Abstract
Two highly conserved cationic amphipathic α-helical motifs, designated lentivirus lytic peptides 1 and 2 (LLP-1 and LLP-2), have been characterized in the carboxyl terminus of the transmembrane (TM) envelope glycoprotein (Env) of lentiviruses . Although various properties have been attributed to these domains, their structural and functional significance is not clearly understood. To determine the specific contributions of the Env LLP domains to Env expression, processing, and incorporation and to viral replication and syncytium induction, site-directed LLP mutants of a primary dualtropic infectious human immunodeficiency virus type 1 (HIV-1) isolate (ME46) were examined. Substitutions were made for highly conserved arginine residues in either the LLP-1 or LLP-2 domain (MX1 or MX2, respectively) or in both domains (MX4). The HIV-1 mutants with altered LLP domains demonstrated distinct phenotypes. The LLP-1 mutants (MX1 and MX4) were replication defective and showed an average of 85% decrease in infectivity, which was associated with an evident decrease in gp41 incorporation into virions without a significant decrease in Env expression or processing in transfected 293T cells. In contrast, MX2 virus was replication competent and incorporated a full complement of Env into its virions, indicating a differential role for the LLP-1 domain in Env incorporation. Interestingly, the replication-competent MX2 virus was impaired in its ability to induce syncytia in T-cell lines. This defect in cell-cell fusion did not correlate with apparent defects in the levels of cell surface Env expression, oligomerization, or conformation. The lack of syncytium formation, however, correlated with a decrease of about 90% in MX2 Env fusogenicity compared to that of wild-type Env in quantitative luciferase-based cell-cell fusion assays. The LLP-1 mutant MX1 and MX4 Envs also exhibited an average of 80% decrease in fusogenicity. Altogether, these results demonstrate for the first time that the highly conserved LLP domains perform critical but distinct functions in Env incorporation and fusogenicity.
The envelope glycoprotein (Env) of lentiviruses is an oligomer of heterodimers (43) that functions primarily to attach virions to target cell surfaces and mediate fusion of viral and cellular membranes in the entry process (14). Env is translated as a 160-kDa polyprotein precursor that is proteolytically cleaved by cellular proteases during its trafficking to the cell surface into a fusion-competent two-subunit form containing gp120 and gp41. The heavily glycosylated, extracellular surface (SU) gp120 subunit interacts noncovalently with the transmembrane (TM) gp41 subunit, and together they form an oligomer that binds the host cellular receptor CD4 and the coreceptor (chemokine receptors, e.g., CXCR4 and CCR5) for virion entry.
The TM protein of lentiviruses is a type I integral membrane protein that bears a fusion peptide at its amino-terminal end that is crucial in mediating virus-cell and cell-cell fusion processes (4, 7). A unique distinguishing feature of lentivirus TM proteins is the presence of a relatively long intracytoplasmic tail (ICT) (∼150 amino acids) in human immunodeficiency virus type 1 (HIV-1), HIV-2, simian immunodeficiency virus (SIV), equine infectious anemia virus, and visna virus. In contrast, oncornaviruses possess a short ICT containing only 20 to 30 amino acids (22). Mutational studies examining the functionality of the ICT by creating frameshifts, deletions, or truncations have shown that ICT sequences can affect basic aspects of viral replication, infectivity, and cytopathicity (15, 21, 29, 51). Truncations of the HIV-1 and SIV ICTs have been found to modulate cell-cell fusion properties of Env, presumably due to alterations in the levels of cell surface Env expression and the conformation of the Env ectodomain (16, 51). Functions of Env internalization (2, 3) via interaction with AP-1 (47) and basolateral release of virus from epithelial cells (30) have also been attributed to the ICT. In the case of SIV, the ICT has been identified as a locus for attenuation (20, 41) of the virus in vivo, in contrast to earlier reports describing the dispensability of the ICT for viral replication in vitro (24).
Despite these numerous functions assigned to the ICT, the precise ICT motifs involved in each of the processes are not clearly defined. To date, only two classes of motifs have been identified in the ICT of lentiviruses. First, endocytic sequence motifs, YXXL and di-leucine sequences (3, 47) present in the ICT of HIV-1 and SIV, are involved in regulating the level of cell surface Env expression and viral pathogenesis (20). Second, computer modeling studies indicate the presence of distinguishing structural motifs in the ICT (17, 34). These include two 20- to 30-residue long cationic amphipathic α-helical domains in the carboxyl terminus of HIV-1 HXB2 gp41 (residues 828 to 855 and 768 to 788; see Fig. 1), designated as lentivirus lytic peptides 1 and 2 (LLP-1 and LLP-2) (34). Consistent with computer modeling predictions, peptide homologs of LLP motifs from SIV have been shown by circular dichroism spectroscopy to form helical structures in solution, with an increase in helicity in the presence of lipids or calmodulin (CaM) (49). Despite the fact that the HIV-1 envelope has a high mutation rate, LLP domain structure is highly conserved among different sequence variants of HIV-1 (45), suggesting that the domains are responsible for functions that are important for the virus.
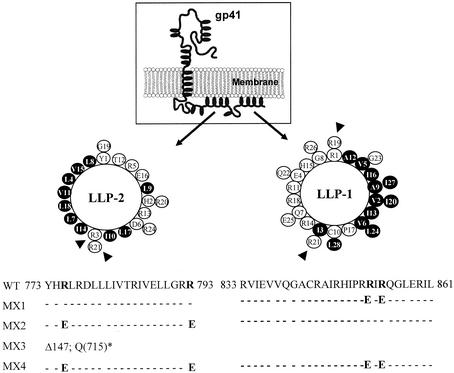
FIG. 1.
Diagram of gp41 with location and amino acid sequences of wild-type (WT) and mutant LLP domains. Mutations in the highly conserved LLP domains were engineered in the proviral clone of ME46. Helical wheel representations of LLP-1 and LLP-2 domains are depicted here with highly conserved arginine residues (shown by arrowheads) that were replaced with glutamate. Hydrophobic amino acids are shown as shaded circles; white circles represent hydrophilic amino acids. Conserved arginine residues in the wild-type sequence that were replaced with glutamate are shown in bold face. In the MX3 mutant, Δ147 refers to a deletion of 147 amino acids from the carboxyl-terminal end of gp41 by replacing the glutamine residue at position 715 [Q(715)] with a stop codon (*).
Based on structural similarities between LLP domains and cytolytic peptides (34), peptide analogs of LLP domains have been shown to have lytic and membrane perturbing effects on prokaryotic and eukaryotic cells (10-12, 33) and are believed to cause fusion of liposomal membranes (11). LLP peptide analogs also demonstrate CaM binding properties leading to a disruption of important CaM-mediated processes (1, 35, 44). These properties are also evident in the context of full-length TM protein (23, 32, 44, 45). Furthermore, the interaction of LLP-2 with α-catenin (25), p115-RhoGEF (50), and prenylated Rab acceptor protein (18) has also been demonstrated. In terms of function, deletion and truncation studies have implicated the LLP domains in a variety of functions, including Fas-mediated apoptosis (32), Env oligomerization (28), stability (27), cell surface expression (6), and incorporation of Env into virus particles (13, 36, 38).
Previous studies involving ICT truncations of various lengths have reported a loss of viral infectivity (15, 21), which was associated in part with defects in the incorporation of Env into virus particles in a cell type-dependent manner (13, 36-38). While several studies have shown that HIV-1 Env is incorporated into the virions via interactions with the Gag matrix (MA) protein, controversy still persists regarding the exact region of ICT that is involved in Env incorporation. Different studies have reported the exclusive involvement of either the LLP-1 domain or the LLP-2 domain in the incorporation of Env into virions (13, 36, 38) via interaction with the Gag MA protein. These functional studies along with the studies that examined the role of the ICT in cell-cell fusion primarily involved ICT deletion analyses. The results from such deletion and truncation analyses may be confounded by Env conformational changes, as observed in the case of both HIV-1 (16) and SIV (51). Thus, simultaneous analysis of LLP-1 and LLP-2 domain functions by using selected point mutations may provide a higher-resolution analysis of the significance of individual LLP domains.
To examine the individual contributions of the highly conserved LLP-1 and LLP-2 domains to Env incorporation and Env-mediated cell-cell fusion, we mutagenized two highly conserved arginine residues within the LLP domains in the context of the proviral genome of a dualtropic primary HIV-1 isolate, ME46 (9). Mutations were engineered to specifically disrupt the functional properties of LLP domains by reducing the net positive charge of the amphipathic helices while maintaining their hydrophobic moment and structural integrity. The biological and biochemical properties of HIV-1 mutants bearing altered LLP domains were then characterized. The results of these studies reveal new fundamental information about the distinct roles of LLP domains in Env incorporation and fusogenicity, indicating diverse functions of the HIV-1 ICT.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
293T cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in Dulbecco's modified Eagle medium (GIBCO, Grand Island, N.Y.) containing 10% fetal bovine serum (FBS), l-glutamine (2 mM), penicillin G (100 U/ml), and streptomycin sulfate (0.1 mg/ml). QT6 cells (a kind gift of R. W. Doms, University of Pennsylvania) were maintained in the same way as the 293T cells. MAGI-R5 cells (HeLa-CD4-long terminal repeat-β-galactosidase, CCR5 and CXCR4 coreceptors; obtained from the National Institutes of Health [NIH] AIDS Research and Reference Reagent Program) were maintained in the same medium as 293T cells in the presence of G418 (0.2 mg/ml), hygromycin B (0.1 mg/ml), and puromycin (1 μg/ml). Human peripheral blood mononuclear cells (PBMC) isolated from fresh heparinized blood by Histopaque-1077 (Sigma Diagnostics, St. Louis, Mo.) density gradient centrifugation were resuspended (106 cells/ml) in RPMI 1640 (GIBCO) supplemented with 20% FBS and stimulated with phytohemagglutinin (PHA) (1 μg/ml) for 3 days. Activated PBMC were subsequently maintained in the presence of human interleukin-2 (10 U/ml) (Boehringer Mannheim, Mannheim, Germany). The lymphoid cell lines H9, CEMx174 (obtained through the NIH AIDS Research and Reference Reagent Program), and MT2 (a kind gift of J. Mellors, University of Pittsburgh) were cultured in RPMI 1640 containing 10% FBS and antibiotics. The proviral clone of ME46 was obtained from a patient with late-stage AIDS and maintained as described previously (9).
Cloning and mutagenesis.
Site-directed mutations were generated in the LLP-1 and LLP-2 domains (see Fig. 1) by using the 3′ half of ME46 in the PCR-ligation-PCR method. Briefly, for each pair of mutations to be generated (affecting arginine residues 851 and 853 in LLP-1 and arginine residues 775 and 793 in LLP-2), two pairs of primers (mutagenic reverse primer and MfeI forward primer and mutagenic forward primer and XhoI reverse primer) were used. The mutagenic reverse and forward primers were designed to encode single arginine-to-glutamate changes at their 5′ ends. The two mutant fragments generated by PCR were blunt-end ligated. The ligated product was then amplified in a second round of PCR by using the MfeI forward primer and the XhoI reverse primer to generate a 1.2-kb fragment from the env coding region containing the desired mutations in LLP-1 or LLP-2. This mutated fragment was then cloned into the 3′ half of the ME46 proviral clone by using the unique restriction enzyme sites MfeI and XhoI to generate LLP-1 (MX1) or LLP-2 (MX2) mutant viruses. MX4 mutant virus bearing alterations in both LLP-1 and LLP-2 domains was generated by PCR amplifying a 1-kb fragment (the MfeI-AlwNI fragment) containing LLP-2 from an MX2 mutant clone and a 200-bp fragment (the AlwNI-XhoI fragment) containing LLP-1 from an MX1 mutant clone. The two fragments were ligated into wild-type proviral DNA digested with MfeI and XhoI. The MX3 mutant, containing a deletion of the carboxyl-terminal 147 amino acid residues of the TM protein, was constructed by the PCR-ligation-PCR method such that three stop codons were inserted sequentially in all three reading frames of the coding sequence for Env, starting at the position corresponding to the glutamine residue at position 715. A single nucleotide was deleted to disrupt the reading frame of env as well. This mutagenesis did not result in any changes in the second exons of tat or rev. All mutations were verified by sequencing the entire Env fragment that was amplified by PCR. A full-length proviral clone of wild-type virus was obtained by EcoRI digestion and subsequent ligation of the two proviral halves. The individual mutated Env fragments were then inserted into the wild-type proviral backbone with the unique restriction enzymes MfeI and XhoI encompassing the mutagenized Env coding sequences. For cloning of wild-type and ICT mutant Env sequences in pCDNA3 vector (Invitrogen, Carlsbad, Calif.) for the cell-cell fusion assay, Env sequences were PCR amplified and the EcoRI and BamHI restriction sites inserted during PCR were used for directional cloning of Env.
Transfections and infections.
Virus stocks of ME46 and ICT mutant derivatives were obtained by transfecting the wild-type and mutant full-length proviral genomes into 293T cells (50 to 80% confluence) by using the calcium phosphate transfection method (CalPhos Mammalian Transfection Kit; Clontech, Palo Alto, Calif.). For measurement of viral replication kinetics, the transfected 293T cells were incubated at 37°C for 48 h and supernatants were collected and filtered through a 0.45-μm-pore-size filter. Samples of filtered cell-free tissue culture supernatant containing similar amounts of p24 were then used to infect 5 × 106 PHA-stimulated PBMC. At an interval of 5 to 6 days, half of the culture medium was replaced with fresh medium containing 2.5 × 106 PHA-stimulated PBMC. Virus production was monitored by measuring the p24 antigen in the culture supernatant with an antigen capture enzyme-linked immunosorbent assay (DuPont, Wilmington, Del.) as per the directions of the manufacturer. CD4+ T-cell lines were transfected by electroporation as described previously (8). Infections of CD4+ T-cell lines were done by using identical 50% tissue culture infective doses (TCID50) of ME46 and mutant derivatives. Briefly, 107 H9 cells were infected at 37°C for 1 h with 1 ml of cell-free virus stock. The cells were then washed and cultured as above. The cells were split 1:4 at an interval of 3 to 5 days, when culture supernatant samples were also taken to assay p24 production.
Single-cycle infectivity assay and viral TCID50 determination.
Supernatants from transiently transfected 293T cells (48 h posttransfection) were filtered, normalized for p24 levels as above, and used to infect MAGI-R5 cells in duplicate (0.8 × 105 cells/well) in a 24-well plate for 12 h at 37°C. The cells were then washed and cultured for 48 h, fixed with glutaraldehyde (0.5%), and stained for β-galactosidase. The number of blue foci was determined for the whole well. For TCID50 determination, a similar protocol was followed. In this case, the infections for each construct were done in triplicate in a 96-well plate with fivefold serial dilutions of the virus. The Reed and Muench method was used for calculating the TCID50 of virus stocks.
Radioimmunoprecipitation.
Methods used for metabolic radiolabeling of transfected 293T cells, preparation of cell lysates, pelleting of labeled virions, and immunoprecipitation of cell- and virion-associated proteins with sera from HIV-1-infected patients (HIV-1 neutralizing sera; obtained from NIH AIDS Research and Reference Reagent Program) were performed as described previously (37). Briefly, log-phase 293T cells (106) were seeded in T25 flasks (Falcon, Franklin Lakes, N.J.). Cells were transfected on the following day with the proviral constructs as described above and incubated for 48 h. The cells were then metabolically labeled overnight at 37°C with 0.5 mCi of [35S]methionine-cysteine protein labeling mix (NEN, Boston, Mass.). Thereafter, cells were washed with phosphate-buffered saline (PBS) and lysed and cell-associated viral proteins were immunoprecipitated by using sera from HIV-1-infected patients. Labeled virus was pelleted from cell-free tissue culture supernatant by centrifugation through a 20% (vol/vol) glycerol cushion at 39,000 rpm in an SW55 Ti rotor (Beckman, Fullerton, Calif.), and immunoprecipitation was performed by using sera from HIV-1-infected patients. Following immunoprecipitation, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a 4 to 15% gradient gel (GIBCO). Dried gels were exposed to film for 1 or more days prior to developing and quantified by a calibrated imaging densitometer (model GS710; Bio-Rad, Hercules, Calif.).
Protein analysis.
Clarified transfection supernatants were centrifuged to pellet viral particles as described above. The viral pellets were then resuspended in PBS for protein analysis by SDS-PAGE and Western blotting. SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.) was used to visualize the bands after blotting with the appropriate monoclonal antibodies (MAbs), mouse anti-HIV-1 p24 (AG3.0), mouse anti-gp41 (T32) (a kind gift of P. Earl, NIH), mouse anti-reverse transcriptase (RT) (8C4), or polyclonal goat anti-gp120 (catalog no. 385). The antibodies to p24, RT, and gp120 were obtained from the NIH AIDS Research and Reference Reagent Program. Individual protein bands were quantitated by using a calibrated imaging densitometer (model GS710; Bio-Rad). Anti-mouse and anti-goat immunoglobulin G (IgG) peroxidase conjugate were purchased from Sigma.
Assays for determination of syncytium induction and cytopathicity.
About 5 × 106 H9 cells in log phase were infected with similar TCID50 of wild-type and MX2 viruses as described above. Following infection, about 50,000 cells (200 μl) were plated in duplicate into a 96-well flat-bottom tissue culture plate (Costar, Cambridge, Mass.) and incubated at 37°C in a 5% CO2 incubator. The remaining cells were cultured in T25 flasks (Costar) and split 1:4 every 3 to 5 days, when cell-free tissue culture supernatant (1 ml) was also sampled for assaying p24 production. The wells were examined for syncytium formation at 3- to 5-day intervals postinfection, and the number and size of syncytia per well were determined for 35 days postinfection. About half of the culture medium was replaced with fresh medium every 3 to 5 days, when cell viability was also determined by using a 50-μl sample of cell suspension. The cells were stained with 0.4% trypan blue (Sigma) for viability assays. Similar syncytium formation assays were also performed with CEMx174 and MT2 cells. For these assays, 30,000 to 50,000 CEMx174 and MT2 cells (150 μl) in log phase were plated in duplicate in a 96-well flat-bottom plate and infected with equal TCID50 (50 μl) of wild-type and LLP-2 mutant (MX2) viruses.
Flow cytometry analysis of HIV-1 surface Env and intracellular p24 expression.
Parallel samples of H9 cells were infected with wild-type virus or with MX2 virus as described above. At 15 days postinfection, the cells were stained for cell surface gp41 with 126-6, Md-1 (obtained from the NIH AIDS Research and Reference Reagent Program), or T32 (mouse anti-gp41) as described previously (37). Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG and FITC-conjugated mouse anti-human IgG were used as secondary antibodies (ICN, Costa Mesa, Calif.). Following incubation for 30 min, cells were washed three times with PBS containing 5% FBS (wash buffer) and fixed in 1% paraformaldehyde for 1 h. Cells were then permeabilized with PBS containing 5% fetal calf serum and 0.5% saponin (permeabilization buffer) for staining intracellular p24 with KC57-RD1 (Coulter Corporation, Miami, Fla.). Following a 30-min incubation, cells were washed three times with permeabilization buffer. Finally, cells were washed twice with wash buffer, resuspended in 1% paraformaldehyde, and analyzed for cell surface gp41 and intracellular p24 expression. A minimum of 50,000 gated-live events were acquired on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) and analyzed with FlowJo batch analysis software (Treestar, San Carlos, Calif.). Env expression was assessed in the p24-positive population of cells, and mean fluorescence intensities of phycoerythrin (PE) and FITC stainings were determined. Ratios of mean fluorescence intensities of p24-PE and Env-FITC were calculated and plotted graphically.
Luciferase-based cell-cell fusion assay.
The cell-cell fusion assay was done as described in detail previously (16). Briefly, effector 293T cells were infected with recombinant vaccinia virus vTF1.1 expressing T7 polymerase (a kind gift of B. Moss, NIH) and transfected via calcium phosphate with Env constructs under the control of the T7 promoter. The levels of Env expression were determined by Western blot analysis with anti-gp41 MAb, and mouse anti-human actin was used to normalize cell lysate (Sigma). Target quail QT6 cells were transfected with CXCR4 and CD4 expression plasmids under the control of the cytomegalovirus promoter and the luciferase gene under the control of the T7 promoter (kind gifts of R. W. Doms, University of Pennsylvania). On the next day, effector cells were mixed with target cells and allowed to fuse for at least 7 h. Fusion was measured by quantification of luciferase in cell lysates as previously described (16). Luciferase activity was monitored with a luminometer (Dynex Technologies, Chantilly, Va.), and the results were normalized to levels of Env expression and total protein amount per well.
RESULTS
Design and construction of ICT mutants.
Mutations were designed in the LLP-1 and LLP-2 domains singly or in combination in the context of a full-length proviral genome to evaluate the contribution of LLP domains to the replication and cytopathic properties of HIV-1. The mutations introduced into the highly conserved LLP-1 and LLP-2 domains of a primary HIV-1 isolate, ME46, are depicted in Fig. 1. The rationale of the site-directed mutagenesis was to abolish specific functions attributed to LLP-1 and LLP-2 domains with minimum structural alterations in the ICT. Thus, based on Clustal analysis, structural predictions, and an in vitro peptide study (46), highly conserved (>90% conservation) positively charged arginine residues (amino acids 851 and 853 in LLP-1 and 775 and 793 in LLP-2) presented on the hydrophilic face of the amphipathic helix (Fig. 1) that are critical for the membrane perturbing and CaM binding functions of the domains were replaced with negatively charged glutamate residues. This mutagenesis resulted in a reduction of the net positive charge of the helix, with no dramatic difference in the calculated hydrophobic moment, as the hydrophobic face of the helix was unaltered. As shown in Fig. 1, four mutants were constructed containing two substitutions in either the LLP-1 (MX1) or the LLP-2 (MX2) domain or in both domains (MX4). In addition to these LLP mutants with site-specific mutations, the MX3 construct was engineered to contain a truncation of the entire ICT, including both LLP domains.
Point mutations in the LLP-1 and LLP-2 domains differentially affect the incorporation of gp120 into virions without evident effects on Env expression or processing.
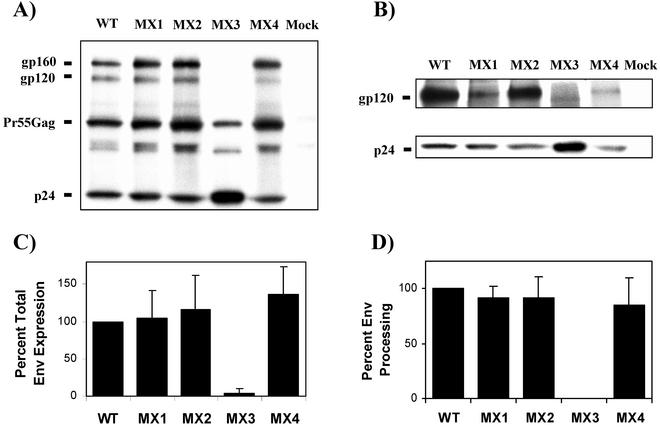
To assess the individual contributions of LLP-1 and LLP-2 domains to the incorporation of Env into virions, the proviral clones bearing LLP mutations were transfected into 293T cells and evaluated for their effects on Env expression, processing, and incorporation into virions by metabolically labeling viral proteins with [35S]methionine-cysteine. Radiolabeled viral proteins were immunoprecipitated from cell lysates (Fig. 2A) and from extracellular viral pellets (Fig. 2B) and resolved by SDS-PAGE (4 to 15% gel). The levels of expression of gp120 and gp160 for all constructs were compared with those of wild-type virus after normalization for p24 expression. Different mutations in the ICT resulted in distinct effects on Env synthesis. As shown in Fig. 2A and C, no dramatic defects in steady-state levels of total Env expression (gp160+gp120/p24) were observed in cell lysates of wild-type virus and MX1, MX2, and MX4 mutant constructs, while the MX3 mutant was evidently deficient in Env expression. Furthermore, the levels of processing of gp160 (Fig. 2D), as determined by the ratio of cell-associated gp120 and total Env (gp120/gp120+gp160), were similar for all Env-expressing mutants, with a slight processing defect in the case of MX4. Figure 2B shows the incorporation of gp120 into virions following the radiolabeling of viral proteins. Interestingly, we observed that the LLP-2 mutant virus MX2 did not exhibit any defects in the incorporation of Env into viral particles while mutations in the LLP-1 domain (MX1) specifically and dramatically inhibited the incorporation of Env into mutant virions. This Env incorporation defect associated with MX1 was dominant and manifested itself even in the presence of a wild-type LLP-2 sequence, suggesting that the Env incorporation function is unique to the LLP-1 domain. Similar to MX1, the MX4 mutant, which contained substitutions in both LLP domains, also showed a decrease in Env incorporation. As expected, no envelope was detectable in the MX3 virus, correlating with the lack of intracellular envelope glycoprotein.
FIG. 2.
Effects of mutations in the LLP domains on Env expression and processing and gp120 incorporation into virions. (A and B) Two days after the transfection of 293T cells with wild-type (WT) and mutant proviral clones, proteins were labeled overnight with [35S]methionine-cysteine, and viral proteins were immunoprecipitated thereafter. Immunoprecipitated viral proteins were resolved on SDS-polyacrylamide gel, and envelope gp160, gp120, and viral p24 from cell lysates (A) and from viral pellets (B) from the same experiment are shown. (C and D) Total Env expression (gp120+gp160/p24) (C) and Env processing (gp120/gp120+gp160) (D) were calculated after quantifying the individual proteins by densitometry. Results were normalized to wild-type levels in each experiment. Mock-transfected cells and supernatant from mock-transfected cultures were included as negative controls. Data are the averages of results from three independent experiments. Error bars show standard deviations.
Thus, these data demonstrate for the first time differential effects of LLP-1 and LLP-2 domains on virion gp120 incorporation. While changes in the LLP-2 domain did not apparently affect Env expression and virion incorporation of gp120, mutations in the LLP-1 domain resulted in an evident defect in Env incorporation despite wild-type levels of Env expression and processing.
The low level of gp120 incorporation into LLP-1 mutants correlates with decreased incorporation of gp41 into virions.
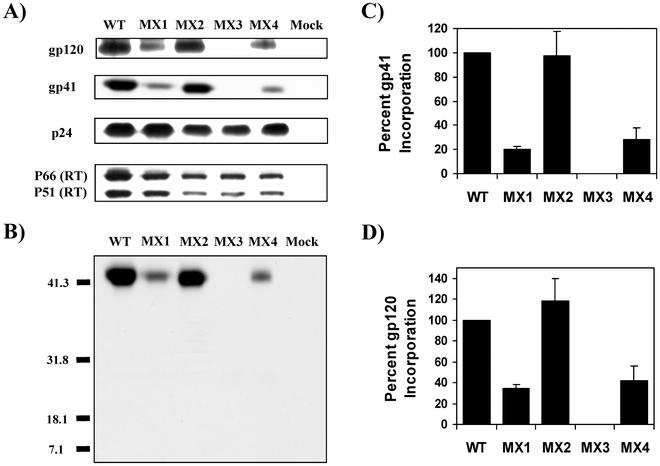
The data presented in Fig. 2 do not exclude the possibility that the apparent decrease in Env incorporation imposed by the mutant LLP-1 may be caused by increased shedding of gp120 from virions after incorporation, possibly due to conformational changes induced in the gp41 ectodomain. To address this possibility, we analyzed the level of gp41 incorporation into LLP mutant virions quantitatively by Western blot analysis with gp41-specific MAb. In parallel, viral proteins gp120, p24, and RT were also assessed in the virus pellet by using specific antibodies (Fig. 3A). For this analysis, the wild-type and mutant proviral clones were transfected into 293T cells, supernatants were collected 3 days posttransfection, and virus was pelleted from the clarified supernatant. Subsequent immunoblot analyses and comparisons of ratios of gp41 and p24 revealed that gp41 incorporation into virions was decreased by an average of 75% in MX1 and MX4 mutants compared with that in wild-type virus (Fig. 3A, B, and C), indicating a true Env incorporation defect and correlating with the reduced levels of gp120 incorporation seen in Fig. 2B. The MX2 mutant exhibited wild-type levels of gp41 incorporation (Fig. 3A, B, and C). In contrast, the MX3 mutant did not show any detectable levels of 24-kDa truncated gp41 in the virions (Fig. 3B). These observations were expected since the lack of gp160 expression seen for MX3 in Fig. 2A logically purports a lack of gp41 expression and incorporation. The evidence for the above-mentioned gp41 incorporation properties of ICT mutant Envs was further supported by gp120-specific immunoblots, wherein similar defects in gp120 incorporation were observed for MX1, MX3, and MX4 mutants while MX2 showed wild-type levels of gp120 incorporation (Fig. 3A and D). Additionally, no evident differences in the levels of RT incorporation were observed in the mutants (Fig. 3A). The above-mentioned data clearly demonstrate a defect in Env incorporation, and not gp120 shedding, that is imposed by mutations in the LLP-1 domain. In contrast, mutations in the LLP-2 domain did not exert any obvious effects on Env incorporation. The fact that the MX4 mutant, in which both LLP domains were mutated, showed levels of Env incorporation similar to those in MX1 suggests that the Env incorporation function is specifically associated with the LLP-1 domain.
FIG. 3.
Western blot analysis of gp41 and gp120 incorporation into virions. Virion lysates were prepared on day 3 from culture supernatants of 293T cells transfected with wild-type (WT) and mutant proviral clones after ultracentrifugation. (A) Samples were transferred to polyvinylidine difluoride membranes and blotted with anti-gp41 MAb (T32), polyclonal goat anti-gp120, anti-p24 MAb (AG3.0), and anti-RT MAb (8C4) (see Materials and Methods). (B) A longer exposure of a Western blot probed with 20 μg of T32/ml was used to confirm the lack of TM expression in the MX3 mutant. Molecular weights are indicated in thousands. (C and D) gp41, gp120, and p24 protein bands were also quantitated by densitometry, and Env incorporation was determined by calculating the ratios of gp41 to p24 (C) and gp120 to p24 (D) and normalizing to wild-type levels. The results shown are representative of results from duplicate experiments. Error bars indicate standard deviations.
Mutations in the LLP-1 domain but not the LLP-2 domain of gp41 affect viral infectivity in a single-cycle assay in MAGI-R5 cells.
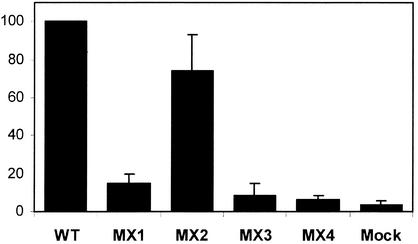
To determine the infectivity of the LLP mutants in a single-round assay, we compared the various ICT mutants in a single-cycle infectivity assay using MAGI-R5 cells. MAGI-R5 cells were infected with tissue culture supernatants from 293T cells transfected with wild-type virus and the respective mutant proviral constructs. Similar amounts of p24 were used to infect MAGI-R5 cells, and the total number of infected cells was determined by staining for β-galactosidase. As shown in Fig. 4, the different ICT mutants segregated into two distinct groups. The LLP-2 mutant virus MX2 obtained from 293T cells was classified as infectious as it exhibited infectivity levels similar to that of the wild-type virus, in agreement with wild-type levels of Env incorporation. In contrast, the MX1, MX3, and MX4 mutants separated into a phenotypic class of decreased infectivity. This assay demonstrated that the infectivity of MX1 was reduced by approximately 85% compared with that of the wild-type virus. Similarly, the MX3 and MX4 mutants also demonstrated dramatically reduced infectivities, with about 90% fewer blue foci than the wild-type virus. The decreased infectivities of MX1, MX3, and MX4 mutants observed in a single-round assay correlated with the mutants' reduced levels of Env incorporation into virus particles. Thus, these data suggest that alterations in the LLP-1 domain, but not the LLP-2 domain, can markedly reduce HIV-1 infectivity presumably by modulating the incorporation of Env into virions.
FIG. 4.
Effects of mutations in the LLP domains on viral infectivity in a single-round assay in MAGI-R5 cells. Supernatants from transfected 293T cells were normalized for p24 levels and used to infect MAGI-R5 cells in duplicate. Cells were stained for β-galactosidase 48 h after infection. Blue foci were counted as the total number per well and normalized to the number of blue foci counted for wild-type virus (WT). Data represent results from at least two independent transfections, and each mutant was analyzed in duplicate. Error bars indicate standard deviations.
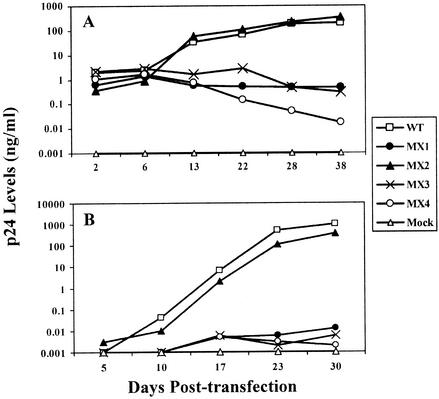
Mutations in the LLP-1 domain but not the LLP-2 domain affect the replication of the virus in primary human PBMC and lymphoid H9 cell samples.
To determine the ability of LLP mutants to establish a productive infection, the replicative capacity of the mutants was assessed with primary human PBMC samples as biologically relevant target cells and with a representative lymphoid CD4+ T-cell line, H9. The ability of the mutants to establish a productive infection and replicate in these cells was assessed by the amount of p24 released into the culture supernatant over a period of 20 to 40 days (Fig. 5). In general, the results of this multiple-round infectivity assay with PBMC and H9 cells were similar to those observed for the single-cycle infectivity assay. The MX2 virus, which incorporated wild-type levels of Env into virions, was replication competent and showed replication kinetics similar to those of the wild-type virus. In contrast, the MX1, MX3, and MX4 mutants were replication defective, in agreement with the observed defects in Env expression or incorporation. The MX1 and MX4 mutants, which exhibited defective Env incorporation, were compromised in their replicative ability in both PBMC and H9 cell samples; minimal replication competence was observed, with about 103- to 104-fold less p24 being released into the culture supernatant than with wild-type virus. Similarly, the MX3 mutant that did not show any detectable Env expression and incorporation was replication deficient. This experiment was performed independently with PBMC from three different donors, and similar replication phenotypes were observed in each experiment (data not shown). In conformity with the Env expression and incorporation data, these results demonstrate differential roles of LLP-1 and LLP-2 domains in viral replication and indicate a critical role for the LLP-1 domain in Env incorporation.
FIG. 5.
Effects of mutations in the LLP domains on viral replication. Mutations in the highly conserved LLP domains were engineered in the proviral clone, and the wild-type (WT) and different mutant clones were transfected into 293T cells to produce viral particles. (A) Two days posttransfection, supernatants from the transfections were filtered through a 0.45-μm-pore-size filter, normalized for p24 levels, and used to infect human PBMC stimulated with PHA. Fresh PHA-PBMC samples were added every 5 days by replacing half of the culture supernatants. Viral replication kinetics were followed by measuring p24 levels in culture supernatants. (B) Additionally, the replication properties of the mutant proviral constructs were compared with those of wild-type virus in H9 lymphoid cells. The cells were split 1:4 every 5 days following transfection, when the culture supernatant p24 levels were also determined.
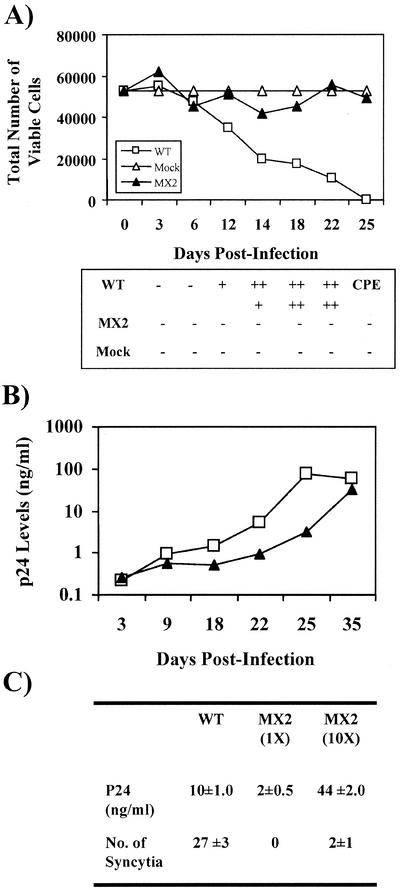
Alterations in the LLP-2 domain modulate the syncytium-inducing ability of Env.
The replication competence of LLP-2 mutant virus provided a unique system to assess the role of LLP-2 in modulating virus-induced cytopathicity. Thus, syncytium formation-dependent cell death induced by MX2 was evaluated in various CD4+ T-cell lines. Wild-type and MX2 virus stocks of equal infectivities were used to infect H9 cells, and cytopathicity was determined by the trypan blue exclusion method (Fig. 6A). Syncytium formation was also scored at different time points postinfection. Interestingly, LLP-2 mutant virus was evidently noncytopathic to H9 cells, as shown in Fig. 6A. At 25 days postinfection, the wild-type virus completely destroyed the cells in culture while cell numbers for MX2 virus-infected cultures remained similar to those for mock-infected cultures. Furthermore, the loss of cytopathicity correlated with a loss of the syncytium-inducing ability of MX2 (Fig. 6A), suggesting that mutations in the LLP-2 domain modulate the syncytium-induced cytopathicity caused by HIV-1. Notably, the decrease in syncytium induction was not a cell type-specific effect, as similar observations of minimal syncytium induction by MX2 were made in MT2 and CEMx174 cells as well (data not shown).
FIG. 6.
Cytopathicity and syncytium induction properties of LLP-2 mutant virus. H9 cells (5 × 106) were infected with the same TCID50 of wild-type (WT) and MX2 virus. About 50,000 infected cells were plated in duplicate in a 96-well flat-bottom tissue culture plate and incubated at 37°C in a 5% CO2 incubator. The remaining infected cells were maintained in T25 flasks and split 1:4 every 3 to 5 days. (A) Following incubation, the wells were scored for syncytium formation at 3- to 5-day intervals postinfection and cell death was determined by the trypan blue exclusion method. The total numbers of live cells were determined and normalized for cell growth by using mock-infected cultures. Induction of up to 5 syncytia per well was graded as +; ++ represents 6 to 10 syncytia, +++ represents 11 to 20 syncytia, and ++++ represents >20 syncytia. Excessive (>90%) cell death in virus-infected cultures is indicated by CPE (cytopathic effect). Data presented here are representative of results from at least three independent infections. (B) A 1-ml sample of supernatant from the infected cells described above was taken from each T25 flask every 3 to 5 days for measurement of tissue culture supernatant p24 levels. (C) H9 cells were also infected with MX2 virus stocks containing infectious doses of the virus that were equal to (1×) or about 10-fold higher than (10×) that of wild-type virus. Syncytia were scored 7 days postinfection, when p24 levels in the culture supernatant were also measured. Results are presented as the averages ± the standard deviations and represent results from three to four independent experiments.
In relation to the syncytium and cytopathicity assay described above, the replicative ability of MX2 was also determined in parallel with cytopathicity and syncytium formation in H9 cells. The culture supernatant p24 levels (Fig. 6B) were determined at 3- to 6-day intervals postinfection. As summarized in Fig. 6B, no major differences in virus production were observed to explain the observed differences in syncytium induction. A minor reduction in the replication of MX2 was observed in H9 cells; however, this decrease was not sufficient to explain the loss of cytopathicity observed for MX2. This loss was evident from a comparison of p24 level in culture supernatant and level of syncytium formation for MX2- and wild-type virus-infected cultures. At 25 days postinfection, MX2-infected T cells demonstrated cell culture supernatant p24 levels that were five to sixfold greater than those of wild-type virus-infected cultures at 18 days postinfection. Despite these higher p24 levels, MX2-infected cultures were defective in syncytium induction, while wild-type virus-infected cultures showed extensive syncytium formation and about 60% cytopathicity. To account for the lower levels of MX2 virus production, H9 cells were infected with MX2 virus stocks containing an infectious dose of the virus that was about 10-fold higher than that of wild-type virus. Syncytia were scored 7 days postinfection, when p24 levels in the culture supernatant were also measured (Fig. 6C). As expected, the wild-type virus-infected cells at 7 days postinfection displayed substantial syncytium formation and about fivefold higher levels of viral infection than cells infected with a similar level of MX2 virus (1×), where no syncytia were observed. Increasing the MX2 virus inoculum 10-fold (10×) yielded supernatant p24 levels that were about fourfold higher than those of the reference wild-type viral infection. Importantly, the MX2 virus failed to induce syncytia despite these higher levels of virus infection. Thus, these results clearly demonstrate for the first time a direct influence of the LLP-2 domain present in the ICT of gp41 in altering the Env-mediated cell-cell fusion process. This defect in cell-cell fusion, however, is in marked contrast to the apparently normal levels of virus-cell fusion by the MX2 Env, as suggested by the normal replication kinetics of MX2 virus and the MAGI assay.
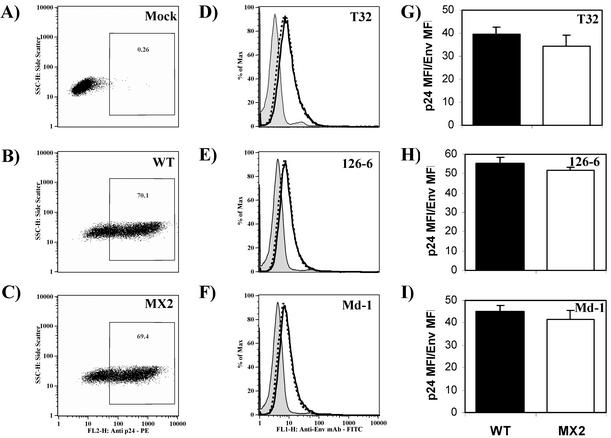
Mutations in LLP-2 causing defects in syncytium induction do not affect the levels of cell surface expression or the conformation of gp41.
To address the question of whether LLP-2 mutations in the ICT affect syncytium formation by decreasing levels of expression of Env on the surface of infected cells, cell surface gp41 expression was assessed by using a panel of MAbs specific to linear and conformational epitopes of HIV-1 Env. H9 cells were infected with wild-type and MX2 viruses, and infected cells were evaluated for the level of cell surface Env expression at 15 days postinfection. Cells were first stained with antibodies specific for gp41 and then fixed, permeabilized, and stained for intracellular p24 to determine the level of infection in cultures. Mock-infected and wild-type virus-infected cells were used to define a gate for the p24-positive population of cells. As determined by the percentage of cells staining positive for intracellular p24, similar levels of infection were achieved for wild-type and MX2 viruses (70% and 69%, respectively [Fig. 7B and C ], compared with 0.3% background staining for p24 in mock-infected cells [Fig. 7A]).
FIG. 7.
Envelope glycoprotein expression on the surface of CD4+ T cells infected with wild-type (WT) or MX2 virus. H9 cells were infected with MX2 virus at infectious doses that were fivefold higher than those of wild-type virus. At 15 days postinfection, cells were simultaneously stained for cell surface envelope glycoprotein and intracellular p24 levels (by using KC57-RD1) and analyzed by flow cytometry. FITC-conjugated goat anti-mouse IgG and mouse anti-human IgG were used as secondary antibodies. A minimum of 50,000 gated-live events were acquired on a flow cytometer and analyzed by using FlowJo batch analysis software. (A to C) The percentage of p24-positive cells was determined for wild-type virus- (B) and MX2- (C) infected cells by using the uninfected control cells (A). (D to F) Env expression was then assessed in p24-gated cells by using antibodies reactive to gp41, T32 (D), 126-6 (E), and Md-1 (F). The solid lines represent the levels of expression of Env on wild-type virus-infected cells, and the broken lines represent the levels of expression on MX2-infected cells. The gray shaded peak in each graph represents the background level of staining for Env with the isotype control, anti-mouse IgG-FITC, or anti-human IgG-FITC in a p24-gated population of cells. Data presented here are representative of results from two independent infections. Mean fluorescence intensities of p24-PE and Env-FITC staining were determined for wild-type virus- and MX2-infected cells. FL2-H and FL1-H, fluorescence channels. (G to I) Ratios of mean fluorescence intensities (MFI) of p24-PE and Env-FITC were calculated as a measure of the levels of Env expressed relative to the level of p24 expressed per infected cell. Ratios are depicted graphically for antibodies T32 (G), 126-6 (H), and Md-1 (I) and are averages of results from two independent infections. Error bars indicate standard deviations.
Env expression levels were then assessed in this p24-gated population of cells. Positive staining for gp41 on wild-type virus- and MX2-infected cell surfaces is shown in Fig. 7D to F. Mouse anti-gp41 MAb T32 (37) (Fig. 7D), reactive with a linear epitope on the ectodomain of gp41, was used to assess the level of expression of gp41 on the cell surface as previously described (37). Panels E and F summarize the reactivities of the 126-6 and Md-1 MAbs to envelope glycoprotein expressed on the surface of wild-type virus- and MX2-infected cells. These antibodies recognize conformational epitopes within oligomeric, but not monomeric, forms of gp41 (5). In general, similar levels of specific-antibody staining were observed for both MX2- and wild-type virus-infected cultures for all the reference envelope-specific antibodies. To further quantify the relative antibody reactivities, the mean fluorescence intensities of p24-PE and Env-FITC stainings were calculated. The ratios of p24 to gp41 are summarized in Fig. 7G to I. The data indicated similar levels of expression of gp41 (T32) (Fig. 7G) on the surface of wild-type and MX2 virus-infected cells. Furthermore, the reactivities of conformation-dependent and oligomerization-dependent (126-6 and Md-1) (Fig. 7H and I) MAbs to MX2- and wild-type virus-infected cultures were similar. The combined envelope-specific antibody reactivity data regarding MX2 demonstrates that mutations in the LLP-2 domain apparently did not affect the levels, conformation, or oligomeric state of the MX2 cell surface Env compared to that of wild-type virus. Thus, the LLP-2-specific effect on syncytium formation does not appear to correlate with obvious changes in cell surface envelope expression.
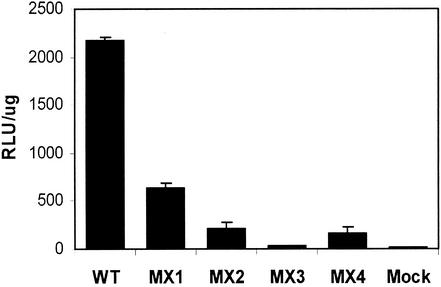
Alterations in both LLP domains can modulate the cell-cell fusion properties of HIV-1 Env.
The results of the syncytium assay for MX2 were further confirmed quantitatively by using a more objective luciferase-based cell-cell fusion assay routinely employed in studying Env-mediated fusion processes. For this assay, 293T cells expressing wild-type and LLP mutant HIV-1 Envs were mixed with target cells expressing CD4 and CXCR4 coreceptor for Env-dependent fusion. About 7 to 10 h after mixing, the cells were lysed and luciferase expression induced by the fusion of target and effector cells was assayed in the cell lysate. Since this assay measures the fusion capacity of HIV-1 Env independently of viral replication, MX1, MX3, and MX4 mutant Envs were also tested for their fusogenic potential. In agreement with the syncytium-defective phenotype of MX2, a decrease of about 90% in MX2 Env fusogenicity (Fig. 8) was observed in comparison with that of wild-type Env. Interestingly, MX1 and MX4 mutant Envs also exhibited a decrease in their cell-cell fusion properties. MX1 Env was about 70% less efficient at induction of cell-cell fusion, while the fusogenic capacity of MX4 Env was decreased by about 90%, a decrease that was similar to that of the MX2 Env. Due to the lack of Env expression in the MX3 mutant (Fig. 2A), no significant cell-cell fusion was observed and the levels of luciferase activity were found to be similar to those of the mock-transfected control. Thus, the MX3 mutant may in fact also suffice as a negative control in such cell-cell fusion assays. As shown for the MX2 mutant in Fig. 7, the expression levels of cell surface Env on 293T cells transfected with MX1, MX2, and MX4 mutants were not significantly affected compared with those of the wild-type construct (data not shown). Altogether, these data indicate a significant role of both LLP domains present in the ICT of HIV-1 Env in cell-cell fusogenicity.
FIG. 8.
Effects of mutations in the LLP domains on cell-cell fusion mediated by Env. ICT mutant Envs from MX1, MX2, MX3, and MX4 were cloned into pCDNA3 vector and transfected along with pCDNA3 vector alone (Mock) into 293T cells, which were also infected with recombinant vaccinia virus vTF1.1 expressing T7 polymerase. These effector cells were mixed with target quail QT6 cells transfected with CXCR4 and CD4 expression plasmids and the luciferase expression vector. The cells were allowed to fuse for at least 7 h, after which fusion was measured by quantitation of luciferase in cell lysates. The results shown are representative of results from two fusion experiments and are presented as relative light units (RLU) per microgram of protein in the cell lysate. Error bars indicate standard deviations. WT, wild type.
DISCUSSION
Previous studies have addressed the role of the ICT of lentiviral TM protein in the infectivity and cytopathicity of HIV-1 by creating truncations and deletions in the ICT (15, 21, 29, 51). These studies demonstrate a functional contribution of the ICT to the cytopathicity and infectivity of the virus. Because of the multiple and diverse effects observed upon truncation of the ICT of gp41, a clear definition of the specific domains in the ICT that are involved in these functions is not available. In the present study, our aim was to assess the functional roles of two structurally similar, highly conserved motifs, LLP-1 and LLP-2 domains, present within the ICT of HIV-1 gp41. In vitro peptide studies were used to rationally design site-specific mutations in the LLP domains such that their known in vitro functions of membrane perturbation and CaM binding were disrupted. Such targeted disruption of the functionality of LLP-1 and LLP-2 domains in the context of a full-length proviral clone has revealed for the first time differential effects of the LLP domains on Env incorporation and Env-mediated syncytium formation.
Arginine modification of LLP-1 by site-directed mutagenesis resulted in a dramatic decrease of about 85% in virion incorporation of mutant Env (Fig. 3), while no evident defect in Env expression or processing was observed (Fig. 2) in 293T cells; the defect in glycoprotein incorporation did not correlate with decreased surface expression of Env as determined by flow cytometry (data not shown). The observed defective Env incorporation associated with mutations in LLP-1 resulted in a corresponding decrease in the infectivity of the virus as determined by a single-round infectivity assay using MAGI-R5 cells (Fig. 4) and a multiple-round replication kinetics assay using both primary human PBMC and a CD4+ T-cell line (Fig. 5). Interestingly, the LLP-2 domain, which is structurally similar to the LLP-1 domain, did not exhibit a role in Env incorporation or infectivity of HIV-1 in our studies using 293T cells, as demonstrated by point substitutions in LLP-2 (MX2). Arginine modifications of the LLP-2 domain yielded a virus that replicated in activated human PBMC and CD4+ T lymphoid cells with kinetics similar to those of the wild-type virus (Fig. 5). Furthermore, the mutations did not apparently alter the expression, stability, or incorporation of the envelope glycoprotein into the virions in 293T cells (Fig. 2 and 3), which was consistent with wild-type levels of infectivity (Fig. 4) and the replication ability of MX2. The MX4 mutant, containing alterations in both LLP-1 and LLP-2 domains, showed levels of Env incorporation that were similar to those of MX1; no additive effect from the presence of a mutant LLP-2 domain in conjunction with a mutant LLP-1 domain was observed. It was noted, though, that MX4 showed slightly increased levels of Env expression and decreased levels of Env processing (Fig. 2) compared with wild-type, MX1, and MX2 clones. A greater decrease in the infectivity of MX4 in the single-cycle infectivity assay was also observed (Fig. 4); the infectivity of MX4 was about 50% lower than that of the MX1 virus. These observations may be attributed to altered Env stability and conformation but require further evaluation.
Taken together, the present studies with point substitutions in LLP domains indicate for the first time a differential role of the LLP-1 domain in uniquely mediating Env incorporation into HIV-1, while similar alterations in the LLP-2 domain did not demonstrate evident effects on the process of Env incorporation. Our observations related to the LLP-1 domain and its role in Env incorporation are supported by similar observations made previously (13, 38) but contrast with the findings of some ICT deletion studies (21, 36) that implicate the LLP-2 domain in the process of Env incorporation. As opposed to dramatic truncations or deletions in the ICT that may significantly alter the conformation of the envelope glycoprotein, our studies are expected to largely preserve the Env conformation as they involve point substitutions in the LLP domains that are predicted to maintain the structural properties of the LLP domains. Preliminary in vitro studies with LLP peptide analogs indicate that the replacement of positively charged amino acid residues in the hydrophilic face of LLP helices with negatively charged residues does not alter the characteristic α-helical properties (data not shown). Mechanistically, one can speculate a direct interaction of the LLP-1 amphipathic helix with the Gag MA protein during the process of Env incorporation. However, the possibility that changes in the LLP-1 domain may modulate the conformation and hence the availability of alternate MA binding sites in the ICT needs to be stringently addressed in future studies to help clarify the significance of the LLP-1 domain in modulating the structure and function of the envelope glycoprotein of HIV-1.
The MX3 mutant that lacks the ICT also demonstrated the significance of a full-length cytoplasmic tail of gp41 in the expression and stability of Env. Mutations were engineered in the ME46 strain of HIV-1 such that the entire ICT, including LLP-1 and LLP-2 domains, was deleted. The ICT deletion resulted in decreased expression or stability of Env (Fig. 2). These of the results of ICT truncation of Env from a primary isolate of HIV-1, ME46, are similar to observations reported previously with various cell-adapted strains of HIV-1 (27, 48). This defective Env expression or stability phenotype may result from alterations in the conformation of Env due to large ICT truncations. This observation reinforces the theory that selective point mutations in the LLP domains may provide a higher-resolution analysis of the functional significance of LLP domains than deletion mutants.
The replication competence of MX2 allowed for a study of the role of the LLP-2 domain in HIV-1-mediated cytopathicity, a phenotype that has also been associated with ICT deletions in the past (29). One of the best-described types of HIV-1-induced cell death is syncytium formation, which is characterized by a high rate of cell-cell fusion events between uninfected CD4-positive cells and infected Env-expressing cells, eventually leading to cell death (42). Thus, levels of syncytium formation and syncytium-induced cell death were determined for MX2-infected CD4+ T cells. Interestingly, the LLP-2 mutant virus was defective in syncytium formation (Fig. 6), thereby defining for the first time a function of the LLP-2 domain in the process of syncytium induction.
The arginine 775 residue that was mutated in LLP-2 is part of a putative endocytic YXXL motif, and the LLP-2 domain has been implicated in regulating cell surface expression of Env. Therefore, we assessed the effect of this arginine replacement on the level of cell surface expression of mutant Env to determine if the mutation resulted in a change in the structural context for presentation and hence function of the YXXL motif. Flow cytometry analyses of cell surface gp41 expression indicated that LLP-2 mutant and wild-type Env were expressed to similar levels on the surface of infected cells (Fig. 7). Despite wild-type levels of cell surface gp41 expression, the ability of MX2 virus to induce cell-cell fusion was apparently compromised. This defect in the ability of Env to mediate cell-cell fusion also manifested itself in the slightly delayed replication kinetics of MX2 in H9 cells, possibly due to a decreased rate of cell-cell transmission of the mutant virus. Thus, our studies demonstrate for the first time an important role of the LLP-2 domain in HIV-1 Env-mediated syncytium induction. It is noteworthy that although the mutations in LLP-2 resulted in a dramatic effect on the cell-cell fusion properties of the virus, the infectivity and thus virus-cell fusion properties of MX2 remained largely unaffected (Fig. 4 and 5). Previous studies of gp41 ectodomain mutants have also distinguished cell-cell fusion and virus-cell fusion phenotypes (7). However, the mechanistic basis for such observations is not available. Thus, the MX2 mutant also provides a unique tool for dissecting mechanistic differences between the processes of virus-cell and cell-cell fusion.
In terms of the mechanism of action of LLP-2 in enhancing cell-cell fusion, one of the possibilities explored was LLP-2-mediated alteration of the structure of Env. Our arginine modifications were designed to minimize alterations in the predicted structure of the LLP domains and the ICT. Only the hydrophilic face of the amphipathic helix was targeted since the hydrophobic face of LLP domains is implicated in functioning as a leucine zipper motif (28). Our analyses of the conformation and the oligomeric form of the LLP-2 mutant Env (Fig. 7) revealed that no major alterations in the conformation or oligomerization of gp41 were induced in the LLP-2 mutant Env. Although more rigorous biochemical analyses of the conformational and oligomeric properties of the LLP-2 mutant Env are required, the reactivities of Md-1 and 126-6 MAbs (Fig. 7) that exclusively recognize conformational epitopes on oligomeric gp41 strongly suggest that the conformation and the multimerization potential of the LLP-2 mutant Env were not significantly modulated. Thus, our analyses suggest that mutations in the LLP-2 domain may alter the intrinsic fusogenicity of Env without exerting any apparent effects on Env expression, conformation, or oligomerization potential.
The defect in syncytium induction by MX2 Env in CD4+ T-cell lines was also replicated in a quantitative luciferase-based cell-cell fusion assay (Fig. 8). This assay also allowed for assessment of the role of the LLP-1 domain in cell-cell fusion, as it is independent of viral replication properties. Interestingly, mutations in the LLP-1 domain were also found to dramatically decrease Env-mediated fusion. The MX1 mutant Env displayed a less severe effect on cell-cell fusion (70% decrease) compared with MX2 and MX4 mutants (90% decrease), which had mutations in the LLP-2 domain. Thus, both LLP domains contribute to Env fusogenicity, albeit to different levels. This defect in cell-cell fusion imposed by mutations in the LLP-1 domain may also contribute in part to the defect in infectivity observed with MX1 and MX4 viruses in MAGI cells (Fig. 4), in addition to the observed decrease in Env incorporation (Fig. 3).
These observations are suggestive of important contributions of two structurally conserved ICT domains to the process of HIV-1 Env-mediated fusion. The possibility that domains in the ICT may function to alter membrane fusion events during the processes of cell-cell fusion is not without precedent, as increasing evidence indicates a contribution of the cytoplasmic tail of the envelope glycoprotein of influenza virus to the process of membrane fusion (26, 31). Mechanistically, based on previous studies with peptide homologs of LLP domains (11, 33), one can predict that the LLP domains directly enhance the process of cell-cell fusion via their membrane perturbing properties. Different motifs in the ectodomain as well as the membrane-spanning domain of gp160 and a tryptophan-rich segment in the gp41 ectodomain (39) are known to be important for cell-cell fusion events (19). It is possible that these LLP domains function similarly to and synergize with the membrane-proximal tryptophan-rich region of gp41 in causing membrane disruption during the process of fusion. Recent studies have shown that the tryptophan-rich region can organize into well-defined amphipathic helices in the presence of a membrane-like environment (40) and that it plays a contributory role in Env-mediated fusion events (39) by inducing positive curvature and disruption of the membrane via the insertion of the indole ring into the membrane-water interface. Alternatively, the CaM binding properties of the LLP domains may also explain their contribution to the process of cell-cell fusion.
Acknowledgments
We thank Eric Freed (National Institute of Allergy and Infectious Diseases, Bethesda, Md.) for helpful discussions during the course of the study. We would also like to acknowledge R. W. Doms and B. Puffer (University of Pennsylvania), Patricia Earl (National Institute of Allergy and Infectious Diseases), Bernie Moss (National Institute of Allergy and Infectious Diseases), and the NIH AIDS Reference and Reagent Program for providing important reagents for our experiments. We also thank Mary White for p24 assays and Jing Jin for technical assistance with vaccinia viruses.
REFERENCES
- 1.Beary, T. P., S. B. Tencza, T. A. Mietzner, and R. C. Montelaro. 1998. Interruption of T-cell signal transduction by lentivirus lytic peptides from HIV-1 transmembrane protein. J. Pept. Res. 51:75-79. [DOI] [PubMed] [Google Scholar]
- 2.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, M. L., P. L. Earl, K. Fargnoli, S. Picciafuoco, F. Giombini, F. Wong-Staal, and G. Franchini. 1989. Identification of the fusion peptide of primate immunodeficiency viruses. Science 244:694-697. [DOI] [PubMed] [Google Scholar]
- 5.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, and A. Jungbauer. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 6.Bultmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 75:5263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J., I. W. Park, A. Cooper, and J. Sodroski. 1996. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J. Virol. 70:1340-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M., M. K. Singh, R. Balachandran, and P. Gupta. 1997. Isolation and characterization of two divergent infectious molecular clones of HIV type 1 longitudinally obtained from a seropositive patient by a progressive amplification procedure. AIDS Res. Hum. Retrovir. 13:743-750. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S. S., S. F. Lee, and C. T. Wang. 2001. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J. Virol. 75:9925-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernomordik, L., A. N. Chanturiya, E. Suss-Toby, E. Nora, and J. Zimmerberg. 1994. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J. Virol. 68:7115-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comardelle, A. M., C. H. Norris, D. R. Plymale, P. J. Gatti, B. Choi, C. D. Fermin, A. M. Haislip, S. B. Tencza, T. A. Mietzner, R. C. Montelaro, and R. F. Garry. 1997. A synthetic peptide corresponding to the carboxy terminus of human immunodeficiency virus type 1 transmembrane glycoprotein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res. Hum. Retrovir. 13:1525-1532. [DOI] [PubMed] [Google Scholar]
- 13.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas, N. W., G. H. Munro, and R. S. Daniels. 1997. HIV/SIV glycoproteins: structure-function relationships. J. Mol. Biol. 273:122-149. [DOI] [PubMed] [Google Scholar]
- 15.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg, D., and M. Wesson. 1990. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers 29:171-177. [DOI] [PubMed] [Google Scholar]
- 18.Evans, D. T., K. C. Tillman, and R. C. Desrosiers. 2002. Envelope glycoprotein cytoplasmic domains from diverse lentiviruses interact with the prenylated Rab acceptor. J. Virol. 76:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O., and M. A. Martin. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270:23883-23886. [DOI] [PubMed] [Google Scholar]
- 20.Fultz, P. N., P. J. Vance, M. J. Endres, B. Tao, J. D. Dvorin, I. C. Davis, J. D. Lifson, D. C. Montefiori, M. Marsh, M. H. Malim, and J. A. Hoxie. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J. Virol. 75:278-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa, H., M. Sasaki, S. Noda, and Y. Koga. 1998. Apoptosis induction by the binding of the carboxyl terminus of human immunodeficiency virus type 1 gp160 to calmodulin. J. Virol. 72:6574-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, E. M., K. H. Lee, and J. W. Kim. 1999. The cytoplasmic domain of HIV-1 gp41 interacts with the carboxyl-terminal region of alpha-catenin. Mol. Cells 9:281-285. [PubMed] [Google Scholar]
- 26.Kozerski, C., E. Ponimaskin, B. Schroth-Diez, M. F. Schmidt, and A. Herrmann. 2000. Modification of the cytoplasmic domain of influenza virus hemagglutinin affects enlargement of the fusion pore. J. Virol. 74:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. F., C. Y. Ko, C. T. Wang, and S. S. Chen. 2002. Effect of point mutations in the N terminus of the lentivirus lytic peptide-1 sequence of human immunodeficiency virus type 1 transmembrane protein gp41 on Env stability. J. Biol. Chem. 277:15363-15375. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. F., C. T. Wang, J. Y. Liang, S. L. Hong, C. C. Huang, and S. S. Chen. 2000. Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J. Biol. Chem. 275:15809-15819. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. J., W. Hu, A. G. Fisher, D. J. Looney, V. F. Kao, H. Mitsuya, L. Ratner, and F. Wong-Staal. 1989. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res. Hum. Retrovir. 5:441-449. [DOI] [PubMed] [Google Scholar]
- 30.Lodge, R., L. Delamarre, J. P. Lalonde, J. Alvarado, D. A. Sanders, M. C. Dokhelar, E. A. Cohen, and G. Lemay. 1997. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J. Virol. 71:5696-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10:1821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Micoli, K. J., G. Pan, Y. Wu, J. P. Williams, W. J. Cook, and J. M. McDonald. 2000. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J. Biol. Chem. 275:1233-1240. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. A., M. W. Cloyd, J. Liebmann, C. R. Rinaldo, Jr., K. R. Islam, S. Z. Wang, T. A. Mietzner, and R. C. Montelaro. 1993. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology 196:89-100. [DOI] [PubMed] [Google Scholar]
- 34.Miller, M. A., R. F. Garry, J. M. Jaynes, and R. C. Montelaro. 1991. A structural correlation between lentivirus transmembrane proteins and natural cytolytic peptides. AIDS Res. Hum. Retrovir. 7:511-519. [DOI] [PubMed] [Google Scholar]
- 35.Miller, M. A., T. A. Mietzner, M. W. Cloyd, W. G. Robey, and R. C. Montelaro. 1993. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retrovir. 9:1057-1066. [DOI] [PubMed] [Google Scholar]
- 36.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piller, S. C., J. W. Dubay, C. A. Derdeyn, and E. Hunter. 2000. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 from human immunodeficiency virus type 1: effects on glycoprotein incorporation and infectivity. J. Virol. 74:11717-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 41.Shacklett, B. L., C. J. Weber, K. E. Shaw, E. M. Keddie, M. B. Gardner, P. Sonigo, and P. A. Luciw. 2000. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J. Virol. 74:5836-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodroski, J., W. C. Goh, C. Rosen, K. Campbell, and W. A. Haseltine. 1986. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature 322:470-474. [DOI] [PubMed] [Google Scholar]
- 43.Sodroski, J. G. 1999. HIV-1 entry inhibitors in the side pocket. Cell 99:243-246. [DOI] [PubMed] [Google Scholar]
- 44.Srinivas, S. K., R. V. Srinivas, G. M. Anantharamaiah, R. W. Compans, and J. P. Segrest. 1993. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 268:22895-22899. [PubMed] [Google Scholar]
- 45.Tencza, S. B., T. A. Mietzner, and R. C. Montelaro. 1997. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res. Hum. Retrovir. 13:263-269. [DOI] [PubMed] [Google Scholar]
- 46.Tencza, S. B., M. A. Miller, K. Islam, T. A. Mietzner, and R. C. Montelaro. 1995. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J. Virol. 69:5199-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adapter. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X., X. Yuan, M. F. McLane, T. H. Lee, and M. Essex. 1993. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 67:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, T., T. A. Mietzner, R. C. Montelaro, and H. J. Vogel. 1995. Characterization of the calmodulin binding domain of SIV transmembrane glycoprotein by NMR and CD spectroscopy. Biochemistry 34:10690-10696. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, H., L. Wang, S. Kao, I. P. Whitehead, M. J. Hart, B. Liu, K. Duus, K. Burridge, C. J. Der, and L. Su. 1999. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr. Biol. 9:1271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]