luciferase expression pattern that mimicked that of endogenous FR-α expression in HeLa, JAR, MG63, HT3, and Caki1 cells and in additional FR-α-positive (JEG3) and FR-α-negative (BG1 and MCF7) cells (Fig. 6). As seen in Fig. 6, insertion of the endogenous FR-α element did not cause an appreciable change in the expression of RSV-luciferase in HeLa, JAR, and JEG3 cells but decreased it 7- to 20-fold in MG63, HT3, Caki1, BG1, and MCF7 cells. In contrast, substitution of the homologous sequence from FR-β did not appreciably alter RSV-luciferase expression in any of the above cell lines (Fig. 6). The transfection efficiencies of RSV-luciferase and the substituted constructs were found to be uniform in all of the cell lines as measured by Southern blot analysis of plasmid DNA in the transfected cells (data not shown).
FIG. 6.
Effect of the 60-bp FR-α cDNA element on RSV-Luc expression in various cell lines. In RSV-Luc, the RSV promoter is upstream of the luciferase reporter gene. In construct 1, the 60-bp FR-α cDNA fragment (nt 234 to 293 relative to the transcription start site) is placed in the 5′ UTR of the reporter in RSV-Luc. Construct 2 is similar to construct 1 except that the 60-bp insert corresponds to the homologous FR-β cDNA sequence. Luciferase activity was measured in cells transfected with RSV-Luc, construct 1, or construct 2. Cells were transfected with 0.5 μg of the plasmid and harvested 48 h later to measure luciferase activity. The data represent ratios to the corresponding values for RSV-Luc. Uniformity in transfection efficiency was ensured in a parallel experiment in which plasmid DNA was purified from the transfected cells at the time of the harvest, digested with HindIII and XbaI, subjected to Southern blot analysis, and probed with 32P-labeled luciferase cDNA (results not shown).
The 60-bp FR-α element causes differential mRNA stability in the nucleus but not in the cytosol.
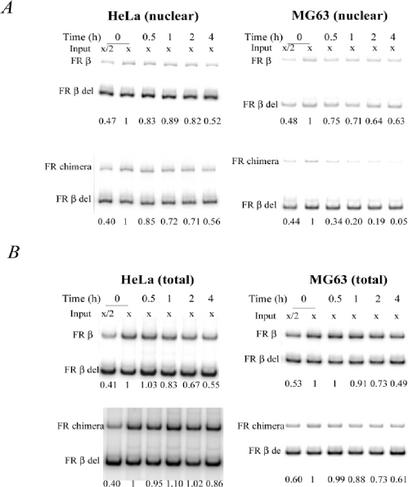
To examine the effect of the 60-bp FR-α element in the nuclear and cytosolic compartments of HeLa and MG63 cells, we transiently transfected the cells with Tet-Off expression plasmids for FR-β and the chimeric FR-β with the 60-bp FR-α element substituted in it. Transient transfections were more suitable for this experiment than a stable transfection system because of the necessity to obtain measurable levels of the chimeric FR mRNA in MG63 cells. In the time course following treatment of cells with Dox to inhibit transcription, the degradation rate of the mRNA for FR-β versus that for the chimeric FR-β was monitored by quantitative RT-PCR by using RNA isolated from the nuclei of transfected HeLa and MG63 cells (Fig. 7A). The time courses for the loss of nuclear mRNAs for FR-β and the FR-β/α chimera were similar in HeLa cells but strikingly different in MG63 cells (Fig. 7A). The nuclear mRNA for FR-β/α was decreased to about one-third of its original level in 30 min following the addition of Dox to MG63 cells (Fig. 7A); in contrast, more than half of the nuclear mRNA for FR-β remained at the end of 4 h (Fig. 7A). This result indicates that the 60-bp element substantially decreased mRNA stability in the nuclei of MG63 cells but not in those of HeLa cells.
FIG. 7.
Effect of the FR-α sequence from nt 234 to 293 on mRNA stability of FR-β. Cells were cotransfected with the Tet-Off plasmid and the expression plasmid pTRE, which contained either FR-β cDNA or chimeric FR in which the FR-α cDNA sequence from nt 234 to 293 was substituted in FR-β. At 48 h posttransfection, the cells were treated with 1 μg of Dox/ml to selectively turn off expression of the transfected FR for the indicated periods of time before extraction of total RNA or nuclear RNA. (A) mRNA stability in the nucleus. Nuclear RNA was purified from isolated nuclei, and mRNA for FR-β or the chimera was quantitated by RT-PCR. For HeLa cells, input x represents 0.25 μg of nuclear RNA; for MG63 cells, input x represents 0.5 μg of nuclear RNA. Before the reverse transcription reaction was initiated, 1 pg of FR-β cRNA with an internal deletion from nt 642 to 822 (FR β del) was added to each reaction tube to serve as an internal standard. The reaction products were electrophoresed on a 6% polyacrylamide gel, and the band intensities were quantified by phosphorimage analysis. Negative controls, which did not generate PCR products (data not shown), included reactions without either RNA or reverse transcriptase. The PCR was carried out for 20 cycles, and the amplification was shown to be in the linear range by using different amounts of input RNA (complete data not shown). The numbers at the bottom of each gel represent band intensities normalized against those of the corresponding cRNA standard; the error from triplicate data sets was ≤15%. (B) Total mRNA stability. For HeLa cells, input x represents 0.25 μg of total RNA; for MG63 cells, input x represents 0.5 μg of total RNA. All other procedures were carried out in the same way as for the nuclear RNA. The numbers at the bottom of each gel represent band intensities normalized against those of the corresponding cRNA standard; the error from triplicate data sets was ≤15%.
The relative time courses for the degradation of FR-β and FR-β/α mRNAs monitored in the total RNA fractions of HeLa and MG63 cells following Dox treatment (Fig. 7B) differed from those observed above for the nuclear RNA fractions. There were relatively small differences between the 4-h time courses for the two mRNAs after Dox treatment in MG63 cells (Fig. 7B). In HeLa cells, on the other hand, even though both the mRNAs were relatively stable, the chimeric mRNA appeared to be somewhat more stable than the FR-β mRNA, reflecting a higher steady-state level of the chimeric mRNA in the cells (Fig. 7B). The relative instability of the FR mRNA in MG63 cells caused by the 60-bp FR-α element was thus found to be restricted to the nuclear compartment.
FR-α expression in HeLa cells is constitutive, and the protein turnover is slow.
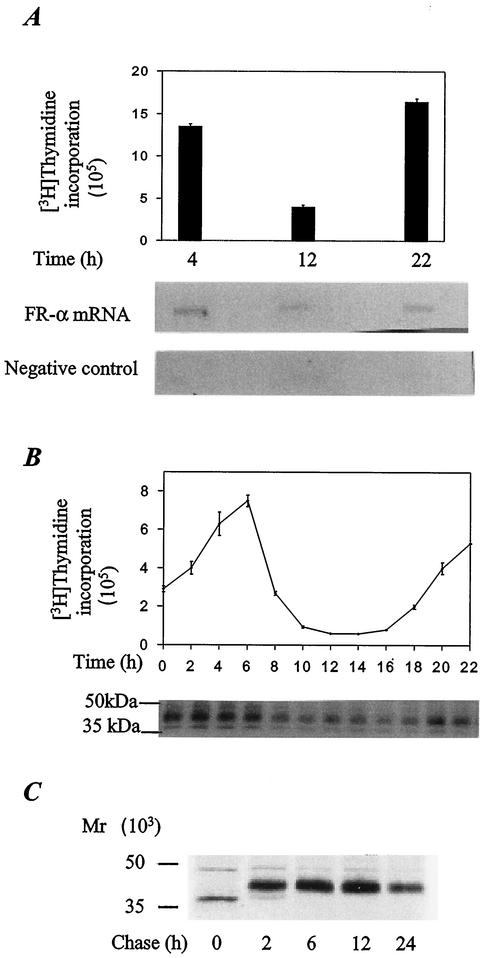
To further understand the basis for the overexpression of FR-α in HeLa cells, transcription and de novo synthesis of FR-α were examined in relation to the cell cycle. From nuclear run-on experiments, comparable rates of transcription of the FR-α gene were observed in the S and G2/M phases (Fig. 8A). Metabolic labeling with [35S]cysteine and immune precipitation analysis were used to measure newly synthesized FR-α at various stages of the cell cycle (Fig. 8B). FR-α appeared to be synthesized throughout the cell cycle, albeit at a two- to threefold slower rate in the G2 and M phases than in the S phase. The half-life of FR-α in HeLa cells determined by [35S]cysteine pulse-chase experiments was >24 h (Fig. 8C). Thus, the stability of FR-α combined with its constant synthesis must contribute to the high level of expression of the receptor in HeLa cells.
FIG. 8.
Constitutive synthesis and stability of FR-α. (A) Relative transcription rates of the FR-α gene in HeLa cells at different stages of the cell cycle. The cells were synchronized at the G1/S boundary by a double thymidine-aphidicolin block. The stage of the cell cycle at the indicated times was determined by [3H]thymidine incorporation (graph). At these times, the nuclei were isolated from cells in a parallel experiment and subjected to nuclear run-on analysis by using [32P]UTP. The labeled RNA was purified and used to probe nitrocellulose filters containing immobilized FR-α cDNA in pCDNA1 or the vector alone (negative control) followed by autoradiography (gels). (B) Relationship between cycle stage and de novo synthesis of FR-α in HeLa cells. The cells were synchronized and the cell cycle was monitored as described in the legend for panel A. Prior to each time point indicated, the cells were metabolically labeled by a 25-min pulse of [35S]cysteine. The labeled FR-α was immunoprecipitated and electrophoresed on a 12% SDS-polyacrylamide gel followed by autoradiography (gel). (C) Turnover of mature FR-α in HeLa cells. The cells were metabolically labeled with a 30-min pulse of [35S]cysteine followed by chase for the indicated periods with unlabeled cysteine. The labeled FR-α was immunoprecipitated and electrophoresed on a 12% SDS-polyacrylamide gel followed by autoradiography.
DISCUSSION
It has been previously noted that overexpression of FR-α in human tumor explants is not associated with FR-α gene amplification or a gross reorganization of the gene (5), and these observations were extended in this study to the model cell lines used. Both from nuclear run-on experiments and from studies of promoter-luciferase reporter constructs, it was also evident that differential transcription rates could not account for the relative levels of FR-α expression in the model cell lines. Interestingly, in the malignant cell lines that either did not express FR-α or that poorly expressed the reporter, the endogenous FR-α gene was transcribed much as in the FR-α-positive cell lines, but in all of the cell lines, the transcription rate of the FR-α gene was weak compared to that of the γ-actin gene. The FR-α promoter activity in all of the cell lines was also much weaker than that of the RSV promoter in transient transfection assays. These results pointed to a posttranscriptional mechanism to explain the tissue specificity of FR-α.
Even though the P1 promoter of the FR-α gene can generate multiple alternately spliced transcripts, the relevant transcript for the posttranscriptional control of FR-α expression observed in this study was clearly the single P4 promoter-generated transcript, since the P4 promoter was much more active than the P1 promoter. In the model cell lines, consistent with this prediction, when the P4 transcript was generated under the control of the strong and universally active RSV promoter by transient transfection, the de novo production of FR-α at both the protein and mRNA levels was greater in the FR-α-positive HeLa and JAR cells than in MG63, Caki1, and HT3 cells. The posttranscriptional mechanism underlying the tissue specificity of FR-α was unrelated to pre-mRNA splicing, since deletion of the introns did not significantly alter the pattern of tissue-specific expression of FR-α under the control of the RSV promoter.
The observation that the relative FR-α protein levels reflected the relative FR-α mRNA levels suggested that the posttranscriptional regulation of FR-α expression among the different cell lines occurred prior to protein synthesis. Furthermore, the relative FR-α mRNA levels in isolated nuclei from the model cell lines corresponded to those in total RNA preparations, which are predominantly cytosolic; this observation suggested that the mechanism determining the tissue specificity of FR-α is related to a nuclear event such as 5′ capping, polyadenylation, or hnRNA or mRNA turnover.
The experiments with chimeric FR-α/FR-β constructs allowed us to narrow the region of the FR-α gene that was responsible for conferring the posttranscriptional tissue specificity to a 60-bp element in the ORF. This sequence element was able to confer the tissue specificity of FR-α not only on FR-β but also on the heterologous luciferase gene, even when placed in the 5′ UTR. These results argued against differential 5′ capping or polyadenylation as a mechanistic basis for the tissue specificity of FR-α. Direct experiments strongly supported a role for this FR-α sequence element in destabilizing transcripts in the nuclei (but not in the cytosol) of FR-α-negative cells in contrast to FR-α-positive cells.
There are only a few reports in the literature suggesting regulation of protein expression by modulation of hnRNA or mRNA stability in the nucleus, and there are apparently none that link such a mechanism to tissue specificity in the expression of a constitutively active gene. For example, in mouse cells containing a dihydrofolate reductase gene amplification, higher expression of the enzyme in growth-stimulated cells relative to that in resting cells was associated with a greater stability of dihydrofolate reductase hnRNA (14). Interleukin-4 decreased the expression of granulocyte-macrophage colony-stimulating factor in murine B cells by destabilizing the primary transcript for granulocyte-macrophage colony-stimulating factor in the nucleus (1). On the other hand, during induction of differentiation in HL60 cells, a decrease in myeloperoxidase expression was associated in part with altered hnRNA levels unrelated to transcription rates but likely due to a failure in RNA processing (31). In many other instances, such as modulation of cellular gene expression by c-myc (22), posttranscriptional regulation in the nucleus is evident but specific mechanisms have not been elucidated; in this and other similar examples (18, 20, 33, 34), the possible levels of gene regulation include hnRNA stability as well as RNA processing. The results of the present study exclude RNA processing as a possible level for the differential regulation of FR-α and provide more direct evidence implicating an instability determinant in the FR-α mRNA or hnRNA that confers differential stability in the context of cell type.
If the differential nuclear mRNA stability of FR-α can account in large part for its tissue specificity, additional factors must contribute to its overexpression, since the gene is relatively poorly transcribed. In this respect, the constitutive transcription and translation of FR-α, as well as the relative stability of the protein (half-life, >24 h) observed in this study, are obviously essential for the high steady-state levels of the protein in the FR-α-positive cells.
In addition to polyadenylation and 5′ capping, the stability of mRNAs in the cytosol is also determined by internal stability determinants. Such elements are known to occur mostly in the 3′ UTRs and include AU-rich sequences as well as stem-loop structures and sequences more specific for one or a few mRNAs (26). Mutational analysis has also suggested the presence of such determinants within the ORF (26). There is, however, a paucity of information about internal mRNA sequences possibly controlling transcript stability in the nuclear compartment. The mRNA sequence element identified in the FR-α ORF is a determinant of transcript stability in the nucleus but not in the cytosol and appears to be a novel cis element. With some possible exceptions, cis elements that govern the half-life of cytosolic mRNAs will, upon insertion into a heterologous RNA, alter its regulatory pattern by decreasing rather than increasing its stability. Therefore, they may be referred to as instability determinants (26). The experiments in this study examining the influence of the 60-bp FR-α sequence on the expression and transcript stability of luciferase and FR-β in a variety of FR-α-positive and FR-α-negative cells indicate that this element is also an instability determinant.
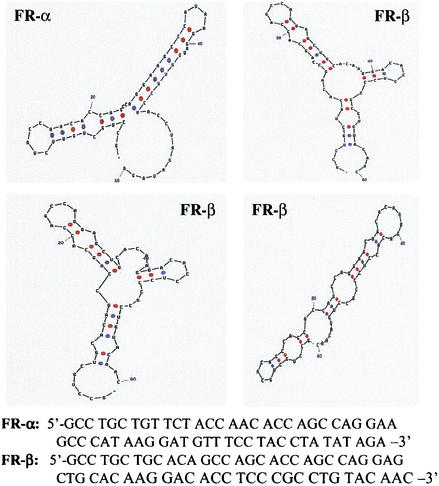
From the aligned sequences of the mRNAs for FR-α and FR-β, there is considerable sequence divergence in the region of the FR-α instability determinant identified in this study. According to the analytical software of Stewart and Zuker (1995-2003, version 3.1 [http://bioinfo.math.rpi.edu/mfold/]), the predicted secondary structures of the FR-α and FR-β RNA fragments corresponding to this region were also different (Fig. 9), offering an explanation for the inability of the FR-β sequence to influence transcript stability. This determinant may serve in the recognition of the FR-α transcript by a tissue-specific nuclear RNA-degrading machinery. Relatively little is known about the enzymes responsible for the many mRNase activities present in mammalian cells, and attempts are being made to isolate and characterize them (4, 10, 13, 16, 26).
FIG. 9.
Theoretically predicted secondary structures of the FR-α mRNA fragment from nt 401 to 460 and the corresponding fragment from the aligned sequence of the FR-β mRNA. The software of Stewart and Zuker (1995-2003, version 3.1 [http://bioinfo.math.rpi.edu/mfold/]) was used for the analysis. The analysis resulted in a single structure for the FR-α fragment and three alternate structures for the FR-β fragment.
There is some evidence for a surveillance mechanism in the nuclei of mammalian cells that degrades defective transcripts (3). The degradation of FR-α mRNA in the nucleus appears to be more selective, and it remains to be known whether other mRNAs share its instability determinant. In the absence of strict transcriptional control of a constitutively and poorly transcribed gene, such as the FR-α gene, mRNA degradation in the nucleus may offer an alternate and effective means of preventing its expression. In cells that can circumvent this nuclear mRNA degradation, the protein product may accumulate to quite high levels, owing to slow turnover rates of the cytosolic mRNA and the mature protein. Tissue-specific gene expression may be attained in this manner without the need for a more complex transcriptional machinery.
Acknowledgments
This work was supported by NCI grants R01CA80183 and R01CA70873 to M.R.
We thank Jenny Zak for typing the manuscript.
REFERENCES
- 1.Akahane, K., and D. H. Pluznik. 1992. Interleukin-4 inhibits interleukin-1 alpha-induced granulocyte-macrophage colony-stimulating factor gene expression in a murine B-lymphocyte cell line via downregulation of RNA precursor. Blood 79:3188-3195. [PubMed] [Google Scholar]
- 2.Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberto, and J. D. Watson. 1989. Molecular biology of the cell, 2nd ed., chapter 9. Garland Publishing, Inc., New York, N.Y.
- 3.Belgrader, P., J. Cheng, X. Zhou, L. S. Stephenson, and L. E. Maquat. 1994. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol. 14:8219-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkard, K. T. D., and J. S. Butler. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Np13p. Mol. Cell. Biol. 20:604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, I. G., T. A. Jones, W. D. Foulkes, and J. Trowsdale. 1991. Folate-binding protein is marker for ovarian cancer. Cancer Res. 51:5329-5338. [PubMed] [Google Scholar]
- 6.Elwood, P. C., K. Nachmanoff, Y. Saikawa, S. T. Page, P. Pacheco, S. Roberts, and K. N. Chung. 1997. The divergent 5′ termini of the alpha human folate receptor mRNAs originate from two tissue-specific promoters and alternative splicing: characterization of the alpha hFR gene structure. Biochemistry 36:1467-1478. [DOI] [PubMed] [Google Scholar]
- 7.Garber, M., S. Panchanathan, R. S. Fan, and D. L. Johnson. 1991. The phorbol ester, 12-O-tetradecanoylphorbol-13-acetate, induces specific transcription by RNA polymerase III in Drosophila Schneider cells. J. Biol. Chem. 266:20598-20601. [PubMed] [Google Scholar]
- 8.Goren, D., A. T. Horowitz, D. Tzemach, M. Tarshish, S. Zalipsky, and A. Gabizon. 2000. Nuclear delivery of doxorubicin via folate-targeted liposomes with bypass of multidrug-resistance efflux pump. Clin. Cancer Res. 6:1949-1957. [PubMed] [Google Scholar]
- 9.Ikeda, E., M. G. Achen, G. Breier, and W. Risau. 1995. Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J. Biol. Chem. 270:19761-19766. [DOI] [PubMed] [Google Scholar]
- 9a.Jackman, A. L., D. S. Theti, A. S. Lornaime, and V. B. Bavetsias. 2000. Exploitation of the α-isoform of folate receptor (α-FR) for the selective delivery of antifolate thymidylate synthase (TS) inhibitors. In Proceedings of the 91st Annual Meeting of the American Association for Cancer Research. American Association for Cancer Research, Philadelphia, Pa.
- 10.Konstantinova, I. M., V. A. Kulichkva, I. N. Evteeva, A. G. Mittenberg, I. V. Volko, J. B. Ermolaeva, and L. N. Gruse. 1999. The specific endoribonuclease activity of small nuclear and cytoplasmic alpha-RNPs. FEBS Lett. 462:407-410. [DOI] [PubMed] [Google Scholar]
- 11.Lacey, S. W., J. M. Sanders, K. G. Rothberg, R. G. W. Anderson, and B. A. Kamen. 1989. Complementary DNA for the folate binding protein correctly predicts anchoring to the membrane by glycosyl-phosphatidylinositol. J. Clin. Investig. 84:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leamon, C. P., and P. S. Low. 2001. Folate-mediated targeting: from diagnostics to drug and gene delivery. Drug Discov. Today 6:36-43. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C. H., P. Leeds, and J. Ross. 1998. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J. Biol. Chem. 273:25261-25271. [DOI] [PubMed] [Google Scholar]
- 14.Leys, E. J., G. F. Crouse, and R. E. Kellems. 1984. Dihydrofolate reductase gene expression in cultured mouse cells is regulated by transcript stabilization in the nucleus. J. Cell Biol. 99:180-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luiten, R. M., S. O. Warnaar, and D. Sanborn. 1997. Chimeric bispecific OC/TR monoclonal antibody mediates lysis of tumor cells expressing the folate-binding protein (Mov 18) and displays decreased immunogenicity in patients. J. Immunother. 20:496-504. [DOI] [PubMed] [Google Scholar]
- 16.Martinez, J., Y. G. Ren, A. C. Thuressou, U. Hellman, J. Astrom, and A. Virtanen. 2000. A 54-kDA fragment of the poly(A)-specific ribonuclease is an oligomeric, processive and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem. 275:24222-24230. [DOI] [PubMed] [Google Scholar]
- 17.Melani, C., M. Figini, and D. Nicosia. 1998. Targeting of interleukin 2 to human ovarian carcinoma by fusion with a single-chain Fv of antifolate receptor antibody. Cancer. Res. 58:4146-4154. [PubMed] [Google Scholar]
- 18.Meskini, R. E., F. Boudouresque, and L. Ouafik. 1997. Estrogen regulation of peptidylglycine α-amidating monooxygenase messenger ribonucleic acid levels by a nuclear posttranscriptional event. Endocrinology 138:5256-5265. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima-Matsushita, N., T. Homma, S. Yu, T. Matsuda, N. Sunahara, T. Nakamura, M. Tsukano, M. Ratnam, and T. Matsuyama. 1999. Selective expression of folate receptor beta and its possible role in methotrexate transport in synovial macrophages from patients with rheumatoid arthritis. Arthritis Rheum. 42:1609-1616. [DOI] [PubMed] [Google Scholar]
- 20.Narayan, P., and H. C. Towle. 1985. Stabilization of a specific nuclear mRNA precursor by thyroid hormone. Mol. Cell. Biol. 5:2642-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peoples, G. E., B. W. Anderson, T. V. Lee, J. L. Murray, A. P. Kudelka, J. T. Wharton, and G. G. Ioannides. 1999. Vaccine implications of folate binding protein, a novel cytotoxic T lymphocyte-recognized antigen system in epithelial cancers. Clin. Cancer Res. 5:4214-4223. [PubMed] [Google Scholar]
- 22.Prendergast, G. C., and M. D. Cole. 1989. Posttranscriptional regulation of cellular gene expression by the c-myc oncogene. Mol. Cell. Biol. 9:124-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratnam, M., H. Marquardt, J. L. Duhring, and J. H. Freisheim. 1989. Homologous membrane folate binding proteins in human placenta: cloning and sequence of a cDNA. Biochemistry 28:8249-8254. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, S. J., K. N. Chung, K. Nachmanoff, and P. C. Elwood. 1997. Tissue-specific promoters of the alpha human folate receptor gene yield transcripts with divergent 5′ leader sequences and different translational efficiencies. Biochem. J. 326:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodolfo, M., C. Melani, and C. Zilocchi. 1998. IgG2a induced by interleukin (IL) 12-producing tumor cell vaccines but not IgG1 induced by IL-4 vaccine is associated with the eradication of experimental metastases. Cancer Res. 58:5812-5817. [PubMed] [Google Scholar]
- 26.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross, J. F., P. K. Chauduri, and M. Ratnam. 1994. Differential regulation of folate receptor isoforms in normal and malignant tissues in vivo and in established cell lines: physiologic and clinical implications. Cancer 73:2432-2443. [DOI] [PubMed] [Google Scholar]
- 28.Ross, J. F., H. Wang, F. G. Behm, P. Mathew, M. Wu, R. Booth, and M. Ratnam. 1999. Folate receptor type beta is a neutrophilic linear marker and is differentially expressed in myeloid leukemia. Cancer 85:348-357. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sudimack, J., and R. J. Lee. 2000. Targeted drug delivery via the folate receptor. Adv. Drug Deliv. Rev. 41:147-162. [DOI] [PubMed] [Google Scholar]
- 31.Tobler, A., C. W. Miller, K. R. Johnson, M. E. Selsted, G. Rovera, and H. P. Koeffler. 1988. Regulation of gene expression of myeloperoxidase during myeloid differentiation. J. Cell. Physiol. 136:215-225. [DOI] [PubMed] [Google Scholar]
- 32.Toffoli, G., C. Cernigo, A. Russo, A. Galls, M. Bagnoli, and M. Boiocchi. 1997. Overexpression of folate binding protein in ovarian cancers. Int. J. Cancer 74:193-198. [DOI] [PubMed] [Google Scholar]
- 33.Vaessen, R. T. M., A. Houweling, and A. J. Van der Eb. 1987. Post-transcriptional control of class I MHC mRNA expression in adenovirus 12-transformed cells. Science 235:1486-1488. [DOI] [PubMed] [Google Scholar]
- 34.Vannice, J. L., J. M. Taylor, and G. M. Ringold. 1984. Glucocorticoid-mediated induction of α1-acid glycoprotein: evidence for hormone-regulated RNA processing. Proc. Natl. Acad. Sci. USA 81:4241-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, H., X. Zheng, F. G. Behm, and M. Ratnam. 2000. Differentiation-independent retinoid induction of folate receptor type β, a potential tumor target in myeloid leukemia. Blood 96:3529-3536. [PubMed] [Google Scholar]
- 36.Ward, C. M. 2000. Folate-targeted non-viral DNA vectors for cancer gene therapy. Curr. Opin. Mol. Ther. 2:182-187. [PubMed] [Google Scholar]
- 37.Weitman, S. D., R. H. Lark, L. R. Coney, D. W. Fort, V. Frasca, V. R. Zurawski, and B. A. Kamen. 1992. Distribution of the folate receptor GP38 in normal and malignant cell lines and tissues. Cancer Res. 52:3396-3401. [PubMed] [Google Scholar]
- 38.Weitman, S. D., A. G. Weinberg, L. R. Coney, V. R. Zurawski, and D. Jennings. 1992. Cellular localization of the folate receptor: potential role in drug toxicity and folate homeostasis. Cancer Res. 52:6708-6711. [PubMed] [Google Scholar]
- 39.Wu, M., W. Gunning, and M. Ratnam. 1999. Expression of folate receptor type alpha in relation to cell type, malignancy, and differentiation in ovary, uterus, and cervix. Cancer Epidemiol. Biomarkers Prev. 8:775-782. [PubMed] [Google Scholar]
- 40.Yan, W., and M. Ratnam. 1995. Preferred sites of glycosylphosphatidylinositol modification in folate receptors and constraints in the primary structure of the hydrophobic portion of the signal. Biochemistry 3:14594-14600. [DOI] [PubMed] [Google Scholar]