Abstract
Transforming growth factor (TGF)-βs are secreted in large latent complexes consisting of TGF-β, its N-terminal latency-associated peptide (LAP) propeptide, and latent TGF-β binding protein (LTBP). LTBPs are required for secretion and subsequent deposition of TGF-β into the extracellular matrix. TGF-β1 associates with the 3rd 8-Cys repeat of LTBP-1 by LAP. All LTBPs, as well as fibrillins, contain multiple 8-Cys repeats. We analyzed the abilities of fibrillins and LTBPs to bind latent TGF-β by their 8-Cys repeats. 8-Cys repeat was found to interact with TGF-β1•LAP by direct cysteine bridging. LTBP-1 and LTBP-3 bound efficiently all TGF-β isoforms, LTBP-4 had a much weaker binding capacity, whereas LTBP-2 as well as fibrillins -1 and -2 were negative. A short, specific TGF-β binding motif was identified in the TGF-β binding 8-Cys repeats. Deletion of this motif in the 3rd 8-Cys repeat of LTBP-1 resulted in loss of TGF-β•LAP binding ability, while its inclusion in non-TGF-β binding 3rd 8-Cys repeat of LTBP-2 resulted in TGF-β binding. Molecular modeling of the 8-Cys repeats revealed a hydrophobic interaction surface and lack of three stabilizing hydrogen bonds introduced by the TGF-β binding motif necessary for the formation of the TGF-β•LAP - 8-Cys repeat complex inside the cells.
INTRODUCTION
Transforming growth factor (TGF)-βs belong to a large TGF-β superfamily of growth and differentiation modulators (for reviews see Kingsley, 1994; Roberts and Sporn, 1996). Three different TGF-β isoforms exist in mammals, namely TGF-βs 1 to 3. TGF-βs effect in numerous biological processes, including the up-regulation of the synthesis of many extracellular matrix proteins, down-regulation of extracellular proteolysis (reviewed in Taipale et al., 1998), and various developmental processes. TGF-βs are also effective in the down-regulation of the immune system.
TGF-βs are secreted from cells as biologically latent protein complexes, in which the disulphide-bound N-terminal dimeric latency-associated peptide (LAP) part is noncovalently associated with the C-terminal disulphide-bound dimeric TGF-β (Gentry et al., 1988; Gray and Mason, 1990). LAP is cleaved from the TGF-β by furin-mediated proteolysis during secretion (Dubois et al., 1995). Before TGF-β can bind to its signaling receptors at the cell surface, it needs to be activated, via disruption of the interaction between TGF-β and its LAP propetide (reviewed in Munger et al., 1997). In nonmalignant cell types, TGF-β is in a large complex, in which a heterologous protein, LTBP (latent TGF-β binding protein), is covalently bound to TGF-β (reviewed in Mangasser-Stephan and Gressner, 1999; Saharinen et al., 1999). LTBPs are required for the secretion and correct folding of TGF-βs (Miyazono et al., 1991). The association with LTBPs results in the storage of latent TGF-β in ECM structures rapidly after secretion. The expression levels of LTBPs decrease upon cell transformation (Dallas et al., 1994; Koski et al., 1999; Koli, Saharinen, Kärkkäinen and Keski-Oja, unpublished data). In certain malignancies the lack of LTBP expression causes retention of TGF-β inside the cells, preventing TGF-β from exerting its biological effects (Eklöv et al., 1993; Mizoi et al., 1993).
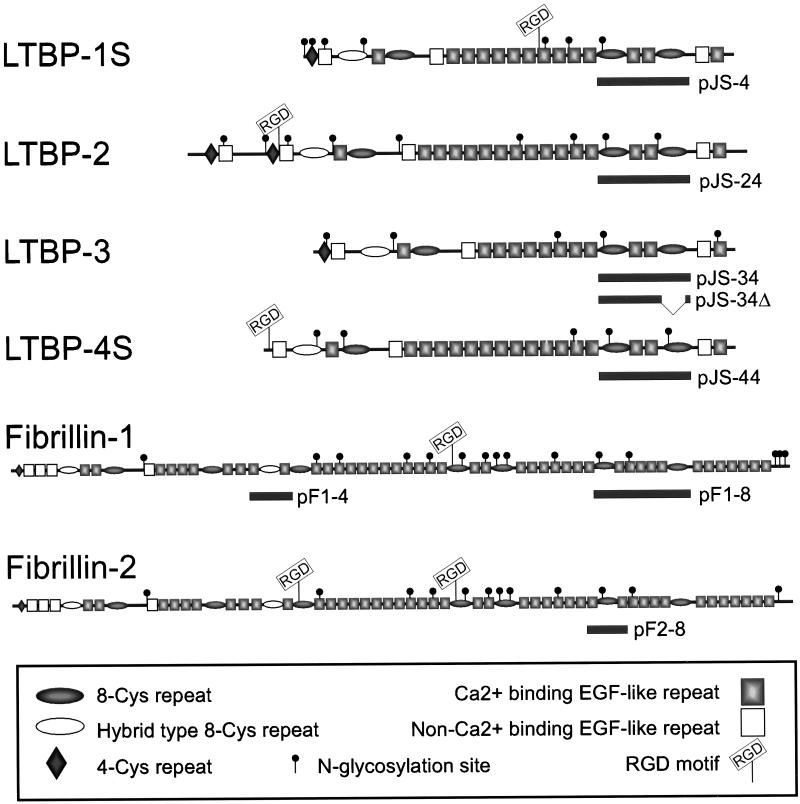
LTBPs form a family of structurally related extracellular matrix (ECM) proteins. LTBPs 1 to 4 are similar in their overall domain structure to fibrillins -1 and -2, which are the major components of the 10 nm ECM microfibrils, often associated with elastic tissue (reviewed in Ramirez and Pereira, 1999). LTBPs and fibrillins are mainly composed of two types of repeated protein domains, namely epidermal growth factor (EGF-like) and 8-Cys-like repeats.1 Fibrillins contain nine repeats and LTBPs contain four 8-Cys repeats each, some of which are also addressed as hybrid domains as being more divergent (see Figure 1). In contrast to the EGF-like repeats, which are abundant in many extracellular matrix proteins, the 8-Cys repeats are found only in LTBPs and fibrillins. The EGF-like repeats mediate various, noncovalent interactions (reviewed in Davis, 1990), whereas covalent protein-protein interactions have been described for the 8-Cys repeats. The best known example is the interaction between LTBP-1 and TGF-β1•LAP, which is mediated by the 3rd 8-Cys repeat of LTBP-1, whereas the other 8-Cys repeats of LTBP-1 are unable to form this covalent interaction (Gleizes et al., 1996; Saharinen et al., 1996). However, the question whether the 8-Cys repeats of fibrillins can also associate covalently with TGF-β•LAPs, which would then be deposited into microfibrils in a latent form, has been unresolved thus far. Other suggested functions for specific 8-Cys repeats include the covalent dimerization of fibrillin-1 and fibrillin-2 proteins (Trask et al., 1999) and the interaction between ECM and LTBP-1 (Unsöld, Hyytiäinen, Bruckner-Tuderman, and Keski-Oja, unpublished data). Noncovalent cell surface interactions between integrins and RGD-motifs in certain 8-Cys repeats of fibrillins have also been found (Pfaff et al., 1996; Sakamoto et al., 1996; D'Arrigo et al., 1998).
Figure 1.
Schematic representation of the human LTBPs 1–4 and human fibrillins -1 and -2. The small (S) isoforms of LTBPs -1 and -4 are illustrated. The solid bars below the proteins describe the area coded by the respective constructs.
We have analyzed here the abilities of the members of the LTBP-fibrillin family to associate with the different isoforms of TGF-β via their 8-Cys repeats. Only a small subset of the 8-Cys repeats was found to possess the ability to associate with the β•LAP. The 8-Cys protein repeats could thus be classified into TGF-β binding and nonbinding types, depending on a specific short sequence motif. Substitution of this motif causes the loss of TGF-β•LAP binding ability, while in a non-TGF-β binding 8-Cys repeat, inclusion of this motif provided binding ability. The interaction involves direct interprotein disulphide bridge formation between the cysteine residues of the β•LAP and an 8-Cys repeat. Molecular models of the 8-Cys repeats indicated that the presence of the motif required for the β•LAP binding resulted in increased surface hydrophobicity and may thus be together with accessible sulfhydryl groups, a prerequisite for the formation of the covalent interaction resulting in the correct secretion and storage of latent TGF-β into the extracellular matrix.
MATERIALS AND METHODS
cDNA Constructs
Constructs pTGFβ1, pLTBP-1, and pLTBP-2 that contained the full-length cDNA for human TGF-β1, human LTBP-1S, and human LTBP-2, respectively, have been described earlier (Hyytiäinen et al., 1998; Saharinen et al., 1996).
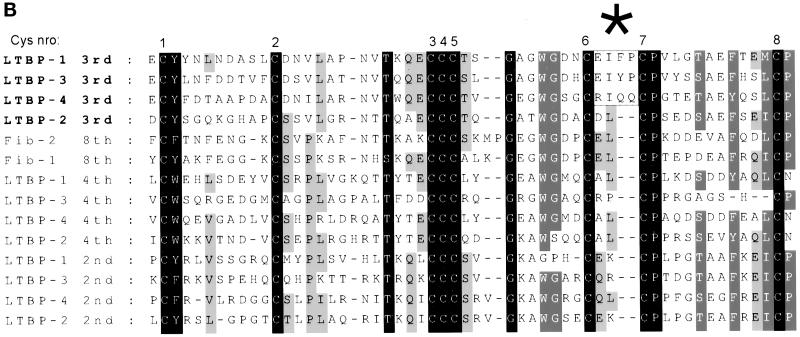
Construct pJS-4 codes for the 3rd 8-Cys repeat, next two following EGF-like repeats and the 4th 8-Cys repeat of LTBP-1 (Saharinen et al., 1996; see Figure 1) and is cloned in pSignal (Saharinen et al., 1996), a eukaryotic secretory expression vector derived from pcDNA3 (InVitrogen, Carlsbad, CA). Analogous constructs were made of human LTBP-2 (pJS-24; amino acids 1397–1656), and LTBP-4 (pJS-44; amino acids 1062–1316) cDNAs by PCR and cloned into pSignal. Construct pJS-34Δ coded for the 3rd 8-Cys repeat and next two following EGF-like repeats mouse of LTBP-3 (amino acids 887-1137), amplified by PCR and cloned as a BamHI–XhoI fragment into pSignal using NIH-3T3 cells random primed cDNA as template. Construct pJS-34Δ contains thus the previously published alternatively spliced variant of mouse LTBP-3 (Yin et al., 1998b). The results obtained with construct pJS-34Δ were later verified with construct pJS-34, which was made using the same primers as with construct pJS-34, but mouse heart cDNA λ library (Clontech HL5002b, Palo Alto, CA) was used as the primary template. During the preparation of the constructs pJS-34 and pJS-34Δ, we found four sequencing errors in the reported sequence of the 3rd 8-Cys repeats of mouse LTBP-3 (GenBank entry L40459) using both the mRNA of NIH-3T3 cells as well as in mouse heart cDNA library. These errors caused four frameshifts in the C-terminus of the 3rd 8-Cys repeat. At the amino acid level, the corrected sequence for the 3rd 8-Cys repeat of mouse LTBP-3 does not contain 9 cysteine residues, as reported earlier (Yin et al., 1995; Yin et al., 1998a). We have cloned the human LTBP-3 cDNA, and the 3rd 8-Cys repeat of human LTBP-3 is identical to the corrected 3rd 8-Cys repeat of mouse LTBP-3 at the amino acid level1 (see Figure 8B).
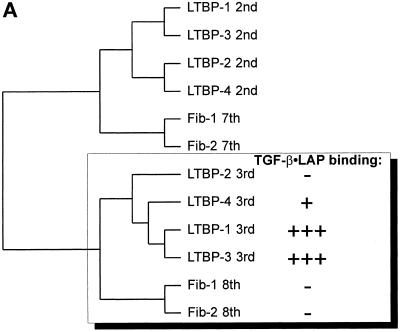
Figure 8.
A) Relationships between the 8-Cys repeats of human LTBPs 1–4 and fibrillins -1 and -2. The amino acid sequences for all known 8-Cys as well as the hybrid domains of human LTBPs and fibrillins were used to make the dendrogram, and the ones most similar to the 3rd 8-Cys repeats of LTBPs are shown. The TGF-β•LAP binding abilities of the 3rd 8-Cys repeats of LTBPs and the 8th 8-Cys repeats of fibrillins are illustrated. B) Multiple sequence alignment of the 8-Cys repeats of human LTBPs and the 8th 8-Cys repeats of fibrillins. The amino acid sequences for all the 8-Cys repeats of human LTBPs and the 8th 8-Cys repeats of fibrillins were aligned. The sequences covering all eight cysteine residues as well as one amino acid on both N- and C-terminal sides were included. The alignment indicates several conserved residues in the 8-Cys repeats. The three TGF-β binding type 8-Cys repeats (3rd ones of LTBPs -1, -3, and -4) are clearly distinguished by the two amino acids insertion between the 6th and 7th cysteine residues (indicated by an asterisk above the alignment).
Constructs pF1–4 (fibrillin-1 amino acids 806–951), pF1–8 (fibrillin-1 amino acids 2057–2401) and pF2–8 (fibrillin-2 amino acids 1928–2083) were generated by amplifying the corresponding region from human fibrillin -1 or -2 cDNA and cloned into pSignal vector as BamHI–XhoI fragments (see Figure 1).
The full-length cDNAs for human TGF-β2 and TGF-β3 were kind gifts from Dr. P. ten Dijke (Ludwig Institute for Cancer Research, Uppsala, Sweden) and Oncogene Science Inc., respectively. The open reading frames of TGF-β2 and TGF-β3 were subcloned into pCl-Neo mammalian expression vector (Promega, Madison, WI) and named as pTGFβ2 and pTGFβ3. Human TGF-β1, in which both codons coding for cysteines 223 and 225 had been mutated to code for serine, was a kind gift of Dr. H. L. Moses (Vanderbilt University Cancer Center, Nashville, TN). The TGF-β1 coding fragment from this construct was transferred to pcDNA3 expression vector generating construct pTGFβ1 C223S-C225S.
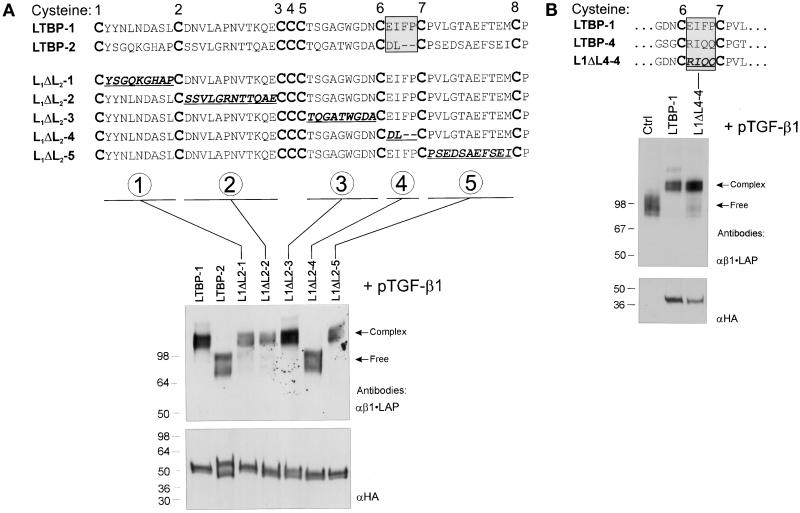
Constructs L1ΔL2 1–5 (illustrated in Figure 6A) were generated using the LTBP-1 construct pJS-4 as backbone. The amino acids between each two successive cysteine residues in the 3rd 8-Cys repeat of human LTBP-1 were changed to code for amino acids in the analogous regions of the 3rd 8-Cys repeat of human LTBP-2 (see Figure 5A). Another chimeric construct, L1ΔL4–4, was made between LTBP-1 and LTBP-4, in which the amino acids between 6th and 7th cysteines of the 3rd 8-Cys repeat of human LTBP-1 (EIFP), were changed to those of the 3rd 8-Cys repeat of human LTBP-4 (RIQQ). In construct L2 GAIN, the region between 6th and 7th cysteines in the 3rd 8-Cys repeat of human LTBP-2 (coding for amino acids DL) was changed to the analogous area in human LTBP-1 (amino acids EIFP). All these constructs containing altered amino acid coding sequences were created by PCR, using the respective wild-type construct as template and cloned in pSignal.
Figure 6.
The region between 6th and 7th Cys-residues of the 8-Cys repeat determines the β•LAP binding ability. A) Loss of function of LTBP-1. The substituted amino acids in chimeric constructs L1ΔL2 1–5 between the 3rd 8-Cys repeats of LTBP-1 and LTBP-2 are underlined. These constructs as well as the wild-type LTBP-1 and LTBP-2 constructs (pJS-4 and pJS-24, respectively) were transfected together with pTGFβ1 cDNA to 293T cells, as indicated above the immunoblot. The secreted proteins were separated by 7.5% PAGE and immunodetected with β1•LAP antibodies. The proteins coded by all LTBP constructs were separated by 4–15% gradient PAGE, and their expression levels were detected by immunoblotting with anti-HA antibody (lower panel). B) The corresponding chimeric construct between LTBP-1 and LTBP-4 retains TGF-β•LAP binding function. Construct L1ΔL4–4, analogous to construct L1ΔL2–4, was transfected with pTGFβ1 cDNA to 293T cells. The conditioned medium was separated by 7.5% PAGE and analyzed for the presence of covalent protein complexes between β1•LAP and L1ΔL4–4 encoded protein. Arrows on the right indicate the free or complexed β1•LAP forms. The secretion of proteins coded by all LTBP constructs were detected by 4–15% gradient PAGE followed by immunoblotting with anti-HA antibody (lower panel).
Figure 5.
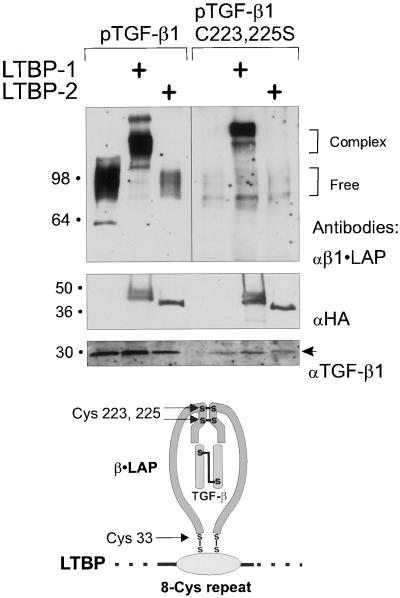
Binding between the 8-Cys repeat of LTBP-1 and Cys-33 of TGF-β1•LAP is mediated by a direct disulfide exchange. Constructs pJS-4 or pJS-24 were transfected with wild-type pTGFβ1 or pTGFβ1 C223,225S to COS-7 cells. The secreted proteins were separated by 7.5% PAGE and detected by immunoblotting with β1•LAP antibodies (top panel). Brackets on the right indicate the free or complexed β1•LAP forms. The expression of proteins coded by LTBP-constructs was verified by 4–15% gradient PAGE followed by anti-HA immunoblot (middle panel). Secretion of TGF-β1 was detected by immunoblotting for TGF-β1 (bottom panel, an arrow). Below the immunoblots is a model illustrating the proposed β1•LAP - 8-Cys repeat interaction.
All PCR generated constructs were sequenced using Pharmacia's ALF Express (Amersham-Pharmacia Biotech, Uppsala, Sweden), Perkin Elmer-Cetus's ABI 373, ABI 377 or ABI 310 (Perkin Elmer-Cetus) automatic DNA-sequencers.
Cell Culture and Transfections
Adenovirus transformed human kidney epithelial cells (293-T, CRL-1573, American Type Culture Collection (ATCC), Rockville, MD), SV-40 transformed African green monkey kidney cells (COS-7, ATCC) and human embryonic lung fibroblasts (CCL-137, ATCC) were cultured in d-MEM supplemented with 10% fetal calf serum (FCS), 100 IU ml−1 penicillin and 50 μg ml−1 streptomycin.
Approximately 7.5 × 105 293T or COS-7 cells were seeded per plate in 6-well plates and transfected the following day with 2 μg of the plasmids indicated using FuGENE6 liposome mediated transfection system (Roche Molecular Biochemicals). Six hrs after transfection the cells were washed, fed with serum-free medium, and the conditioned medium was collected after 60 h.
CCL-137 cells were transfected as postconfluent cell layers. Before transfection the cells were changed to d-MEM containing 10% FCS, and transfection was carried out for 24 h using FuGENE6 reagent and 12 μg of pTGFβ1 per 100 mm diameter dish. Subsequently, the cells were washed and changed to serum-free medium, which was harvested after 7 days.
Antibodies
Polyclonal rabbit antibodies against human LTBP-1 (Ab39), TGF-β1•LAP (immunoprecipitating antibody Lt2), and TGF-β3•LAP (Ab95) were kind gifts of Dr. C.-H. Heldin (Ludwig Institute for Cancer Research, Uppsala, Sweden), and used as purified IgG. Mouse monoclonal antifibrillin-1 antibodies mAb 201 and mAb 69 were kind gifts of Dr. Lynn Sakai (Shriners Hospital, Oregon). Affinity purified rabbit antihuman TGF-β1•LAP and human LTBP-2 antibodies have been described previously (Hyytiäinen et al., 1998; Taipale et al., 1994). Polyclonal anti human TGF-β2 sc-20 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antihemagglutinin antibody 12CA5 was from BabCO (Richmond, CA). Immunodetection was performed as described earlier (Saharinen et al., 1996). Conditioned medium samples were nonreduced in SDS-PAGEs in order to detect the covalent complexes, except when using the anti-HA antibody to detect the expression of HA-tagged constructs.
Sequence Analysis and Molecular Modeling
The multiple sequence alignment was carried out using the Clustal W 1.74 program and corrected by hand. In the sequence alignments, the human LTBP-3 sequence was used (Genbank accession number AF135960; Saharinen, Koski, and Keski-Oja, unpublished data). The molecular models were built using the Insight II version 98 (Molecular Simulations Inc., San Diego, CA), using an SGI Origin 2000 computer (Center for Scientific Computing, Espoo, Finland). The NMR solved structure of the 8th 8-Cys repeat of human fibrillin-1 (Yuan et al., 1997, PDB accession number 1APJ, 7th structure out of 21 structures in entry 1APJ) was used as a template. The identity percentages between the amino acids in the 8th 8-Cys repeat of fibrillin-1 and the modeled 8-Cys repeats of LTBPs were 37–46%. All the indels were modeled by searching from PDB-loop database. The preliminary models were soaked in a waterbox, extending at least 9 Å beyond the 8-Cys repeat. The energy minimizations were done by gradually diminishing the fixations of the model between successive minimization steps with the steepest descent followed by conjugate gradient algorithms using the Discover v. 2.98 module and Amber forcefield.
RESULTS
Overexpression of TGF-β1 in Human Fibroblasts Results in Complex Formation with LTBP-1, but Not with LTBP-2 or Fibrillin-1
Endogenous LTBP-1 forms a covalent complex with TGF-β1•LAP in fibroblasts (Taipale et al., 1994). To analyze if fibrillin-1 or LTBP-2 are also capable of covalent complex formation with β1•LAP, we transfected postconfluent fibroblasts with TGF-β1 cDNA. Fibroblasts were used as a model, because all LTBPs and fibrillins are expressed in confluent fibroblast cultures (Kanzaki et al., 1990; Moren et al., 1994; Saharinen et al., 1998; Sakai et al., 1986; Zhang et al., 1994; Unsöld, Hyytiäinen, Bruckner-Tuderman, and Keski-Oja, unpublished data; see Figure 2A). In nontransfected cells, only very small amounts of secreted endogenous TGF-β1•LAP, all in large latent complex, was detected. This is consistent with previous results, showing that most cultured cell types secrete more LTBP-1 than TGF-β1 (Taipale et al., 1994). In longer exposures, also minor amounts of complexed endogenous LTBP-1 was detected. Endogenous LTBP-2 and fibrillin-1 were always in the uncomplexed, free form.
Figure 2.
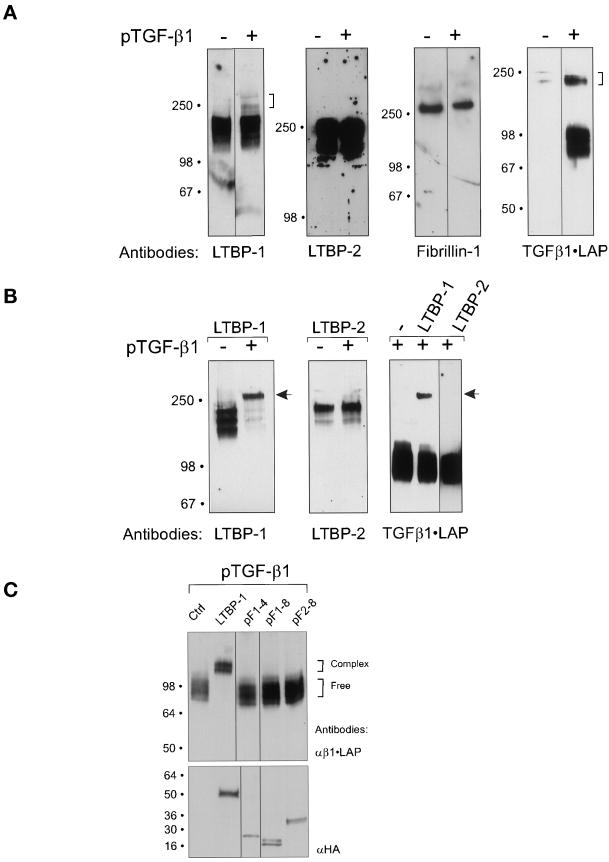
A) Overexpressed TGF-β1•LAP forms covalent complexes with endogenous LTBP-1, but not with LTBP-2 or fibrillin-1 in human fibroblast culture. Confluent human embryonic lung fibroblast cultures were transfected with pTGFβ1. Proteins secreted into fibroblast conditioned medium were separated in SDS-PAGEs. The concentrations of acrylamide in SDS PAGES were the following: 7,5% for LTBP-1 blot, 5% for LTBP-2 blot, 7,5% for fibrillin-1 blot, 4–15% gradient for β1•LAP blot. The formation of β1•LAP complexes with endogenous LTBPs -1 or -2 or fibrillin-1 was detected by immunoblotting. Transfection of TGF-β1 cDNA is indicated above the immunoblots, and the antibodies used are below the immunoblot. Brackets on the right indicate the complexed β1•LAP forms. B) Overexpressed LTBP-1 binds covalently β1•LAP, whereas LTBP-2 does not. pLTBP-1 and pLTBP-2 were transfected together with pTGFβ1 cDNA to 293T cells, and proteins secreted into the conditioned medium were separated by 7.5% PAGEs. The secreted proteins were detected by immunoblotting with antibodies for LTBP-1, LTBP-2, or β1•LAP as indicated, in order to detect the covalent LTBP - β1•LAP complexes. The transfected cDNAs are indicated above the immunoblots, and the antibodies used are below them. Arrows indicate the large latent β1•LAP - LTBP-1 complexes. C) β1•LAP does not associate with 8-Cys repeats of fibrillin-1. Expression constructs containing certain 8-Cys repeats of fibrillins were transfected together with pTGFβ1 to 293T cells, as indicated on the top of the figure. LTBP-1 construct pJS-4 (Figure 1) was used as positive control for a protein capable of forming covalent complexes with β1•LAP. Secreted proteins were separated by 7.5% PAGE and analyzed for β1•LAP by immunoblotting (upper panel). Brackets on the right indicate the free and complexed β1•LAP forms. Expression levels of the proteins coded by all HA-tagged fibrillin and LTBP-1 constructs were verified by 4–15% PAGE followed by immunoblotting with HA-antibody from reduced samples (lower panel).
Upon TGF-β1 overexpression, β1•LAP saturated the endogenous TGF-β binding LTBPs, and most β1•LAP was in the small latent complex, which is consistent with our previous results (Saharinen et al., 1996). Due to the low transfection efficiency of the primary fibroblasts, most endogenously expressed LTBP-1 was in the free form, however, the mobility of a fraction of LTBP-1 was retarded, indicating complex formation with the overexpressed β1•LAP (brackets in Figure 2A). Similar larger molecular weight complexes were not detected with LTBP-2 or fibrillin-1, indicating that neither LTBP-2 nor fibrillin-1 were able to form the covalent complexes with β1•LAP. In addition, the apparent molecular weight of the large latent β1•LAP complex was smaller than that expected for a possible fibrillin-1 - β1•LAP complex. The antibodies for LTBPs-1 and -2 are polyclonal against a large protein fragment, excluding the possibility that steric hindrance could prevent the detection of the β•LAP - LTBP-2 complexes. However, the fibrillin-1 antibody mAb 201 recognizes a specific epitope in the C-terminus. We therefore repeated the experiment by using another fibrillin-1 antibody mAb 69 against an epitope in the N-terminal region with same results, confirming that fibrillin-1 is unable to associate covalently with β1•LAP.
Overexpressed LTBP-2 Does Not Associate with TGF-β1•LAP
We analyzed next whether overexpressed LTBP-2 could associate covalently with overexpressed TGF-β1•LAP in a cell system that does not express endogenous LTBPs. pLTBP-1 or pLTBP-2 as well as pTGFβ1 constructs were transfected to 293T cells, and LTBP-1 as well as LTBP-2 were immunoblotted from conditioned medium. When both LTBP-1 and TGF-β1 were overexpressed, LTBP-1 was seen predominantly in complex with β1•LAP (Figure 2B). On the contrary, when both LTBP-2 and TGF-β1 were overexpressed, no change in the migration of LTBP-2 was observed, indicating the inability of LTBP-2 to form a covalent complex with TGF-β1•LAP.
The results were confirmed by immunoblotting for β1•LAP. When TGF-β1 was overexpressed alone, it was exclusively in the small latent complex. The concurrent expression of LTBP-1 resulted a portion of β1•LAP to be recruited to a complex with LTBP-1. Due to lower expression levels of full-length LTBP-1 than TGF-β1, the majority of the β1•LAP was in free form. On the contrary, when expressed with LTBP-2, all β1•LAP remained in the small latent complex. These results were verified by using another cell line, COS-7.
In LTBP-1, the 3rd 8-Cys repeat mediates the covalent binding to β1•LAP, and cDNA constructs containing this domain have earlier been found to associate very efficiently with β1•LAP (Saharinen et al., 1996; Gleizes et al., 1996). The inability of the 8-Cys repeats of LTBP-2 to associate with β1•LAP was confirmed by multiple LTBP-2 constructs containing 3rd or 3rd and 4th 8-Cys repeats. None of the proteins encoded by these constructs was able to form covalent complexes when overexpressed with TGF-β1•LAP.
8-Cys Repeats Containing Fibrillin -1 or -2 Fragments Do Not Complex with Overexpressed TGF-β1•LAP
The inability of overexpressed 8-Cys repeats of fibrillin-1 to form a covalent complex with TGF-β1•LAP was confirmed by using two constructs, pF1–4 and pF1–8, containing different regions of fibrillin-1 in an overexpression system. These constructs included the 8th 8-Cys repeat of fibrillin-1, which is most similar to the TGF-β binding 3rd 8-Cys repeat of LTBP-1 (see Figure 8A). Fibrillin-1 constructs were transfected with TGF-β1 cDNA to 293T cells and β1•LAP was immunoblotted from the conditioned medium (Figure 2C). Neither of the fibrillin-1 construct encoded proteins was able to associate with β1•LAP, unlike the protein from the LTBP-1 control construct pJS-4.
We also analyzed the complex forming ability of the 8th 8-Cys repeat of fibrillin-2 construct pF2–8, which encodes the 8th 8-Cys repeat and one EGF-like repeat on both sides. The protein expressed from pF2–8 was accordingly unable to form covalent complexes with coexpressed β1•LAP.
Neither Fibrillin-1 nor LTBP-2 Coimmunoprecipitates with TGF-β1•LAP
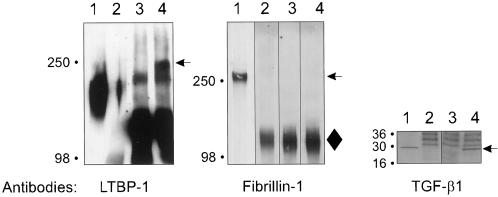
In addition to covalent interaction between TGF-β1 and its binding proteins, we analyzed the possibility of a noncovalent interaction between β1•LAP and members of the LTBP-fibrillin family. Postconfluent fibroblast conditioned medium was immunoprecipitated by β1•LAP antibodies followed by immunoblotting using either LTBP -1, -2 or fibrillin-1 antibodies. LTBP-1, but not fibrillin-1, coprecipitated with β1•LAP (Figure 3). No LTBP-2 was detected in the immunoprecipitated material either (our unpublished results). These results further verify the lack of high stringency interactions, covalent or noncovalent, between β1•LAP and fibrillin-1 or LTBP-2. The secretion of TGF-β1 in the conditioned medium as well as in the immunoprecipitates was verified using a TGF-β1 immunoblot (Figure 3, third panel).
Figure 3.
Endogenous fibrillin does not coprecipitate with endogenous β1•LAP from human fibroblasts. β1•LAP was immunoprecipitated from the conditioned medium lung fibroblasts. The immunoprecipitates were separated by 5% PAGE (LTBP-1 and fibrillin-1) or 4–15% gradient PAGE (TGF-β1) and analyzed for β1•LAP complexed proteins by immunoblotting. Lanes in each panel are: 1) fibroblast conditioned medium; 2) conditioned medium mock-precipitated with nonimmune serum; 3) d-MEM immunoprecipitated with β1•LAP antibodies; 4) fibroblast conditioned medium immunoprecipitated with β1•LAP antibodies. Arrows on the right indicate the latent β1•LAP - LTBP-1 complexes, fibrillin-1 and TGF-β1 dimer. A diamond (♦) on the right of fibrillin-1 immunoblot indicates the migration of a background band.
All TGF-β•LAP Isoforms Associate with the 3rd 8-Cys Repeats of LTBPs -1 and -3, but Not with LTBP-2
Using constructs containing only the 3rd 8-Cys repeats of different LTBPs, β1•LAP binding ability was confirmed to reside in the 3rd 8-Cys repeat of LTBPs -1, -3, and -4. Protein from an analogous LTBP-2 construct was negative in β1•LAP binding (our unpublished results). Together with results presented in Figure 2, only three of the studied 8-Cys repeats of fibrillins and LTBPs were observed to posses the TGF-β1•LAP binding function.
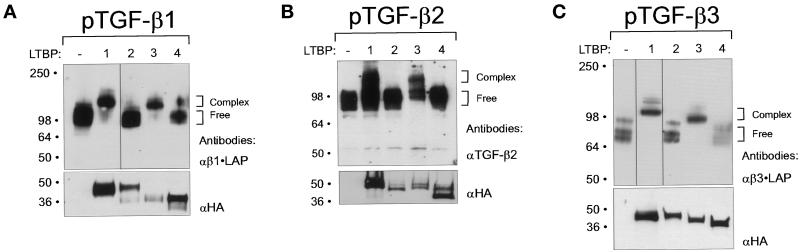
LTBP - TGF-β•LAP interactions have previously been studied using the TGF-β1 isoform. We analyzed whether the other mammalian TGF-β isoforms, β2 and β3, have similar ability to associate covalently with LTBPs. For this purpose, expression constructs of all LTBPs, containing the 3rd 8-Cys repeat, two following EGF-like repeats and the 4th 8-Cys repeat were used (Figure 1). Although the association between TGF-β binding LTBPs and β1•LAP takes place via the 3rd 8-Cys repeat, we included also other protein domains of LTBPs in these constructs to yield larger mobility differences between the complexed and free β•LAPs. The LTBP constructs were transfected together with cDNAs for TGF-βs to 293T cells. For the detection of TGF-β•LAPs -1 and -3, antibodies against the LAP regions were used. Since a portion of TGF-βs is not correctly cleaved by proteolysis during secretion (Gentry et al., 1988; Dubois et al., 1995), this allowed the detection of both complexed and free β2•LAP using antibodies against the growth factor part of TGF-β2, which was necessary because of the unavailability of β2•LAP antibodies. All β•LAP isoforms were found to be very efficiently complexed with the LTBP-1 and LTBP-3 derived proteins (Figure 4 A, B, and C). The β•LAP binding ability of the LTBP-4 fragment was much less efficient than that of LTBP-1 or LTBP-3 fragments. Only minor traces of covalent complexes between the protein coded by the LTBP-4 construct and β1•LAP, but not with the other β•LAP isoforms, were detected. Protein encoded by LTBP-2 construct could not form covalent complexes with any of the β•LAP isoforms.
Figure 4.
Association of TGF-β•LAP isoforms 1, 2, and 3 with LTBPs 1–4. Expression constructs covering the 3rd 8-Cys repeats of LTBPs (LTBP-1: pJS-4, LTBP-2: pJS-24, LTBP-3: pJS-34Δ and LTBP-4: pJS-44, illustrated in Figure 1), were transfected together with the three human TGF-β isoforms to 293T cells. The secreted proteins were separated by 7.5% PAGE and detected by immunoblotting with antibodies against β1•LAP (A), TGF-β2 (B), or β3•LAP (C) in order to detect covalent β•LAP - LTBP-fragment complexes (upper panels). The secretion of proteins coded by LTBP constructs was verified by 4–15% gradient PAGE followed by immunoblotting with anti-HA antibody (lower panels). Brackets on the right indicate the free and complexed β•LAP forms. Note the higher molecular weight bands in Figure 4C (lanes 1–3), which most likely represent the unprocessed TGF-β3•LAP, where the TGF-β and LAP parts are not proteolytically processed.
Binding between the 8-Cys Repeat and TGF-β1•LAP Is Mediated by a Direct Disulphide Bond between the Cys-33 of TGF-β1•LAP and the 8-Cys Repeat
The covalent interaction between the 3rd 8-Cys repeat of LTBP-1 and TGF-β1•LAP is dependent on the Cys-33 of β1•LAP (Saharinen et al., 1996) and can be disrupted by reducing agents. The other cysteines 223 and 225 of β1•LAP are required for the dimerization of the β1•LAP (Gentry et al., 1988). It is also known that the cysteine residues in the 8-Cys repeat are all in oxidized form (Gleizes et al., 1996; Reinhardt et al., 1996). However, it is not known whether the cysteine residues required for 8-Cys - β1•LAP complexes are involved in inter- or intramolecular disulphide bridges. In the intermolecular disulphide bridge model, one or both of the Cys-33s of the β1•LAP dimer are forming a disulphide bridge with unknown cysteine(s) of the 8-Cys repeat. In the intramolecular cysteine disulphide bridge model, all the cysteines of the TGF-β binding 8-Cys repeat and β1•LAP are involved in intramolecular disulphide bridges. However, the molecules would be folded in such a way that they would be kept together like two closed circles. The complex would thus resist the nonreducing sample denaturation in SDS-PAGEs.
To analyze β1•LAP - 8-Cys repeat interaction, we prevented the β1•LAP dimerization by mutating the cysteines 223 and 225 to serine residues. This construct was transfected with LTBP-1 or LTBP-2 constructs pJS-4 and pJS-24, respectively, into COS-7 cells. The LTBP-2 construct was used as a control to exclude nonspecific disulphide binding of the mutated TGF-β1 and 8-Cys repeats. The complex formation was analyzed from the conditioned medium by immunoblotting with β1•LAP antibodies (Figure 5). The mutated β1•LAP expressed alone or with the LTBP-2 construct was not detected in β1•LAP immunoblots due to the inability of the antibodies to recognize monomeric β1•LAP, possibly due to misfolding of monomeric β1•LAP, as observed from an immunoblot using reduced conditioned medium of cells transfected with wild-type TGF-β1 (our unpublished results). The mutated β1•LAP protein was found to retain the ability to form covalent LTBP-1 complexes. This interaction recruited both copies of the monomeric β1•LAP, as indicated by both the observed mobility of the complex and by its detection in the immunoblot. These results suggest that the interaction between β1•LAP and LTBP-1 is mediated by two direct cysteine disulphide bridges between the molecules (see proposed structure in Figure 5).
Chimeric Constructs between LTBP-1 and LTBP-2 Suggests the Motif Required for TGF-β•LAP Binding of the 8-Cys Repeats
In order to analyze the regions in 8-Cys repeats providing the TGF-β•LAP binding ability, we made chimeric constructs between the 3rd 8-Cys repeats of LTBP-1 and LTBP-2. In each chimeric construct, the amino acid residues between two successive cysteine residues of the 3rd 8-Cys repeat of LTBP-1 were changed to those of the analogous region in LTBP-2 (Figure 6A). These constructs were transfected with TGF-β1 cDNA to 293T cells, and the secretion of covalent β1•LAP - LTBP complexes to conditioned medium was detected by immunoblotting. The protein coded by the chimeric construct L1ΔL2–4, in which the region between the 6th and 7th Cys-residues was replaced, was unable to covalently associate with β1•LAP. All other proteins encoded by the chimeric constructs retained the ability to associate with β1•LAP.
To verify that the observed loss of β1•LAP binding function of construct L1ΔL2–4 is not due to misfolding of the 8-Cys repeat, we made a chimeric construct L1ΔL4–4, analogous to L1ΔL2–4, in which the sequence between the 6th and 7th cysteine residues was replaced with analogous sequence from LTBP-4 (see Figure 6B). The ability of the protein coded by this construct to associate covalently with β1•LAP was analyzed as with the L1ΔL2 constructs. Unlike the construct L1ΔL2–4 encoded protein, the protein coded by chimeric construct L1ΔL4–4 between LTBP-1 and LTBP-4 yielded covalent complexes with β1•LAP (Figure 6B) with almost the same efficiency as with the protein coded by wild-type LTBP-1 construct, supporting the importance of the region between the 6th and 7th cysteine residues as the region distinguishing TGF-β binding and nonbinding type 8-Cys repeats
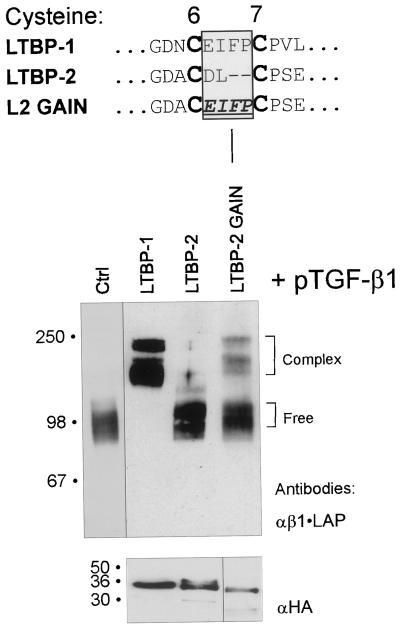
Construction of a Gain of TGF-β•LAP Binding Form of LTBP-2
The function of the critical TGF-β binding region of 8-Cys repeats was further analyzed by creating a gain-of-function type cDNA construct, L2GAIN, in which the backbone was the non-TGF-β•LAP binding LTBP-2 construct pJS-24. The two amino acids between the 6th and 7th Cys-residues were replaced by the analogous four amino acid peptide from LTBP-1 (see Figure 7). This construct was then transfected to 293T cells together with TGF-β1 cDNA, and the conditioned medium was analyzed as above. Although the secretion level of the chimeric L2GAIN construct encoded protein was somewhat lower than that of protein from both LTBP-1 and LTBP-2 wild-type construct (Figure 7), the protein encoded by L2GAIN construct was capable of complex formation with β1•LAP, providing the gain of TGF-β1•LAP binding function.
Figure 7.
Replacement of the region between the 6th and 7th cysteine residues in the 3rd 8-Cys repeat of LTBP-2 with that of LTBP-1 results in gain-of-function of TGF-β-β1•LAP binding. Wild-type LTBP -1 and -2 constructs, pJS-4 and pJS-24, respectively, as well as construct L2GAIN were transfected together with TGF-β1 cDNA to 293T cells. The proteins from conditioned medium were separated by 7.5% PAGEs and analyzed for covalent complex formation between the proteins coded by LTBP constructs and β1•LAP. Brackets on the right indicate the free or complexed β1•LAP forms. Secretion of the proteins coded by LTBP constructs was verified by 4–15% gradient PAGEs followed by immunoblotting with anti-HA antibody (lower panel).
Multiple Sequence Alignment Distinguishes between the TGF-β Binding and Nonbinding Types of 8-Cys Repeats
In order to elucidate which 8-Cys repeats contain the TGF-β binding motif and to study the relatedness of the 8-Cys repeats, a multiple sequence alignment as well as a dendrogram of the 8-Cys repeats were done. For simplicity, the hybrid domains were excluded from the figures as well as all the other 8-Cys repeats of fibrillins, except the 7th and 8th 8-Cys repeats, which are the most similar to the 8-Cys repeats in LTBPs.
In a dendrogram of the 8-Cys repeats (Figure 8A), the 3rd 8-Cys repeats of LTBPs -1 and -3 are most similar to each other, with the weaker TGF-β•LAP binding 3rd 8-Cys repeat of LTBP-4 being their closest relative. The non-TGF-β binding type 3rd 8-Cys repeat of LTBP-2, as well as the 8th 8-Cys repeats of fibrillins are more distant. It should also be noted that all other 8-Cys repeats of LTBPs are even more distant to the 3rd 8-Cys repeats of LTBPs than the 8th 8-Cys repeat of fibrillins. The primary sequences of the 8-Cys repeats, and especially the 3rd 8-Cys repeats of LTBPs and the 8th 8-Cys of fibrillins are quite conserved (Figure 8B). However, the three TGF-β binding type 8-Cys repeats, namely the 3rd 8-Cys repeats of LTBPs -1, -3, and -4 differ clearly from all other 8-Cys repeats, in the respect of the insertion of two additional amino acid residues between the 6th and 7th Cys-residues. This TGF-β binding motif is not present in any other 8-Cys repeats or hybrid domains of LTBPs or fibrillins, including the ones excluded from Figure 8B.
Molecular Models of the 3rd 8-Cys Repeats of Human LTBPs -1, -2, and -3 Give Insight into the Structural Features Required for the Covalent Binding to TGF-β1•LAP
We used molecular modeling to analyze the structural implications of the presence of the TGF-β binding motif in 8-Cys repeats. We modeled the 3rd 8-Cys repeats from the human LTBPs -1 and -2, as well as the 8-Cys repeats having the modifications as in constructs L1ΔL2 1–5 and L2GAIN using the NMR structure of the 8th 8-Cys repeat of fibrillin-1 as template (Yuan et al., 1997).
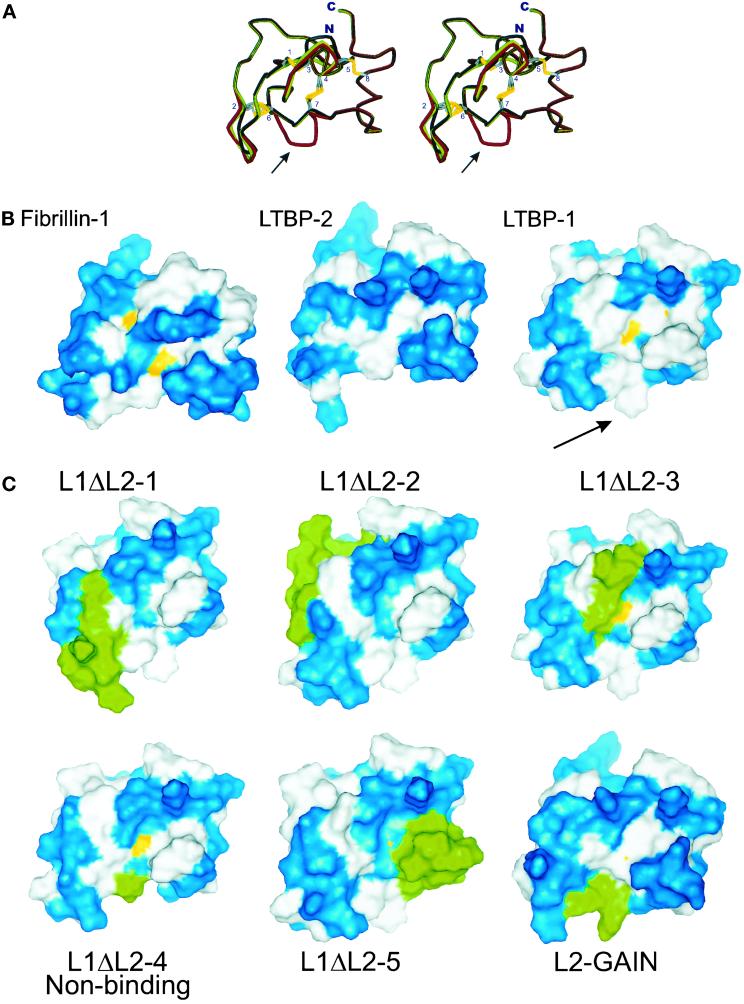
The backbone of the previously published structure of the 8th 8-Cys repeat of fibrillin is aligned with the models for the 3rd 8-Cys repeat of LTBPs -1 and -2 in the stereo image (Figure 9A). The largest difference in the backbone alignment was in the TGF-β binding determinant region between the 6th and 7th cysteine residues of the model for the 3rd 8-Cys repeat of LTBP-1. Within this region, the insertion of two amino acids caused a loss of a small β-strand and introduced a bending in the structure (indicated by an arrow in Figure 9A). This bend resulted in a loss of altogether three hydrogen bonds that are present both in the structure for the 8th 8-Cys repeat of fibrillin and in the model for the 3rd 8-Cys repeat of LTBP-2. These hydrogen bonds are formed between the Asp/Glu and second Cys-residues in the non-TGF-β binding motif [DE]-L located between cysteines 6 and 7 (see Figure 8B) and with the side chains of the amino acids between cysteines 5 and 6. The disulphide bridges between cysteines 4 and 7 in both the structure for the 8th 8-Cys repeat of fibrillin-1 and the model for the 3rd 8-Cys repeat of LTBP-1 were exposed to solvent (Figure 9B). In the model for the 3rd 8-Cys repeat of LTBP-2, this disulphide bridge was buried. In the model for the 3rd 8-Cys repeat of LTBP-1, the surface hydrophobicity was increased considerably, as compared with the non-TGF-β binding models. The whole side of the 3rd 8-Cys repeat of LTBP-1 was hydrophobic and especially, the hydrophobic region caused by the exposed aromatic side chain of the phenylalanine residue in the region between 6th and 7th cysteine residues is easily noted (Figure 9B, indicated by an arrow).
Figure 9.
Molecular modeling of 8-Cys repeats. A) Stereoimage of the superpositioned structure of the 8th 8-Cys repeat of fibrillin-1 and the models for the 3rd 8-Cys repeat of LTBPs -1 and -2. The backbone of the previously published structure of the 8th 8-Cys repeat of fibrillin (Yuan et al., 1997) is illustrated in black, the molecular models for the 3rd 8-Cys repeat of LTBP-1 and LTBP-2 in red and green, respectively. The side chains of cysteine residues are shown with the numbering referring to the ordinals of the cysteine residues. At the bottom of the illustration is the region between the 6th and 7th Cys-residues, where the backbone alignment between the TGF-β binding and nonbinding type 8-Cys repeats is lost (indicated by an arrow). B) The solvent accessible surfaces for the models of the 3rd 8-Cys repeats of LTBPs. The structures of TGF-β binding (LTBP-1) and nonbinding type (Fibrillin-1, LTBP-2) 8-Cys repeats are shown. Blue: hydrophilic amino acids; white: hydrophobic amino acids; yellow: SH groups of cysteine residues exposed to solvent. The hydrophobic area caused by the insertion of two amino acid between the 6th and 7th Cys-residues of the TGF-β binding type 8-Cys repeat of LTBP-1 is marked by an arrow. C) Solvent accessible surfaces of the models coded by L1ΔL2 1–5 and L2GAIN constructs. The structures of the 8-Cys repeat chimeras, described in Figure 6A, 6B, and 7, were modeled. The modified amino acids in these models are shown in green, hydrophobic in white, hydrophilic in blue and the SH-groups of Cys-residues in yellow.
We modeled also the 8-Cys repeats coded by the chimeric constructs L1ΔL2 1–5 and L2GAIN (Figure 9C). The sulfhydryl groups of the disulphide bridge between cysteine residues 4–7 were less exposed in some of the chimeric models than in the wild-type 3rd 8-Cys repeat of LTBP-1 model. However, the increased surface hydrophobicity caused by the introduction of an aromatic amino acid between the 6th and 7th cysteine of the chimeric models correlated with the TGF-β•LAP binding ability. The three hydrogen bonds involving the residues between the 6th and 7th cysteines were present in the non-TGF-β biding type 8-Cys repeats coded by construct L1ΔL2–4, while they were lost in all the models of TGF-β binding chimeric 8-Cys repeats. Taken together, the models for TGF-β binding and nonbinding type 8-Cys repeats suggest the interaction with the β•LAP takes place via a hydrophobic interaction.
DISCUSSION
In the present study we have focused on the molecular mechanism of association of TGF-β•LAP with the 8-Cys repeats of its binding proteins LTBPs as well as their relatives, fibrillins. We and others have previously shown that TGF-β1•LAP associates covalently with the 3rd 8-Cys repeat of LTBP-1 (Saharinen et al., 1996, Gleizes et al., 1996). This association was the first-ever characterized function for these novel protein domains. However, interesting questions remained, namely what determines the TGF-β1•LAP binding ability of the certain 8-Cys repeat of LTBP-1, are the other TGF-β isoforms capable of the similar covalent association, and whether the 8-Cys repeats of the other members of the LTBP-fibrillin family have similar capability to associate with TGF-β•LAP. In the current study we have analyzed the abilities of a large number of 8-Cys repeats to associate with TGF-β. We found that of the members of LTBP-fibrillin family, only LTBPs -1 and -3, and to lesser extent LTBP-4, were able to associate with β•LAP via their 3rd 8-Cys repeats. Although there are 9 copies of the 8-Cys repeats in both fibrillins, none of those tested was able to interact with β•LAP. Unexpectedly, we also found that LTBP-2 was unable to form this interaction. Experiments to detect any noncovalent associations between β1•LAP and LTBP-2 or fibrillin-1 were also negative, phenomena known with other growth factors, like HGF and FGFs and their respective binding proteins (reviewed in Taipale and Keski-Oja, 1997). These results show that although there are altogether 34 known 8-Cys repeats in the LTBP-fibrillin family with quite conserved sequences, only a minor subset of them was able to associate with β•LAP. The inability of LTBP-2 to associate with β•LAP supports the suggested function of LTBP-2 as being an integral microfibrillar protein and functionally more related to fibrillins than the other LTBPs (Gibson et al., 1995; Bashir et al., 1996; Gibson et al., 1995; Hyytiäinen et al., 1998). Taken together, these results encourage search for other functions for the majority of the abundant 8-Cys repeats of LTBPs and fibrillins. The Marfan syndrome is a genetic disorder resulting from mutations in fibrillin-1 gene. Several Marfan cases have been reported to result from mutations in the 8-Cys repeats (reviewed in Ramirez et al., 1999), emphasizing other biological functions for the 8-Cys repeats.
All three mammalian β•LAP isoforms were able to associate with the 3rd 8-Cys repeats of LTBPs -1 and -3, while β1•LAP, but not the other β•LAP isoforms, could only weakly associate with LTBP-4. Since multiple LTBPs are expressed in many tissues, the weak interaction between β1•LAP and LTBP-4 is most likely often overcome in vivo by the simultaneous expression of LTBPs -1 or -3 and their efficient interaction with β•LAPs. Thus LTBP-4 seems to have a less important role in depositing TGF-β to the extracellular matrix than LTBPs -1 and -3. Interestingly, large proportions of LTBP-4 have been found to contain heterologous protein(s) associated with its 3rd 8-Cys repeat (Saharinen et al., 1998), and recently we have identified an alternatively spliced form of LTBP-4 from many different tissues, lacking the 3rd 8-Cys repeat, which thus provides a new regulatory mechanism for the deposition of the heterologous protein(s) associated with its 8-Cys repeat (Koli, Saharinen, Kärkkäinen, and Keski-Oja, unpublished data).
The interaction between β1•LAP and the 3rd 8-Cys repeat of LTBP-1 involves oxidized cysteine residues in the 8-Cys repeat and requires the Cys-33 residue of β1•LAP (Saharinen et al., 1996). We found that the interaction between the Cys-33 of β•LAP and the 8-Cys repeat is a direct disulphide bond reshuffling, which involves both of the Cys-33s of β•LAP dimer and yet unknown cysteines in the 8-Cys repeat. The interaction of an 8-Cys repeat with β•LAP thus results in a rearrangement of at least one disulphide bridge in the 8-Cys repeat. Since all TGF-β isoforms contain cysteines at analogous positions, one might expect that all β•LAP - LTBP interactions are mechanistically similar to the β1•LAP - LTBP-1 interaction studied here. The question whether the other growth factors of the TGF-β superfamily could be covalently deposited to the ECM by an analogous mechanism via an interaction with the 8-Cys repeats seems thus to include the requirement of a cysteine residue in their propeptide parts analogous to the Cys-33 of β1•LAP.
The sequence conservation between the 8-Cys repeats of fibrillins and LTBPs, especially among the 3rd 8-Cys repeats of LTBPs and 8th 8-Cys repeats of fibrillins is quite high. In addition to the identical patterning of the cysteine-residues, also several other residues are conserved or replaced by similar residues among these repeats. Using a chimeric protein approach, we localized the area between the 6th and 7th Cys-residues of a TGF-β binding 8-Cys repeat as a determinant of the binding ability. Replacement of this motif by the analogous region in the 3rd 8-Cys of LTBP-2 in construct L1ΔL2–4 disrupted the association with β•LAP. The motif was also verified by using chimeric constructs L1ΔL4–4 and L2 GAIN, which both had β1•LAP binding ability. Notably, the efficient TGF-β•LAP binding of LTBPs -1 and -3 could be explained by the conserved sequence between the 6th and 7th cysteine residues of the 3rd 8-Cys repeat, coded by sequences EIFP and EIYP, respectively. The same TGF-β binding motif is conserved between all known mammalian LTBPs -1 and -3, whereas in LTBP-4, which possesses a much weaker TGF-β•LAP binding ability, this sequence is more divergent (RIQQ). Replacement of the wild-type sequence EIFP in the 3rd 8-Cys repeat of LTBP-1 with that of LTBP-4, resulted in slightly decreased β1•LAP binding ability. We thus propose that the 8-Cys repeats can be divided to TGF-β binding and nonbinding types, based both on the presence of the inserted sequence motif between the 6th and 7th Cys-residues and high sequence similarity to the 3rd 8-Cys repeats of LTBPs.
After the mapping of the TGF-β binding motif of the 8-Cys repeats, the question remained how this short motif contributes to TGF-β binding ability. We used molecular modeling to explore the actual structural changes caused by the small TGF-β binding motif on the 8-Cys repeats. Although the obtained models are somewhat speculative because of the lack of experimental structural information of the TGF-β binding type 8-Cys repeats, the models suggest a clear difference between TGF-β binding and nonbinding types. An increased hydrophobic surface, extending from the vicinity of the TGF-β binding motif was present in the TGF-β binding type 8-Cys repeats. This hydrophobic surface was not present in any of the models for non-TGF-β binding type 8-Cys repeats. Instead, the hydrophobic area was present in all of the modified TGF-β binding type models of the 8-Cys repeats. One of the TGF-β binding type 8-Cys repeat, namely the 3rd 8-Cys repeat of LTBP-1 has been previously modeled using the structure of the 8th 8-Cys repeat of fibrillin-1 as template (Yuan et al., 1997). Although the FP-insertion between cysteines 6 and 7 in that 8-Cys repeat was not included in their model, the similar increased surface hydrophobicity along the 8-Cys repeat was observed and the authors speculated of the potential importance of the change in surface hydrophobicity as well as the FP-insertion in binding to β•LAP. Structural information of the β•LAP is not available, however, the present findings strongly suggest that the association of β•LAP with the TGF-β binding type 8-Cys repeats in the secretory pathway involves hydrophobic interactions. The possible hydrophobic interaction site of β•LAP could cause the inefficient secretion and misfolding, observed for small latent TGF-β•LAP, in the absence of TGF-β binding LTBPs (Miyazono et al., 1991, Miyazono et al., 1992, Mizoi et al., 1993). Although the interaction with TGF-β is formed by disulphide bridges, the availability of the SH-groups, as deduced from the models, did not correlate with the TGF-β binding ability of the 8-Cys repeats, suggesting the possibility that the fold undergoes structural changes upon interacting with β•LAP. The models also showed that the non-TGF-β binding type 8-Cys repeat possesses three hydrogen bonds, which stabilize the fold and may thus hinder the association with TGF-β•LAP. The lack of these hydrogen bonds in the TGF-β binding type 8-Cys repeats may be important for these domains making them more flexible and thus allowing them to adopt a new conformation upon associating with TGF-β•LAP in the secretory pathway. Together the molecular models suggest that the increased surface hydrophobicity of the TGF-β binding types of 8-Cys repeats together with more flexible structure is critical for their ability to interact with the TGF-β•LAP.
The deposition of TGF-β in the extracellular matrix in a latent form has wide biological implications. TGF-β itself is a multipotent growth factor, which employs the unique postsecretory latency concept. The LTBPs, as being required for the correct folding and secretion of TGF-β, target TGF-β to extracellular structures. Our results show that the storage of TGF-β is dependent on a specific sequence determinant on the surface of the 8-Cys repeats of TGF-β binding LTBPs. The accumulation of TGF-β to extracellular microfibrillar structures ensures that the whole TGF-β system is capable of providing focused and fast responses via activation of the stored growth factor.
ACKNOWLEDGMENTS
We thank Prof. Mark Johnson (Department of Biochemistry, Åbo Academi), Prof. Kari Alitalo (Cancer/Molecular Biology Laboratory, Haartman Institute, University of Helsinki), Dr. Juha Rouvinen (Department of Chemistry, University of Joensuu) and Dr. Marc Baumann (Institute of Biomedicine, University of Helsinki) for critical comments, and Drs. Kimmo Mattila (Center for Scientific Computing, Espoo, Finland) and Sakari Jokiranta (Department of Bacteriology and Immunology, Haartman Institute, University of Helsinki) for advice in the molecular modeling as well as Sami Starast for fine technical assistance.
This work has been supported by The Academy of Finland, Sigrid Juselius Foundation, Finnish Cultural Foundation, Finnish Cancer Organizations, Maud Kuistila Foundation, Instrumentarium Science foundation, Ella and Georg Ehrnrooth Foundation, Novo Nordisk Foundation, Paulo Foundation, Ida Montin Foundation, Biocentrum Helsinki, Helsinki University Central Hospital, HUCH Institute, Oskar Öflund Foundation, and the University of Helsinki.
Footnotes
The 8-Cys repeats of LTBPs and fibrillins are also known as TGF-β binding protein-like domains (TB-domains) and as cysteine-rich domains (CR-domains).
REFERENCES
- Bashir MM, Han MD, Abrams WR, Tucker T, Ma RI, Gibson M, Ritty T, Mecham R, Rosenbloom J. Analysis of the human gene encoding latent transforming growth factor-β-binding protein-2. Int J Biochem Cell Biol. 1996;28:531–542. doi: 10.1016/1357-2725(95)00167-0. [DOI] [PubMed] [Google Scholar]
- Dallas SL, Park-Snyder S, Miyazono K, Twardzik D, Mundy GR, Bonewald LF. Characterization and autoregulation of latent transforming growth factor-β (TGF-β) complexes in osteoblast-like cell lines: production of a latent complex lacking the latent TGF-β-binding protein. J Biol Chem. 1994;269:6815–6821. [PubMed] [Google Scholar]
- D'Arrigo C, Burl S, Withers AP, Dobson H, Black C, Boxer M. TGF-β1 binding protein-like modules of fibrillin-1 and -2 mediate integrin-dependent cell adhesion. Connect Tissue Res. 1998;37:29–51. doi: 10.3109/03008209809028898. [DOI] [PubMed] [Google Scholar]
- Davis CG. The many faces of epidermal growth factor repeats. New Biol. 1990;2:410–419. [PubMed] [Google Scholar]
- Dubois CM, Laprise MH, Blanchette F, Gentry LE, Leduc R. Processing of transforming growth factor-β 1 precursor by human furin convertase. J Biol Chem. 1995;270:10618–10624. doi: 10.1074/jbc.270.18.10618. [DOI] [PubMed] [Google Scholar]
- Eklöv S, Funa K, Nordgren H, Olofsson A, Kanzaki T, Miyazono K, Nilsson S. Lack of the latent transforming growth factor-β binding protein in malignant, but not benign prostatic tissue. Cancer Res. 1993;53:3193–3197. [PubMed] [Google Scholar]
- Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor-β to the mature polypeptide. Mol Cell Biol. 1988;8:4162–4168. doi: 10.1128/mcb.8.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MA, Hatzinikolas G, Davis EC, Baker E, Sutherland GR, Mecham RP. Bovine latent transforming growth factor-β1-binding protein 2: molecular cloning, identification of tissue isoforms, and immunolocalization to elastin-associated microfibrils. Mol Cell Biol. 1995;15:6932–6942. doi: 10.1128/mcb.15.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-β binding protein-1 that mediates bonding to the latent transforming growth factor-β1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- Gray A, Mason A. Requirement for activin A and transforming growth factor-β1 pro-regions in homodimer assembly. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- Hyytiäinen M, Taipale J, Heldin CH, Keski-Oja J. Recombinant latent transforming growth factor-β binding protein 2 assembles to fibroblast extracellular matrix and is susceptible to proteolytic processing and release. J Biol Chem. 1998;273:20669–20676. doi: 10.1074/jbc.273.32.20669. [DOI] [PubMed] [Google Scholar]
- Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin CH. TGF-β1 binding protein: a component of the large latent complex of TGF-β1 with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- Kingsley D. The TGF-β superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- Koski C, Saharinen J, Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-β binding protein-1 (LTBP-1) in a cell type-specific manner. J Biol Chem. 1999;274:32619–32630. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- Mangasser-Stephan K, Gressner AM. Molecular and functional aspects of latent transforming growth factor-β binding protein: just a masking protein? Cell Tissue Res. 1999;297:363–370. doi: 10.1007/s004410051364. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-β1-binding protein in the assembly and secretion of TGF-β1. EMBO J. 1991;10:1091–2101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Thyberg J, Heldin CH. Retention of the transforming growth factor-β1 precursor in the Golgi complex in a latent endoglycosidase H-sensitive form. J Biol Chem. 1992;267:5668–5675. [PubMed] [Google Scholar]
- Mizoi T, Ohtani H, Miyazono K, Miyazawa M, Matsuno S, Nagura H. Immunoelectron microscopic localization of transforming growth factor-β1 and latent transforming growth factor-β1 binding protein in human gastrointestinal carcinomas: qualitative difference between cancer cells and stromal cells. Cancer Res. 1993;53:183–190. [PubMed] [Google Scholar]
- Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, ten Dijke P, Miyazono K, Heldin CH. Identification and characterization of L.T.B.P-2, a novel latent transforming growth factor-β binding protein. J Biol Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- Munger JS, Harpel JG, Gleizes PE, Mazzieri R, Nunes I, Rifkin DB. Latent transforming growth factor-β: structural features and mechanisms of activation. Kidney Int. 1997;51:1376–1382. doi: 10.1038/ki.1997.188. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Reinhardt DP, Sakai LY, Timpl R. Cell adhesion and integrin binding to recombinant human fibrillin-1. FEBS Lett. 1996;384:247–250. doi: 10.1016/0014-5793(96)00325-0. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Gayraud B, Pereira L. Marfan syndrome: new clues to genotype-phenotype correlations. Ann Med. 1999;31:202–207. doi: 10.3109/07853899909115979. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Pereira L. The fibrillins. Int J Biochem Cell Biol. 1999;31:255–259. doi: 10.1016/s1357-2725(98)00109-5. [DOI] [PubMed] [Google Scholar]
- Reinhardt DP, Keene DR, Corson GM, Poschl E, Bachinger HP, Gambee JE, Sakai LY. Fibrillin-1: organization in microfibrils and structural properties. J Mol Biol. 1996;258:104–116. doi: 10.1006/jmbi.1996.0237. [DOI] [PubMed] [Google Scholar]
- Roberts A, Sporn M. The Molecular and Cellular Biology of Wound Repair, ed R. Clark. New York: Plenum Press; 1996. Transforming growth factor-β; pp. 275–308. [Google Scholar]
- Saharinen J, Hyytiäinen M, Taipale J, Keski-Oja J. Latent transforming growth factor-β -binding proteins (LTBPs): structural extracellular matrix proteins for targeting TGF-β action. Cytokine Growth Factor Rev. 1999;10:99–117. doi: 10.1016/s1359-6101(99)00010-6. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-β with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–353. [PMC free article] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new latent transforming growth factor-β binding protein, LTBP-4. J Biol Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol. 1986;103:2499–2509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Broekelmann T, Cheresh DA, Ramirez F, Rosenbloom J, Mecham RP. Cell-type specific recognition of RGD- and non-RGD-containing cell binding domains in fibrillin-1. J Biol Chem. 1996;271:4916–4922. [PubMed] [Google Scholar]
- Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-β1 associates to fibroblast extracellular matrix via latent TGF-β binding protein. J Cell Biol. 1994;124:171–181. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Saharinen J, Keski-Oja J. Extracellular matrix-associated transforming growth factor-β: role in cancer cell growth and invasion. Adv Cancer Res. 1998;75:87–134. doi: 10.1016/s0065-230x(08)60740-x. [DOI] [PubMed] [Google Scholar]
- Trask TM, Ritty TM, Broekelmann T, Tisdale C, Mecham RP. N-terminal domains of fibrillin 1 and fibrillin 2 direct the formation of homodimers: a possible first step in microfibril assembly. Biochem J. 1999;340:693–701. [PMC free article] [PubMed] [Google Scholar]
- Yin W, Fang J, Smiley E, Bonadio J. 8-Cysteine TGF-BP structural motifs are the site of covalent binding between mouse LTBP-3, LTBP-2, and latent TGF-β1. Biochim Biophys Acta. 1998a;1383:340–350. doi: 10.1016/s0167-4838(98)00003-x. [DOI] [PubMed] [Google Scholar]
- Yin W, Smiley E, Bonadio J. Alternative splicing of LTBP-3. Biochem Biophys Res Commun. 1998b;245:454–458. doi: 10.1006/bbrc.1998.8456. [DOI] [PubMed] [Google Scholar]
- Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor-β binding protein gene (LTBP-3) J Biol Chem. 1995;270:10147–10160. doi: 10.1074/jbc.270.17.10147. [DOI] [PubMed] [Google Scholar]
- Yuan X, Downing AK, Knott V, Handford PA. Solution structure of the transforming growth factor-β binding protein-like module, a domain associated with matrix fibrils. EMBO J. 1997;16:6659–6666. doi: 10.1093/emboj/16.22.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Apfelroth SD, Hu W, Davis EC, Sanguineti C, Bonadio J, Mecham RP, Ramirez F. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J Cell Biol. 1994;124:855–863. doi: 10.1083/jcb.124.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]