Abstract
An important step in the herpesvirus life cycle is the switch from latency to lytic reactivation. The RTA transcription activator of Kaposi's sarcoma-associated herpesvirus (KSHV) acts as a molecular switch for lytic reactivation. Here we demonstrate that KSHV RTA recruits CBP, the SWI/SNF chromatin remodeling complex, and the TRAP/Mediator coactivator into viral promoters through interactions with a short acidic sequence in the carboxyl region and that this recruitment is essential for RTA-dependent viral gene expression. The Brg1 subunit of SWI/SNF and the TRAP230 subunit of TRAP/Mediator were shown to interact directly with RTA. Consequently, genetic ablation of these interactions abolished KSHV lytic replication. These results demonstrate that the recruitment of CBP, SWI/SNF, and TRAP/Mediator complexes by RTA is the principal mechanism to direct well-controlled viral gene expression and thereby viral lytic reactivation.
Regulation of cellular gene expression requires carefully choreographed binding by multiple transcription cofactors. A group of these cofactors are involved in the regulated alteration of chromatin structure, termed chromatin remodeling. These cofactors include the SWI/SNF complex, which disrupts nucleosomes in vitro and facilitates transcription factor binding in an ATP-dependent manner, and histone acetyltransferase and histone deacetylase, which act through covalent modification of histone tails (23, 25, 32, 46). Several types of activators, including nuclear receptors, C/EBPβ, c-Myc proto-oncoprotein, and erythroid Krüppel-like factor (EKLF), have been shown to physically or functionally interact with SWI/SNF complexes and histone acetyltransferase-histone deacetylase (3, 11, 24, 31, 44, 47). Recent studies indicate that chromatin remodeling is not an inherent feature of transcriptional activators but rather an important event required for subsequent transcription preinitiation complex assembly or a defining step in the transcriptional initiation process.
RNA polymerase II is found in a large holoenzyme complex containing several general transcription factors and the Mediator (32). Mediator is a large complex composed of polypeptides that range in size from 10 to 240 kDa. Several mammalian Mediator activities were discovered that specifically supported (TRAP/SMCC, ARC, DRIP, and Srb/Mediator) or repressed (NAT) the function of activators (30, 32). This complex functions as an interface between sequence-specific transcription factors and the general transcriptional apparatus. For example, the TRAP complex interacts with p53, VP16, NF-κB, and E1A to recruit RNA polymerase II and general transcription factors to form a functional preinitiation complex at the promoter (20). More specifically, the TRAP220 subunit of this complex is known to interact with nuclear receptors, including the thyroid receptor, vitamin D receptor, estrogen receptor, and glucocorticoid receptor; the TRAP150β subunit is likely an integrator of the E1A and RAS signaling pathways; and the TRAP80 subunit interacts directly with the p53 and VP16 activation domains (6, 18, 21, 40, 48). Thus, TRAP/Mediator/SMCC, a multifunctional complex, contains diverse subunits that serve as specific targets for distinct activators.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8, is thought to be an etiologic agent of Kaposi's sarcoma (9). It is also associated with two diseases of B-cell origin, primary effusion lymphoma and an immunoblast variant of Castleman's disease (5, 7). The genomic sequence indicates that KSHV is a gamma herpesvirus that is closely related to Epstein-Barr virus, herpesvirus saimiri, rhesus monkey rhadinovirus, and murine gammaherpesvirus 68 (2, 9, 35, 37, 43).
An important step in the herpesvirus life cycle is the switch from latency to lytic replication. KSHV RTA has been shown to play a central role in the switch of the viral life cycle from latency to lytic replication. Ectopic expression of RTA is sufficient to disrupt viral latency and activate lytic replication to completion (15, 29, 42). As a typical transcription activator, KSHV RTA contains an N-terminal basic DNA-binding domain and a C-terminal acidic activation domain. Its N-terminal DNA-binding domain is well conserved with that of Epstein-Barr virus RTA and other gammaherpesvirus RTA homologs and shows a sequence-specific DNA-binding activity (8, 27, 38). While it is less conserved, a carboxyl acidic activation domain exhibits strong transactivation activity in the heterologous context with the Saccharomyces cerevisiae GAL4 transcription factor (16, 28). It has been shown that RTA activates the expression of numerous viral genes in the KSHV lytic cycle, including its own promoter, polyadenylated nuclear (PAN) RNA, ORF57, vOX-2, viral G protein-coupled receptor, and vIRF1 (10, 12, 13, 22, 36, 38).
While the detailed mechanism of RTA-mediated transcription activation remains unclear, several pieces of evidence suggest that RTA activates its target promoter activity through both direct binding to the specific sequence and interaction with various cellular transcriptional factors. In fact, numerous cellular proteins, including Oct-1, Stat3, novel cellular protein MGC2663, CBP, and RBP Jκ, have been found to interact with and synergize with RTA (16, 17, 26, 36, 45).
Despite extensive studies of RTA-mediated transcriptional activation of viral lytic genes, details of the mechanism are mostly unknown. To delineate the molecular mechanism of RTA-mediated lytic gene expression, we purified proteins that bound to RTA. Mass spectrometry demonstrated that RTA recruits cellular SWI/SNF and TRAP/Mediator complexes through its carboxy-terminal short acidic sequence. Recruitment of SWI/SNF and TRAP/Mediator complexes by RTA into the viral lytic promoters is essential for their gene expression and thus for KSHV reactivation. Furthermore, genetic ablation of these interactions abolishes KSHV viral lytic replication. These results demonstrate that the molecular mechanisms that underlie RTA-mediated transcriptional activation require a large number of transcriptional cofactors and that their actions ultimately direct well-controlled viral gene expression and thereby viral lytic reactivation.
MATERIALS AND METHODS
Cell culture, transient transfection, immunoprecipitation, and immunoblot.
Detailed procedures were described in previous reports (16, 17). The antibodies used for the chromatin immunoprecipitation assay and immunoblot analysis were purchased from Santa Cruz Biotechnology (Brg-1, sc-10678; Ini1, sc-9751; BAF170, sc-9742; BAF155, sc-9747; TBP, sc-421; TRAP240, sc-12013; TRAP230, sc-5374; TRAP220, sc-5334; TRAP100, sc-5338; TRAP95, sc-5366; Cdk8, sc-1521; CBP, sc-369; and p300, sc-584).
Protein purification and mass spectrometry.
To identify RTA-binding proteins, [35S]methionine- and [35S]cysteine-labeled Raji cells (1 × 107 cells) or 20 liters of Raji cells were resuspended with lysis buffer (0.15 M NaCl, 0.5% Nonidet P-40, and 50 mM HEPES buffer [pH 8.0]) containing protease and phosphatase inhibitors. Precleared lysates were mixed with glutathione beads containing glutathione S-transferase (GST) and GST-RTA fusion protein for 4 h, and the beads were washed extensively with lysis buffer. Proteins bound to glutathione beads were eluted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein bands isolated from SDS-PAGE were analyzed by ion-trap mass spectrometry, and the amino acid sequence was determined by tandem mass spectrometry and database search.
GST pulldown assays.
RTA and TRAP subunits were in vitro transcribed and translated with a T7-coupled transcription-translation system (Promega, Madison, Wis.). The labeled proteins were incubated with GST fusion protein-saturated resin in binding buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, and 0.1% NP-40 supplemented with protease inhibitors). The reaction mixture was incubated at 4°C for 2 h. The beads were then washed four times with binding buffer, SDS-PAGE sample buffer was added, and the proteins were analyzed by SDS-PAGE and visualized by PhosphorImager (BAS-1500; Fuji Film Co., Tokyo, Japan).
Flow cytometry.
Cells (5 × 105) were washed with complete medium and stained with unconjugated K8.1 primary antibody, followed by fluorescein isothiocyanate-conjugated secondary antibody at 4°C. After a final wash, the cells were fixed with 2% paraformaldehyde, and flow cytometry was performed with a fluorescence-activated cell scan/sorter (Becton Dickinson, Mountain View, Calif.).
ATPase assay.
Reactions were carried out at 30°C with 2 μM ATP in 12 mM sodium HEPES (pH 7.9)-60 mM KCl-7 mM MgCl2-60 ng of bovine serum albumin per μl-6% glycerol, as described previously (34). ATP hydrolysis was measured with affinity-purified resin in the presence or absence of 20 nM plasmid DNA (5.6 kb). A PhosphorImager was used to quantitate the ratio of inorganic phosphate to ATP at each time point.
In vitro transcription, TRAP/Mediator purification, and immunodepletion of TRAP/Mediator from nuclear extracts.
Detailed procedures for in vitro transcription, TRAP/Mediator purification, and immunodepletion of TRAP/Mediator were described previously (4).
Chromatin immunoprecipitation.
The chromatin immunoprecipitation assay was performed according to the manufacturer's (Upstate Biotech) instructions with several modifications. Briefly, T75 culture dishes were treated with 1% formaldehyde for 10 min at room temperature. After a brief sonication, immunoprecipitation was performed with the appropriate antibody. After several washes, the immunocomplexes were eluted with 50 mM Tris (pH 8.0)-1 mM EDTA-1% SDS at 65°C for 10 min, adjusted to 200 mM NaCl, and incubated at 65°C for 5 h to reverse the crosslinks. After successive treatments with 10 μg of RNase A and 20 μg of proteinase K per ml, the samples were extracted with phenol-chloroform and precipitated with ethanol. One tenth of the immunoprecipitated DNAs was analyzed by PCR with the primer sets for Rp (nucleotides 71221 to 71550, KSHV GenBank accession number U75698), Mp (nucleotides 81661 to 81920), vOX-2p (nucleotides 127684 to 127911), PANp (nucleotides 28727 to 29043), vIRFp (nucleotides 85589 to 85709), gBp (nucleotides 8341 to 8642), and POLp (nucleotides 10602 to 11162). Amplifications (26 cycles) were performed in the presence of 5 μCi of [α-32P]dCTP, and the PCR products were analyzed in 5% polyacrylamide gels. For the chromatin reimmunoprecipitation (reChip) assays, after washing of protein G-Sepharose beads from the primary immunoprecipitation, the complexes were eluted by incubation with 10 mM dithiothreitol at 37°C for 30 min and diluted to 40 times the original volume. Eluates were reimmunoprecipitated with the second antibody. The anti-acetyl histone H3 antibody was purchased from Upstate Biotechnology.
Generation of riboprobe templates and RNase protection assay.
RNA (100 ng/reaction) extracted from KSHV-infected BCBL1 cells was used as the template for cDNA synthesis initiated by random hexamer primers, followed by PCR with a GeneAmp kit (Perkin-Elmer Cetus, Foster City, Calif.) and appropriate primers for the amplification of KSHV-specific DNA fragments. The reverse transcription (RT)-PCR conditions, design of PCR primers, and ligation of the amplified DNA fragments into the pEF1/Myc-His A vector were detailed previously (19). The subclone designations and nucleotide sequence based from KSHV GenBank accession number U75698 were as follows: RTA (nucleotides 68349 to 68680), vIRF-1 (nucleotides 18618 to 18857), and ORF57 (6779 to 6977). A riboprobe template set specific for KSHV RTA, ORF57, and vIRF1 was assembled from EcoRI-linearized and purified subclones. All riboprobe syntheses were driven by T7 bacteriophage RNA polymerase with [α-32P]UTP (Amersham, Arlington Heights, Ill.) as the labeling nucleotide. Probe bands were visualized by autoradiography by with a PhosphorImager.
RESULTS
Identification of RTA binding proteins.
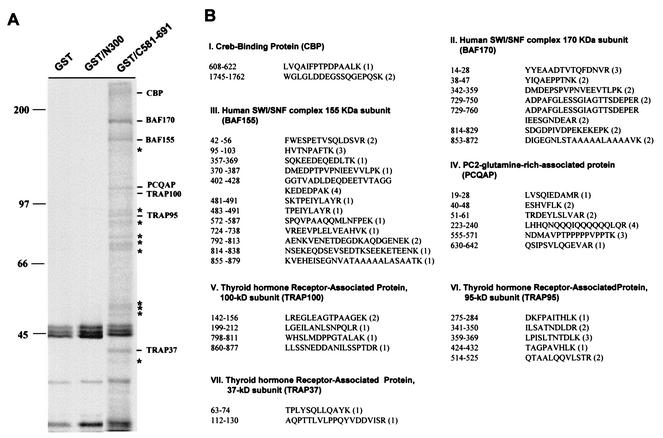
To identify cellular proteins interacting with RTA, bacterially expressed GST-RTA fusion proteins were used as an affinity column for 35S-labeled lysates of Raji B cells. GST-RTA(N1-300) contains the amino-terminal DNA binding domain and leucine zipper motif of RTA (amino acids 1 to 300), and GST-RTA(C581-691) contains the carboxy-terminal activation domain of RTA (amino acids 581 to 691). Approximately 20 polypeptides with molecular masses ranging from 20 to 300 kDa were found to interact with GST-RTA(C581-691). None of these cellular proteins interacted with GST and the GST-RTA(N1-300) fusion protein under the same conditions (Fig. 1A).
FIG. 1.
Purification and identification of RTA-binding proteins. (A) Identification of RTA-binding proteins. Glutathione-Sepharose beads containing 5 μg of GST, GST-RTA(N1-300), or GST-RTA(C581-691) fusion protein were mixed with lysates of 35S-labeled Raji B cells. RTA-binding proteins were resolved in SDS-PAGE and autoradiographed with a PhosphorImager. Asterisks indicate the proteins that were not identified by mass spectrometry. Sizes are shown in kilodaltons. (B) Mass spectrometry analysis of RTA-binding proteins. The peptide sequence of each protein isolated by the mass spectrometry analysis is presented with its published amino acid sequence number. The number in parentheses indicates the frequency with which a particular peptide sequence was identified from the mass spectrometry analysis.
To further characterize these cellular proteins, they were purified from 20 liters of Raji cells. The resulting proteins were subjected to mass spectrometry and then matched to known sequences (Fig. 1B). Surprisingly, all of the polypeptides were part of three groups of cellular transcription cofactors: (i) CBP histone acetyltransferase, (ii) SWI/SNF complex (BAF 170 and 155), and (iii) TRAP/Mediator complex (TRAP100, -95, and -35 and PCQAP) (Fig. 1B). The identification of CBP from the mass spectrometry analysis not only confirmed our previous finding (16) but also indicated the effectiveness of our purification procedure. Because some of the proteins had relatively low concentrations and molecular weights near that of the GST-RTA(C581-691) fusion protein (approximately 50 kDa), the cellular polypeptides with molecular masses of 50 to 70 kDa could not be analyzed further by mass spectrometry (Fig. 1A, asterisks).
Interaction of RTA with SWI/SNIF and TRAP/Mediator complexes.
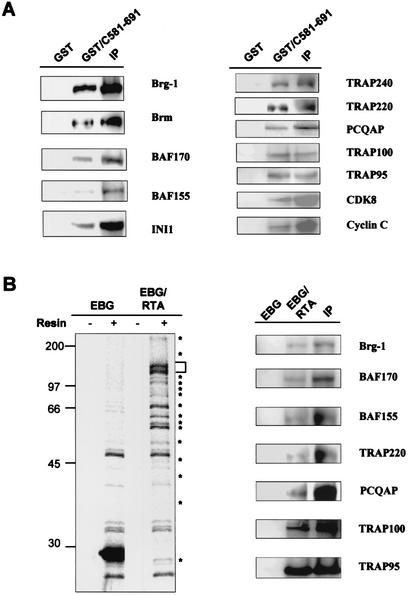
To confirm the interaction of RTA with SWI/SNF and TRAP/Mediator complexes, we repeated the GST pulldown and performed immunoblot analysis with antibodies to SWI/SNF and TRAP/Mediator subunits. This showed that the subunits of SWI/SNF and TRAP/Mediator complexes tested were detected in the GST-RTA complexes, whereas none of them were present in the GST complex (Fig. 2A). Furthermore, CDK8 and cyclin C were also readily detected in the GST-RTA complexes but not in the GST complexes (Fig. 2A).
FIG. 2.
Interaction between RTA and cellular transcription cofactors. (A) In vitro interaction between RTA and cellular transcription cofactors. Lysates of Raji B cells were mixed with GST and GST-RTA(C581-691) or used for immunoprecipitation (IP) with the antibodies to cellular transcription cofactors indicated at the right side of the figure. Polypeptides present in GST fusion complexes and immunoprecipitation complexes were separated by SDS-PAGE, followed by immunoblot assay with the same antibodies. (B) Interaction between RTA and cellular transcription cofactors. [35S]methionine/cysteine-labeled 293T cells transfected with the EBG or EBG-RTA vector were used for glutathione-Sepharose affinity chromatography (resin). The bound GST fusion complexes were separated by SDS-PAGE followed by autoradiography (left panel). The bracket indicates the GST-RTA protein, and asterisks indicate the proteins associated with GST-RTA. Lysates were also used for glutathione-Sepharose affinity chromatography or Immunoprecipitation with antibodies to cellular transcription cofactors as indicated at the right side of the figure. Polypeptides present in the GST fusion and immunoprecipitation complexes were separated by SDS-PAGE, followed by immunoblot assay with the same antibodies (right panel). Based on the comparison between GST-RTA pulldown and immunoprecipitation, 12.5% of Brg1, 7.2% of BAF170, 5.4% of BAF55, 4% of TRAP220, 4.1% of PCQAP, 7.3% of TRAP, and 70% of TRAP95 were copurified by GST-RTA protein.
To further confirm an interaction of RTA with SWI/SNF and TRAP/Mediator complexes, we transfected mammalian expression vectors containing either GST (EBG vector) or GST-full-length RTA (EBG-RTA vector) into 293T cells and labeled these with [35S]methionine and [35S]cysteine. Radioactively labeled RTA complexes were purified with a glutathione-Sepharose column and separated by SDS-PAGE (Fig. 2B). This analysis showed that full-length RTA interacted in living cells with multiple cellular proteins that had molecular weights similar to those of GST-RTA complexes (C581-691). Subsequent immunoblot assays with antibodies to specific SWI/SNF and TRAP/Mediator subunits further demonstrated that RTA interacted with the SWI/SNF complex and the TRAP/Mediator complex in living cells (Fig. 2B). These results demonstrate that the carboxyl region of RTA interacts efficiently with SWI/SNF and TRAP/Mediator. In addition, since the binding assay was performed in the absence of an RTA-dependent promoter sequence, RTA likely interacts with cellular transcription cofactors independently of the DNA template.
Specific region of RTA required for SWI/SNF and TRAP interaction.
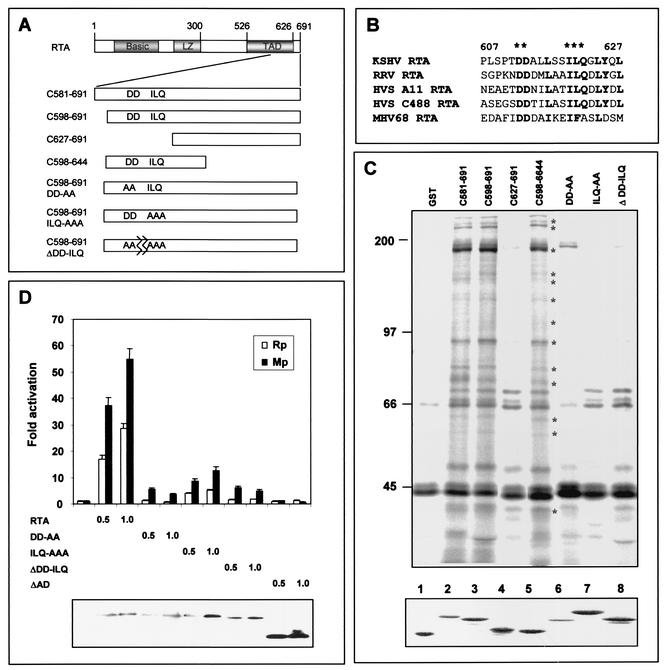
To define the specific region of RTA required for the interaction with SWI/SNF and TRAP/Mediator complexes, additional GST-RTA fusion proteins, GST-RTA(C598-691), GST-RTA(C627-691), and GST-RTA(C598-644), were generated (Fig. 3A). Pulldown assays showed that the GST-RTA(C598-691) and GST-RTA(C598-644) fusion proteins interacted with the SWI/SNF and TRAP complexes as efficiently as GST-RTA(C581-691) (Fig. 3C). In contrast, GFT-RTA(C627-691) was not capable of interacting with the SWI/SNF and TRAP/Mediator complexes under the same conditions (Fig. 3C). Immunoblot analysis confirmed that cellular proteins associated with GST-RTA(C598-691) and GST-RTA(C598-644) were part of the SWI/SNF and TRAP/Mediator complexes (data not shown). This indicates that a sequence of 47 amino acids of RTA is necessary for interacting with SWI/SNF and TRAP complexes.
FIG. 3.
Identification of region of RTA required for interaction with cellular transcription cofactors and their role in RTA-mediated transcriptional activation. (A) Schematic diagram of RTA mutants. RTA contains an N-terminal basic domain, an internal leucine zipper (LZ) motif, and a C-terminal transcription activation domain (TAD). Each RTA mutant is described in detail in the text. (B) Conserved sequences in the carboxyl transcription activation domain of gamma-2 herpesvirus RTA homologs. Asterisks indicate the conserved amino acid sequences that were mutated. (C) The conserved sequence at the carboxyl activation domain of RTA is necessary for interaction with cellular transcription cofactors. [35S]methionine/cysteine-labeled Raji B cells were mixed with GST, GST-RTA, and GST-RTA mutants. 35S-labeled polypeptides associated with GST fusion proteins were separated by SDS-PAGE, followed by autoradiography. A similar amount of each GST fusion protein was used in this assay (bottom panel). Asterisks indicate the cellular proteins associated with GST-RTA fusion proteins. (D) The conserved sequence at the carboxy-terminal region of RTA is required for efficient transcriptional activation of the RTA (Rp) and ORF57 (Mp) promoters. 293T cells were transfected with an expression vector containing RTA or one of its mutants together with the Rp-luciferase (open rectangle) or Mp-luciferase (solid rectangle) reporter. The RSV-β-galactosidase vector was included as a transfection control. Luciferase activity was measured at 48 h posttransfection, and luciferase values were normalized by β-galactosidase activity. Luciferase activity is represented as the average of three independent experiments. Error bars indicate the standard error. The expression levels of RTA and its mutants are shown (bottom panel).
Our previous report showed that the sequence between amino acids 607 and 626 likely plays an important role in RTA-mediated transcriptional activation (16). Although there is poor sequence homology of the carboxyl transcriptional activation domain among RTA homologs of gamma-2 herpesviruses, several amino acids between 607 and 626 are highly conserved. These are the two glutamic acids at residues 612 and 613 and the three residues ILQ at 619 to 621 (Fig. 3B). In particular, the ILQ sequence has also been shown to be conserved in the activation domain of other viral and cellular transcription factors, including VP16, SP1, CTF, and E1A (28).
To test whether these sequences are involved in the interaction with SWI/SNF and TRAP complexes, we generated GST-RTA fusions containing mutations at these sequences as follows. The two glutamic acids at residues 612 and 613 were replaced with alanines to generate GST-RTA(C598-691 DD-AA), the ILQ residues at 619 to 621 were replaced with alanines to generate GST-RTA(C598-691 ILQ-AAA), and 10 amino acids at residues 612 to 621 were deleted to generate GST-RTA(C598-691 ΔDD-ILQ) (Fig. 3A and 3C). Interestingly, GST-RTA(C598-691 ILQ-AAA) migrated in SDS-PAGE with a slightly altered rate, suggesting a conformation change (bottom of Fig. 3C). Similar amounts of GST-RTA fusion proteins were used for the pulldown assay with 35S-labeled Raji cell lysates. We found that all three point mutations or deletions at the conserved sequences of the RTA carboxyl region abolished its interaction with SWI/SNF and TRAP/Mediator complexes (Fig. 3C). This result was further confirmed by immunoblot assay with antibodies to SWI/SNF and TRAP/Mediator complexes (data not shown). These results indicate that a region of 47 amino acids of RTA is necessary for interacting with SWI/SNF and TRAP/Mediator complexes and that the conserved sequences of this region appear to play an important role in this interaction.
Role of SWI/SNF and TRAP/Mediator complexes in RTA-mediated transcriptional activation.
To elucidate the role of the RTA interaction with SWI/SNF and TRAP/Mediator complexes, we examined the level of transcriptional activity of RTA(DD-AA), RTA(ILQ-AAA), and RTA(ΔDD-ILQ) mutants, which do not bind to SWI/SNF and TRAP complexes, as shown in Fig. 2B. As a control, we included RTA ΔAD, which contains a deletion of the carboxy-terminal transcriptional activation domain (amino acids 598 to 691) (16). While wild-type RTA activated its own promoter (Rp) and the ORF57 promoter (Mp) by 30- to 50-fold, the RTA(DD-AA), RTA(ILQ-AAA), and RTA(ΔDD-ILQ) mutants exhibited a marked reduction of transcriptional activation activity (Fig. 3D). The RTA(ΔDD-ILQ) mutant in particular exhibited very little transcriptional activation activity compared to wild-type RTA, whereas the RTA ΔAD deletion mutant had no activity (Fig. 3D). RTA and its mutants were expressed at equivalent levels (Fig. 3D). These results indicate that mutations at the conserved sequences which cripple the interaction with SWI/SNF and TRAP/Mediator complexes drastically reduce the RTA-mediated activation of Rp and Mp promoter activity, suggesting an important role of SWI/SNF and TRAP complexes in RTA-mediated transcription of Rp and Mp.
SWI/SNF complex facilitates RTA-mediated transcriptional activation.
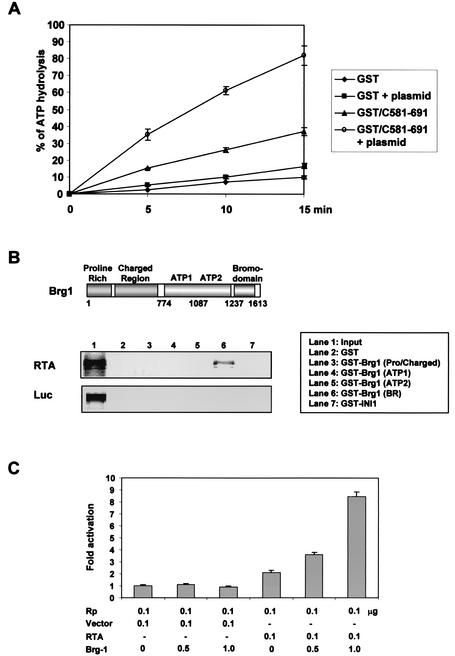
Since RTA interacts with the chromatin-remodeling SWI/SNF complex that contains DNA-dependent ATPase activity, we examined ATPase activity in the affinity-purified RTA complexes. After incubation with DNase and ethidium bromide to exclude any potential contamination by chromosomal DNA, crude extracts from Raji B cells were mixed with GST or GST-RTA(C581-691). After extensive washing, affinity-purified GST and GST-RTA(C581-691) complexes were assayed for ATPase activity. Purified GST-RTA(C581-691) complexes contained an approximately threefold-higher basal level of ATPase activity than purified GST complexes (Fig. 4A). In addition, this ATPase activity was significantly enhanced by the addition of template plasmid DNA (Fig. 4A). In contrast, GST complexes did not contain a significant level of ATPase activity, nor did the addition of template plasmid DNA augment ATPase activity (Fig. 4A). These results demonstrate that purified RTA complexes contain a strong DNA-dependent ATPase activity that appears to be derived from the SWI/SNF complex.
FIG. 4.
ATPase activity in RTA complexes and the role of Brg1 in RTA-mediated transcriptional activation. (A) The presence of ATPase activity in RTA complexes. Raji B-cell lysates were mixed with GST or GST-RTA(C581-691). After extensive washing, GST and GST-RTA(C581-691) complexes were used for ATP hydrolysis in the presence or absence of 20 nM plasmid DNA. The reaction from each time point was separated on polyethyleneimine-cellulose thin-layer chromatography plates, and the ratio of inorganic phosphate to ATP was quantitated by PhosphorImager. (B) Interaction of RTA with the carboxyl region of Brg1. The GST-Brg1 fusion protein contains each domain of Brg1 depicted in the top panel. [35S]methionine-labeled RTA and luciferase proteins from in vitro translation were mixed with GST or GST-Brg1 fusion protein. After extensive washing, 35S-labeled polypeptides associated with GST or GST-Brg1 fusion protein were separated by SDS-PAGE, followed by autoradiography. Lane 1 indicates 10% input amount of RTA or luciferase protein used for the binding assay. (C) The role of Brg1 in RTA-mediated transcriptional activation. SW13 cells were transfected with expression vector and reporter vector as indicated below the figure. Luciferase activity was measured at 48 h posttransfection, and luciferase values were normalized by β-galactosidase activity. Luciferase activity is represented as the average of three independent experiments. Error bars indicate the standard error.
The human SWI/SNF complex consists of multiple components, including BAF47, BAF50, BAF155, BAF170, BAF250, INI1, and Brg1 (33). The carboxyl domain of Brg1 in particular has been shown to interact with numerous cellular factors, including pRb, p107, and p130 (41). Thus, we examined whether RTA potentially targeted Brg1 for a direct interaction. To test this, the individual domains of Brg1 were fused in frame into GST. GST-Brg1(Pro/charged) protein contains the amino-terminal proline-rich and charged regions; the GST-ATP1 and GST-ATP2 proteins contain the first and second portions of ATPase region, respectively; and GST-Brg1-BR contains the carboxyl E7-like sequence and Bromo domain (Fig. 4B). In addition, GST-INI1, containing a full-length INI1, was included as a control. [35S]methionine-labeled RTA and luciferase proteins from in vitro translation were mixed with GST, GST-Brg1, and GST-INI1 fusion proteins. After extensive washing, polypeptides associated with GST, GST-Brg1, and GST-INI1 fusion proteins were separated by SDS-PAGE, followed by autoradiography (Fig. 4B). The resulting gel showed that GST-Brg1-BR interacted efficiently with RTA, whereas GST, GST-INI1, and other GST-Brg1 fusion proteins did not. The interaction between RTA and GST-Brg1-BR appeared to be specific because the GST-Brg1-BR fusion protein showed no apparent interaction with [35S]methionine-labeled luciferase under the same conditions (Fig. 4B). These results indicate that, as seen with other cellular factors (41), KSHV RTA is capable of interacting with the carboxy-terminal domain of Brg1.
To investigate the functional role of the interaction between the SWI/SNF complex and RTA, we cotransfected expression vectors containing RTA and Brg1 cDNA together with the RTA promoter (Rp) luciferase reporter vector into SW13 adrenal carcinoma cells, which lack a functional Brg1 gene. In the absence of a functional Brg1, RTA induced basal Rp promoter activity (Fig. 4C). In contrast, coexpression of Brg1 strongly enhanced RTA activity, resulting in over eightfold induction of Rp promoter activity (Fig. 4C). This result suggests that the efficient activation of Rp promoter activity by RTA requires a functional SWI/SNF complex.
TRAP/Mediator complex is essential for RTA-mediated transcription in vitro.
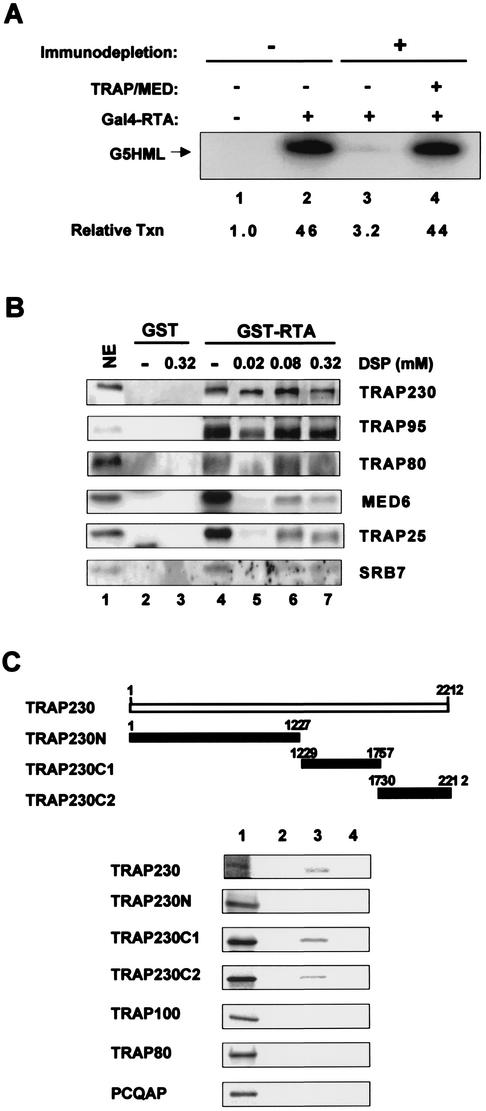
To establish the functional consequences of an interaction of RTA with the TRAP/Mediator complex, we first asked if TRAP could function as a coactivator for RTA-mediated transcription in an in vitro transcription reaction with nuclear extracts. We have shown previously that TRAP25 antibody efficiently depletes the majority of TRAP/Mediator complex from the nuclear extract, leaving RNA polymerase II and other general transcription factors unaffected (4). Thus, we tested the intact nuclear extracts and TRAP/Mediator-depleted nuclear extracts for the ability of TRAP/Mediator to support transcriptional activation induced by Gal4-RTA, which contains the C-terminal activation domain of RTA (16, 28). In addition, the G5HML plasmid has five copies of a Gal4 binding site upstream of a hybrid core promoter and permits measurement of the transcriptional activity of the Gal4-RTA fusion (4).
In the intact nuclear extracts, Gal4-RTA protein activated in vitro transcription of G5HML up to 46-fold (Fig. 5A). By striking contrast, Mediator-depleted extracts had only a basal level of Gal4-RTA transcriptional activation (Fig. 5A, lane 3). The reconstitution of purified TRAP/Mediator complex into Mediator-depleted nuclear extracts completely restored Gal4-RTA transcription activity (Fig. 5A, lane 4). These results demonstrate that the reduction of Gal4-RTA transcription activity in TRAP/Mediator-depleted nuclear extracts is due solely to the absence of TRAP/Mediator rather than other coimmunoprecipitated factors and that the TRAP/Mediator complex plays an important role in RTA-mediated transcriptional activation.
FIG. 5.
Interaction of RTA with TRAP/Mediator complex. (A) The essential role of TRAP/Mediator in in vitro RTA-mediated transcription. Each in vitro transcription reaction mixture contained 20 μg of nuclear extract (lanes 1 and 2, untreated; lanes 3 and 4, anti-TRAP25 antibody-depleted nuclear extract [ΔMED]). Gal4-RTA(C581-691) protein was used as an activator, and 50 ng of pG5HML template was added as a template. Relative transcription (Txn) levels, determined by PhosphorImager analysis, are indicated. (B) RTA interacts directly with the TRAP230 subunit when bound to the TRAP/Mediator complex. One-half milliliter of HeLa nuclear extract was incubated with 10 μg of GST or GST-RTA(C581-691) for 7 h at 4°C and washed with phosphate-buffered saline. The bound proteins were exposed to increasing amounts of the crosslinker DSP in dimethylsulfoxide or dimethyl sulfoxide only for 10 min at room temperature. Lane 1, nuclear extract (NE, 5% of starting nuclear extract); lanes 2 and 4, affinity purification of GST and GST-RTA, respectively, before DSP treatment; lanes 3 and 7, 0.32 mM DSP; lane 5, 0.02 mM DSP; lane 6, 0.08 mM DSP. The crosslinking reagent was quenched by addition of Tris (pH 7.5) to a final concentration of 50 mM. After extensive washing with 8 M urea (lanes 3, 5, 6, and 7), polypeptides present in GST and GST-RTA(C581-691) complexes were subjected to the immunoblot assay with antibodies specific to TRAP230, TRAP95, TRAP80, MED6, TRAP25, and SRB7 subunits. (C) Interaction of RTA with TRAP230. [35S]methionine-labeled TRAP230, TRAP230N, TRAP230C1, TRAP230C2, TRAP100, TRAP80, and PCQAP proteins from in vitro translation reactions were mixed with GST (lane 2), GST-RTA(C598-691) (lane 3), or GST-RTA(C598-691ΔDD-ILQ) (lane 4). After extensive washing with lysis buffer, 35S-labeled polypeptides associated with GST fusion proteins were separated by SDS-PAGE, followed by autoradiography. Lane 1 indicates 10% of an input amount of each protein used for the binding assay.
To gain further insight into the mechanism of action of the TRAP/Mediator complex in RTA transcription, we investigated the potential interaction of RTA with individual subunits of the TRAP/Mediator complex. Purified TRAP/Mediator complex was mixed with GST or GST-RTA(C581-691) and exposed to increasing amounts of the chemical crosslinker dithiobis succinimidyl propionate (DSP) in dimethyl sulfoxide or dimethyl sulfoxide only for 10 min. Crosslinking reagent was quenched by addition of Tris (pH 7.5), followed by extensive washes with 8 M urea. Polypeptides present in GST and GST-RTA(C581-691) complexes were subjected to an immunoblot assay with numerous antibodies specific for the components of the TRAP/Mediator complex.
The resulting blots showed that while RTA interacted with TRAP230, TRAP95, TRAP80, MED6, TRAP25, and SRB7 subunits after treatment with the crosslinking agent, TRAP230 exhibited a saturated level of binding activity to GST-RTA(C581-691) even at the lowest concentration of DSP tested (Fig. 5B, lane 5). In contrast, the amounts of other TRAP subunits binding to GST-RTA(C581-691) gradually increased as the DSP concentration was increased (Fig. 5B). None of the TRAP subunits tested was shown to bind to GST protein under the same conditions, indicating the specificity of the RTA interaction with TRAP subunits (Fig. 5B). This also suggests that TRAP230 has a higher affinity for RTA protein than other TRAP subunits.
To further characterize this, GST, GST-RTA(C598-691), and GST-RTA(C598-691 ΔDD-ILQ) fusion proteins were mixed with [35S]methionine-labeled TRAP subunits, TRAP230, TRAP100, TRAP80, and PCQAP, followed by autoradiography. In addition to full-length TRAP230, three fragments of TRAP230, TRAP230N, TRAP230C1, and TRAP230C2, were included in this assay. The results showed that the carboxyl activation domain of RTA interacted efficiently with TRAP230, TRAP230C1, and TRAP230C2 but not with TRAP230N, TRAP100, TRAP80, and PCQAP (Fig. 5C). In contrast, the RTA(ΔDD-ILQ) mutant did not interact with full-length TRAP230 and its C-terminal fragments under the same conditions (Fig. 5C). This result shows an activation domain-dependent interaction of RTA with the TRAP230 subunit.
Recruitment of cellular transcriptional cofactors into RTA-dependent promoters.
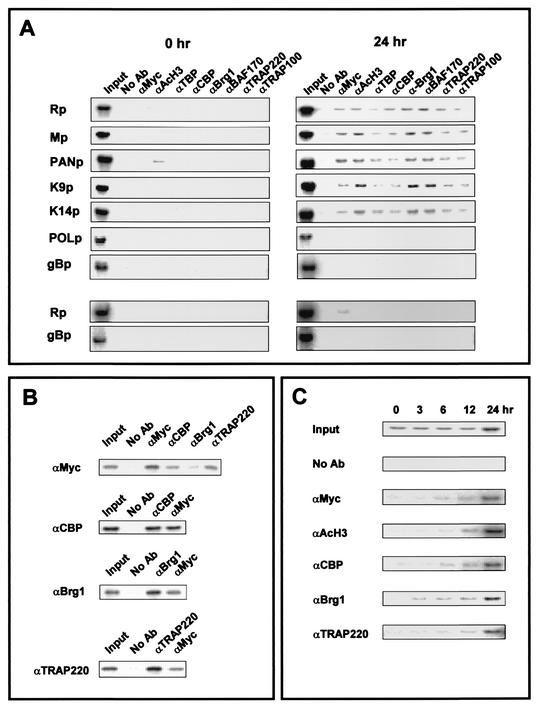
RTA activates expression from the promoters of several KSHV genes, including its own promoter (Rp), ORF57 (Mp), polyadenylated nuclear RNA (PANp), vOX2 (vOX2p), and vIRF1 (vIRFp), but RTA cannot activate expression of KSHV DNA polymerase (POLp) and glycoprotein B (gBp). To investigate whether RTA recruits cellular transcriptional cofactors into the RTA-dependent promoters, we performed chromatin immunoprecipitation experiments. For this assay, we used a KSHV-infected BCBL1 cell line (TRExBCBL1) in which Myc epitope-tagged wild-type RTA or the RTA ΔAD mutant gene was integrated into the chromosomal DNA under the control of a tetracycline-inducible promoter. Treatment of these cells with doxycycline strongly activated RTA and RTA ΔAD expression (data not shown).
KSHV-infected TRExBCBL1 cells with or without doxycycline treatment were subjected to the chromatin immunoprecipitation assay. RTA-dependent viral promoters (Rp, Mp, PANp, vOX2p, and vIRFp) and RTA-independent viral promoters (POLp and gBp) were tested for RTA binding and the recruitment of cellular transcription cofactors. Twenty-four hours after doxycycline treatment, wild-type RTA was readily detected on the RTA-dependent promoters (Rp, Mp, PANp, vOX2p, and vIRFp) but not on the RTA-independent promoters (POLp and gBp) (Fig. 6A). Additionally, the recruitment of TBP, CBP, Brg1, BAF170, TRAP220, and TRAP100 as well as acetylated histone was strongly detected on the RTA-dependent promoters (Rp, Mp, PANp, vOX2p, and vIRFp), whereas it was not detected on the RTA-independent promoters (POLp and gBp) (Fig. 6A). In the absence of RTA expression, however, none of the cellular transcription cofactors were recruited to viral promoters, indicating that RTA is necessary for the recruitment of cellular transcription cofactors (Fig. 6A). This finding was further supported by the results from TRExBCBL1 cells expressing the RTA ΔAD mutant. Because of the presence of the amino-terminal DNA binding region, RTA ΔAD was detected on the Rp promoter (Fig. 6A, bottom two panels). However, due to the lack of the carboxyl activation domain on the RTA ΔAD mutant, none of these cellular transcriptional cofactors were recruited onto the Rp promoter (Fig. 6A, bottom two panels). Finally, a reChip assay further confirmed that RTA interacted with and recruited CBP, Brg1, and TRAP220 onto the Rp promoter (Fig. 6B).
FIG. 6.
Recruitment of cellular transcription cofactors to RTA-dependent promoters. (A) Chromatin immunoprecipitation assay of RTA-dependent and RTA-independent promoters. KSHV-infected TRExBCBL1-RTA cells or TRExBCBL1-RTA(ΔDD-ILQ) cells (bottom two panels) were collected without treatment or after 24 h of treatment with 1 μg of doxycycline per ml. Chromatin immunoprecipitation assays were performed with antibodies against the protein indicated at the top. PCR products corresponding to each viral promoter were generated from an aliquot (1/10) of total immunoprecipitated material (Input). (B) ReChip assay. TRExBCBL1-RTA cells were stimulated for 24 h with 1 μg of doxycycline per ml and subjected to the chromatin immunoprecipitation assay as described above. After washing the protein-G-Sepharose beads from the primary immunoprecipitation shown on the left panel, the complexes were eluted by incubation with 10 mM dithiothreitol at 37°C for 30 min and diluted to 40 times the original volume. Eluates were reimmunoprecipitated with the second antibody indicated at the top of figure, followed by PCR amplification. PCR products were generated from an aliquot (1/10) of total immunoprecipitated material (Input). (C) Time course of chromatin immunoprecipitation assay. TRExBCBL1-RTA cells were stimulated with doxycycline for 1, 3, 6, 12, and 24 h and then used for the chromatin immunoprecipitation assay as described above. PCR products were generated from an aliquot (1/10) of total immunoprecipitated material (Input).
Recent reports indicate that cellular transcription cofactors act in both a sequential and combinatorial manner to reorganize chromatin templates and to activate transcription (14, 32, 33). To test the potential ordered recruitment of cellular cofactors to the Rp promoter, TRExBCBL1-RTA cells were treated with doxycycline and then subjected to the chromatin immunoprecipitation assay (Fig. 6C). The results showed that RTA was recruited to the Rp promoter at a rate similar to that of CBP, Brg1, and TRAP220. The occupancy of these proteins on the Rp promoter was detected immediately after doxycycline treatment and increased gradually up to 24 h (Fig. 6C). Thus, these results suggest that the multiple complexes containing RTA and cellular transcription cofactors are assembled and then recruited to the Rp promoter at a similar rate.
Recruitment of TRAP/Mediator and SWI/SNF complexes is required for RTA-mediated KSHV lytic reactivation.
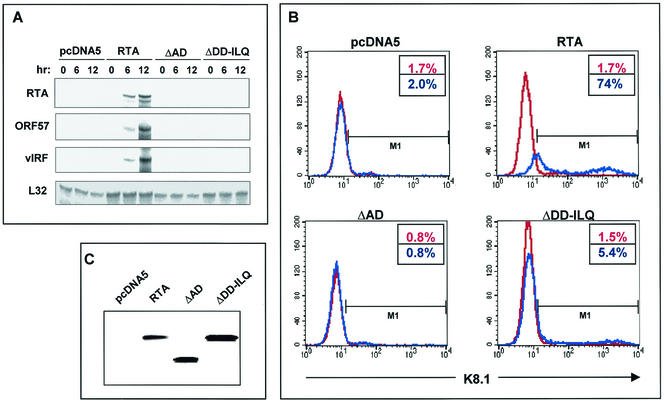
RTA has been shown to be sufficient to induce a complete cycle of KSHV lytic reactivation. To assay the induction of KSHV gene expression by wild-type RTA and its mutants, we treated TRExBCBL1-cDNA5, TRExBCBL1-RTA, TRExBCBL1-RTA ΔAD, and TRExBCBL1-RTA(ΔDD-ILQ) cells with 1 μg of doxycycline per ml. As shown in Fig. 3, RTA(ΔDD-ILQ), which contains a deletion of amino acids 612 to 621, does not interact with TRAP/Mediator and SWI/SNF complexes and lacks transcriptional activation. Despite its complete loss of transcriptional activation activity, the RTA ΔAD mutant still contains intact DNA binding activity and thus functions as a dominant negative mutant that suppresses KSHV lytic reactivation (28).
For the RNase protection assay, we chose to analyze three genes (RTA, ORF57, and vIRF1) having RTA-dependent expression. Also, a probe for the gene encoding ribosomal protein L32, which serves as a housekeeping gene, was included as a control. Total RNA was harvested at 0, 6, and 12 h after doxycycline (1 μg/ml) treatment and analyzed by the RNase protection assay. Within a short period of doxycycline treatment, rapid, robust expression of the RTA, ORF57, and vIRF1 genes was detected in TRExBCBL1-RTA cells, and the expression of these genes was further increased at 12 h of treatment (Fig. 7A). By striking contrast, the expression of RTA, ORF57, and vIRF1 was undetectable in TRExBCBL1-cDNA5, TRExBCBL1-RTA ΔAD, and TRExBCBL1-RTA(ΔDD-ILQ) cells under the same conditions (Fig. 7A). Similar levels of L32 transcript were detected in all these cells (Fig. 7A).
FIG. 7.
Recruitment of TRAP/Mediator and SWI/SNF complexes is required for RTA-mediated KSHV lytic reactivation. (A) Activation of KSHV RTA, ORF57, and vIRF1 gene expression by RTA and its mutants. After doxycycline treatment for 0, 6, and 12 h, RNA was extracted from KSHV-infected TRExBCBL1-cDNA5, TRExBCBL1-RTA, TRExBCBL1-RTA ΔAD, and TRExBCBL1-RTA(ΔDD-ILQ) cells, and 5 μg of total RNA was subjected to RNase protection assay analysis with 32P-labeled riboprobe templates. The protected RNA fragments were separated by 5% PAGE and visualized with a PhosphorImager. (B) Level of KSHV lytic reactivation. Three days after stimulation with (red) or without (blue) doxycycline, cells [TRExBCBL1-cDNA5, TRExBCBL1-RTA, TRExBCBL1-RTA ΔAD, and TRExBCBL1-RTA(ΔDD-ILQ)] were fixed with paraformaldehyde and reacted with K8.1 rabbit serum, followed by fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin secondary antibody. Blue numbers and red numbers in the box indicate the percentages of K8.1-positive cells with and without stimulation, respectively. The data were reproduced in three independent experiments. (C) Equivalent amounts of RTA and its mutant proteins. Three days after stimulation with doxycycline, cell lysates were immunoblotted with an anti-Myc antibody to detect the Myc-tagged RTA protein.
To further address the role of the RTA interaction with the TRAP/Mediator and SWI/SNF complexes, we examined the level of KSHV lytic reactivation of TRExBCBL1-cDNA5, TRExBCBL1-RTA, TRExBCBL1-RTA ΔAD, and TRExBCBL1-RTA(ΔDD-ILQ) cells. The reactivation assay used a polyclonal antibody that recognizes the K8.1 envelope glycoprotein expressed with late kinetics. This protein is specific to lytically infected cells, since it is expressed only on cells that are committed to viral reactivation. Approximately 1.7% of TRExBCBL1-cDNA5 cells showed spontaneous surface expression of K8.1 before doxycycline stimulation and did not increase K8.1 surface expression after 3 days of doxycycline stimulation (Fig. 7B). In contrast, TRExBCBL1-RTA cells showed a dramatic increase in K8.1 surface expression: more than 74% of these cells underwent lytic reactivation after 3 days of stimulation (Fig. 7B). However, both TRExBCBL1-RTA(ΔDD-ILQ) and TRExBCBCL1-RTA ΔAD cells showed little or almost no increase in K8.1 surface expression after 3 days of doxycycline stimulation (Fig. 7B). In particular, TRExBCBCL1-RTA ΔAD cells showed extremely low K8.1 expression with or without stimulation, which was likely due to the dominant negative activity of the RTA ΔAD mutant (Fig. 7B). Wild-type RTA, RTA(ΔDD-ILQ), and RTA ΔAD were expressed at equivalent levels in KSHV-infected TRExBCBL1 cells (Fig. 7C). In addition, expression of RTA(DD-AA) and RTA(ILQ-AAA) also showed little or no increase in K8.1 surface expression under the same conditions (data not shown). Thus, these results show a strong correlation between the ability of RTA to activate viral lytic reactivation and the ability to recruit SWI/SNF and TRAP/Mediator complexes.
DISCUSSION
Chromatin remodeling acts as an essential and key element of transcriptional regulation by controlling the ability of many transcription factors to access the genome (23, 25, 32, 46). An ATP-dependent SWI/SNF complex and a histone acetyltransferase family protein appear to act synergistically to establish a local chromatin structure that is permissive for subsequent events. In addition, TRAP/SMCC/Mediator, which is a large complex composed of polypeptides ranging in size from 10 to 240 kDa, functions as an interface between sequence-specific transcription factors and the general transcriptional apparatus (30, 32).
Using affinity purification and mass spectrometry analysis, we found that KSHV RTA recruits numerous cellular transcription cofactors, including CBP, SWI/SNF complex, and TRAP/Mediator complex. While most but not all subunits of SWI/SNF and TRAP/Mediator complexes were isolated from our analysis, additional coimmunoprecipitation and immunoblot assays unambiguously demonstrated that RTA interacts with these complexes. Furthermore, our chromatin immunoprecipitation assay showed that RTA also recruits a p160 family member, NcoA-1/steroid receptor coactivator-1 (SRC-1), to its own promoter (unpublished results). Transcriptional activation by numerous cellular factors, specifically nuclear receptors, also requires multiple cellular cofactors that act in both a sequential and combinatorial manner to reorganize chromatin templates and to modify and recruit basal transcription factors and RNA polymerase II (20, 32, 33). These cofactors include the p160 family, SWI/SNF complex, CBP/p300/PCAF, TRIP/DRIP/ARC complex, and TRAP/SMCC/Mediator complex. This suggests that RTA resembles cellular nuclear receptors in the recruitment of cellular transcription cofactors and in the initiation of gene expression. Thus, our findings indicate that the molecular strategies that underlie RTA-mediated KSHV reactivation require the actions of a large number of transcriptional cofactors, which reorganize viral promoter chromatin structure and further recruit basal transcriptional factors to initiate well-controlled viral gene expression.
Previous reports have demonstrated that numerous cellular proteins, including Oct-1, RBP Jκ, and the novel cellular protein MGC2663, interact with RTA and that this interaction is mediated through the amino-terminal DNA binding domain and the central leucine zipper motif of RTA. Despite low sequence homology at the carboxyl transcriptional activation domain among RTA homologs of gamma-2 herpesviruses, this region has been shown to play a critical role in transcription activation of KSHV genes and thereby induction of viral reactivation. Furthermore, we have demonstrated that mutations of two glutamic acids at residues 612 and 613 and the ILQ sequence at residues 619 to 621 of RTA cripple the interaction of RTA with SWI/SNF and TRAP/Mediator complexes and that the loss of interaction with cellular transcription cofactors results in a drastic reduction in RTA-mediated transcriptional activity and thereby KSHV reactivation. This indicates that a specific interaction of RTA with the SWI/SNF and TRAP/Mediator complexes plays an important role in RTA-mediated transcription.
Of note, while the loss of interaction with these cellular cofactors results in a drastic reduction in RTA-mediated gene expression, a minor level, less than 3%, of RTA activity still remained in viral reactivation. This low level of RTA activity may be derived from other cellular factors, including Oct-1, Stat3, novel cellular protein MGC2663, RBP Jκ, or other proteins not yet characterized. Finally, we also found that, similar to KSHV RTA, the herpesvirus saimiri and gammaherpesvirus 68 RTA homologs interact with CBP, SWI/SNF, and TRAP/Mediator, suggesting that an interaction with these cellular cofactors is a general mechanism for gamma-2 herpesvirus reactivation (unpublished results). Further study of the synergistic contribution of these cellular cofactors to viral gene expression will provide the detailed molecular mechanism of gamma-2 herpesvirus replication and assist in the development of antiviral therapeutic agents.
Upon external signals, such as chemical treatment, RTA gene expression is initiated, and subsequently RTA recruits a number of transcription coactivator complexes (such as CBP, SWI/SNF complex, TRAP/mediator complex) and basic transcription machinery onto its own promoter and other RTA-dependent promoters. A recent report (39) showed that a complete preinitiation complex, including phosphorylated RNA polymerase II, is assembled at the human antitrypsin promoter long before the recruitment of transcription cofactors, suggesting that chromatin reconfiguration is a defining step of the initiation process, acting after the assembly of the RNA polymerase II machinery. In contrast, chromatin remodeling prior to preinitiation complex assembly is required at the beta interferon gene promoter (1). Further study is necessary to define the specific order of transcription factor assembly on the RTA promoter.
An important step in the herpesvirus life cycle is the switch from latency to lytic replication, and this switch should be well controlled at the molecular and cellular level. In this study, we have demonstrated that KSHV RTA recruits CBP and SWI/SNF chromatin remodeling complex and TRAP/Mediator transcriptional regulatory complex into viral promoters through interactions with a short acidic sequence in its carboxyl-terminal region. The recruitment strongly correlates with and presumably is essential for RTA-dependent viral gene expression and thereby viral lytic reactivation. Thus, these results not only have important implications for understanding the molecular basis of KSHV RTA action but also suggest a basis for the synergistic contributions of cellular transcription cofactors to gamma-2 herpesvirus reactivation.
Acknowledgments
We especially thank S. Gygi at the Harvard Mass Spectrometry facility for mass spectrometry analysis and Yun Kyoung Kang, Etsuko Uno, and Soichiro Yamamura for providing TRAP 230N, TRAP230,and TRAP95 DNAs. We also thank J. Macke for critical reading of the manuscript.
This work was partly supported by U.S. Public Health Service grants CA82057, CA91819, AI38131, and RR00168 and ACS grant RPG0010201. J. Jung is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 4.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, S. W., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 12.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 13.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 14.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 15.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 18.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobbs, M. V., W. O. Weigle, D. J. Noonan, B. E. Torbett, R. J. McEvilly, R. J. Koch, G. J. Cardenas, and D. N. Ernst. 1993. Patterns of cytokine gene expression by CD4+ T cells from young and old mice. J. Immunol. 150:3602-3614. [PubMed] [Google Scholar]
- 20.Ito, M., and R. G. Roeder. 2001. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12:127-134. [DOI] [PubMed] [Google Scholar]
- 21.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 22.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 24.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 25.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 26.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the Epstein-Barr virus R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 30.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 31.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 33.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 34.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 35.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 40.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 41.Strober, B. E., J. L. Dunaief, S. Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576-1583. [DOI] [PMC free article] [PubMed]
- 42.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin, H. W. T., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallberg, A. E., K. E. Neely, A. H. Hassan, J. A. Gustafsson, J. L. Workman, and A. P. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA(ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 47.Yoshinaga, S. K., C. L. Peterson, I. Herskowitz, and K. R. Yamamoto. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598-1604. [DOI] [PubMed] [Google Scholar]
- 48.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]