Abstract
Translation of picornavirus RNAs is mediated by internal ribosomal entry site (IRES) elements and requires both standard eukaryotic translation initiation factors (eIFs) and IRES-specific cellular trans-acting factors (ITAFs). Unr, a cytoplasmic RNA-binding protein that contains five cold-shock domains and is encoded by the gene upstream of N-ras, stimulates translation directed by the human rhinovirus (HRV) IRES in vitro. To examine the role of Unr in translation of picornavirus RNAs in vivo, we derived murine embryonic stem (ES) cells in which either one (−/+) or both (−/−) copies of the unr gene were disrupted by homologous recombination. The activity of picornaviral IRES elements was analyzed in unr+/+, unr+/−, and unr−/− cell lines. Translation directed by the HRV IRES was severely impaired in unr−/− cells, as was that directed by the poliovirus IRES, revealing a requirement for Unr not previously observed in vitro. Transient expression of Unr in unr−/− cells efficiently restored the HRV and poliovirus IRES activities. In contrast, the IRES elements of encephalomyocarditis virus and foot-and-mouth-disease virus are not Unr dependent. Thus, Unr is a specific regulator of HRV and poliovirus translation in vivo and may represent a cell-specific determinant limiting replication of these viruses.
After infection of a susceptible cell, translation of the picornavirus plus-strand RNA genome is controlled by the internal ribosome entry site (IRES). This cis-regulatory RNA element of about 450 nucleotides folds into complex and highly conserved secondary structures and facilitates translation by direct binding of ribosomes to an internal site of the viral RNA. IRES elements have been found in various viral RNAs and also in cellular mRNAs (for reviews, see references 1 and 16).
Internal initiation of picornavirus translation seems to require all canonical eukaryotic initiation factors (eIFs) also involved in cellular cap-dependent translation (28, 33), except for the actual cap-binding protein eIF4E. For the majority of eukaryotic mRNAs that are translated by a ribosome-scanning mechanism (13, 18, 38), the initiation of translation is the most important point of regulation in the overall process of protein synthesis (29). Modulation of the activity of these eIFs alters the general rate of protein synthesis, and the signal transduction routes leading to the eIFs are becoming clear now (36), supporting the idea that translational control substantially contributes to the regulation of gene expression in eukaryotes.
For picornaviruses, the initiation of translation also appears to be a major point of control. In vitro studies revealed that additional noncanonical translation initiation factors are involved in picornavirus IRES-mediated translation (1, 41). Accordingly, there is significant genetic evidence that picornavirus IRES elements contain determinants of cell specificity. Analysis of poliovirus IRES mutants showed that translation defects could be cell type specific, since a decreased translation capacity of mutant templates was evident in cell extracts of neuronal origin, but not in HeLa cells (26). Cell-specific determinants were recently demonstrated to exist in the poliovirus 5′-untranslated region (UTR) by experiments using viruses with chimeric genomes. When the poliovirus IRES was replaced with that of human rhinovirus (HRV), neuropathogenicity in a mouse model was abrogated (15). The differential translation of wild-type and attenuated Sabin vaccine strains of poliovirus in different cell types indicates that cellular factors influencing translational activity may be differentially expressed, consistent with the idea that factors distinct from the standard initiation factors determine picornavirus translation efficiency (for review, see reference 1). The general idea that IRES elements may allow fine-tuning of gene expression was also supported for cellular IRES elements. The ornithine decarboxylase and p58PISTLRE IRES elements specifically function during the G2/M period (8, 37), the Ultrabithorax and Antennapedia IRES activities exhibit a high degree of developmental regulation in transgenic Drosophila (46), and stage- and tissue-specific modulation of FGF2 and c-myc IRES activities was observed in transgenic mice (9, 10). Although the molecular basis for cell-specific activity of IRES elements is not known, it is likely that cell type-specific proteins are involved in modulation of both picornaviral and cellular IRES-directed translation.
Five cellular trans-acting factors (ITAFs) specific for picornaviral IRES elements have been identified, four of which are RNA-binding proteins: the polypyrimidine tract-binding protein (PTB) (24, 25, 32); the poly(rC)-binding protein 2 (PCBP2) (3, 12, 44); the autoantigen La (30); the Unr protein (20), an RNA-binding protein with five cold shock domains that is encoded by the gene upstream of N-ras (11, 23, 27); and ITAF45, a cell-cycle-dependent protein homologous to Mpp1 (murine proliferation-associated protein) (34). Interestingly, functional in vitro assays revealed that some picornavirus IRES elements require a specific combination of two or three of these ITAFs for an efficient translational activity: PTB plus ITAF45 for the IRES of foot-and-mouth disease virus (FMDV) (34), PTB plus PCBP2 for the poliovirus IRES (21), and PTB plus Unr plus PCBP2 for the HRV IRES (20). An attractive hypothesis is that specific translation initiation factors assembled on IRES elements regulate translation initiation in the same way as specific transcription factors bound to promoters regulate transcription (40). An exception is the encephalomyocarditis virus (EMCV) IRES, which appears to depend only on PTB in vitro. PTB is an abundant and ubiquitously expressed protein and is not expected to confer cell-specific translational regulation alone. Accordingly, the EMCV IRES displays a broad range of activities in different cell lines (5) and in transgenic mice (10).
Very few studies have focused on the in vivo role of ITAFs in the regulation of viral or cellular IRES activities. In this study, we have developed an in vivo approach to evaluate the significance of unr's contribution to IRES-mediated translation. We generated mouse embryonic stem (ES) cell lines with gene-targeted deletion of unr and examined whether the unr−/− cells are defective in internal initiation of translation directed by picornaviral IRES elements. The advantage of this strategy is that primary cells are used, providing a physiological environment for the analysis of regulatory interactions that is as close as possible to the natural conditions. Thus, this strategy relies on the effect of endogenous cellular unr, but not on unr overexpressed in cell lines. We report here that the type I IRES-dependent translation of HRV and poliovirus RNA is strongly dependent on Unr expression in ES cells. In contrast, the activities of the type II IRES elements of EMCV and FMDV IRES elements were not affected by the absence of Unr expression. These results indicate that Unr functions as a specific regulator of HRV and poliovirus IRES-directed translation in vivo.
MATERIALS AND METHODS
ES cell culture.
ES cells were grown in Dulbecco's modified Eagle's medium supplemented with 15% heat-inactivated fetal bovine serum, 2 mM glutamine, 150 μM monothioglycerol (Sigma), and 1,000 U of leukemia inhibitory factor per ml (ESGRO).
Generation of unr−/− cell clones.
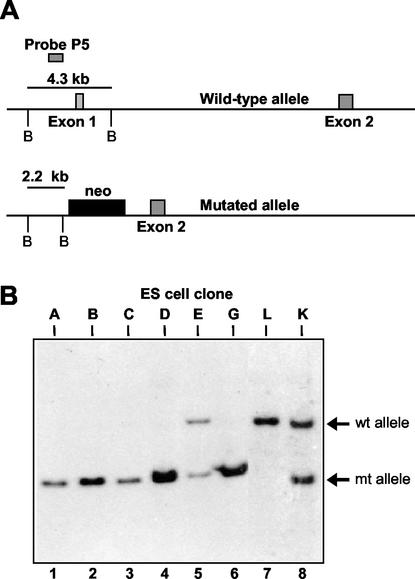
Heterozygous unr+/− mice were intercrossed, and ES cell lines were established from individual blastocyst-stage embryos as described previously (39). Briefly, blastocysts were cultured on irradiated feeders in a 24-well tissue culture well in ES media. After 4 to 6 days blastocysts were picked, disaggregated, and redispersed onto fresh feeders (1 well of a 24-well dish). Undifferentiated colonies that grew were picked 1 to 2 weeks later and replated on fresh feeders. Exponentially growing cells were passaged, grown to near confluency, and frozen. The genotype was determined by Southern blotting with a unr probe corresponding to exon 1. This probe detects 4.3- and 1.6-kb bands representing the 5′ ends of the wild-type and mutant alleles, respectively.
RNA isolation and analysis.
To eliminate possible contamination of the ES cells with feeder cells, ES cells were trypsinized, seeded on gelatinized plates, and incubated for 60 min at 37°C. Nonadherent ES cells were then harvested for RNA extraction. Total RNA was prepared according to the guanidium-thiocyanate-CsCl protocol (45). RNase protection assays were carried out as previously described (6), with the murine unk probe (unr, N-ras, Ki-ras). Northern blot analysis was performed as described previously (42). Briefly, 15 μg of total RNA per lane was electrophoresed through 1.2% agarose gels containing formaldehyde and transferred in 150 mM NH4Ac onto an uncharged nylon membrane (Amersham Hybond N). Hybridization was performed overnight in 50% formamide at 42°C for DNA probes and at 60°C for RNA probes. Signals were detected and quantified with a Fuji Bioimager (BAS 1000). The Rluc probe was a 960-bp HindIII-BamHI fragment corresponding to the Renilla luciferase coding sequence. The Fluc probe was a 600-bp NcoI-EcoRI fragment corresponding to the 5′end of the firefly luciferase coding sequence. 32P-radiolabeled probes were generated with the Stratagene random primer labeling kit Prime-It II. The β-actin probe was the 600-bp PstI-Taq-I fragment corresponding to the 5′ end of the murine cDNA cloned into the Bluescript plasmid. Linearized plasmid was used as template for in vitro transcription to generate 32P-radiolabeled probe.
Western blot analysis.
For Western blots, cell lysates were prepared from ES cells made devoid of feeders by incubation on gelatinized plates as described above. Samples were fractionated on 10% denaturing gels, transferred to Immobilon-P membranes (Millipore), and probed with antibodies. Anti-unr rabbit polyclonal antibodies (23) were used at a 1:350 dilution, and anti-Flag M5 and anti-α-tubulin mouse monoclonal antibodies (Sigma) were used at 1:600 and 1:10,000 dilutions, respectively. Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antibodies (Amersham) were used as the secondary antibodies. Blots were developed with the Amersham ECL enhanced chemiluminescence system.
Plasmids.
Most of the dicistronic constructs used in this study have already been described previously (9, 10, 19). The two luciferase open reading frames (ORFs) coding for Renilla luciferase (Rluc) and firefly luciferase (Fluc) are controlled by the cytomegalovirus immediate-early (CMV-IE) promoter and are separated by either a hairpin (pCRHL), the EMCV IRES, or the poliovirus IRES element (pCREL or pCRpolL, respectively). The plasmid pHRVL was derived from pCREL by replacing the EMCV IRES with the HRV-2 IRES between the two luciferase genes. To generate pHRVL, the full-length 5′ UTR sequence of HRV-2 (nucleotides 1 to 612) was excised from pGEM2HRV2 as a SacI-NcoI fragment, and the HRV-2 SacI (blunt ended)-NcoI fragment was ligated together with the NcoI-XhoI firefly luciferase fragment into pCREL previously digested with BamHI (blunt ended)-XhoI. Plasmid pD128 (32) contains downstream of the CMV-IE promoter a dicistronic expression unit composed of the chloramphenicol acetyltransferase (CAT) ORF, the FMDV IRES, and the Fluc ORF. In the control plasmid pDmt, a hairpin was inserted between CAT and Fluc. pSGluc contains the firefly luciferase coding sequence inserted at the BglII site of pSG5 (Stratagene).
To construct pSGFlag-unr, an EcoRI-BclI (blunt ended) Unr cDNA fragment (nucleotides 1008 to 2750) was ligated to EcoRI (blunt ended)-BamHI sites of pSG5 to generate plasmid pSG3′unr. A BamHI-EcoRI fragment, containing the 5′ Unr cDNA (nucleotides 414 to 1008) in which the ATG initiating codon was substituted for by a BamHI site, was isolated from pRSET-Hisunr (43). The EcoRI-BamHI Unr fragment was ligated, along an EcoRI-BamHI Flag epitope tag, to the EcoRI site of pSG3′unr to generate the final construct, pSGFlag-unr. From this plasmid, a full-length unr gene (−exon 5) with a Flag tag added at the amino terminus is expressed.
DNA transfection.
ES cells (105 cells) in six-well pregelatinized petri dishes were routinely transfected with 1.5 μg of plasmid and 4.5 μl of Fugene 6 reagent (Boehringer-Roche) according to the manufacturer's recommendations. Twenty-four hours following transfection, cells were assayed for reporter gene activity. Adherent ES cells were washed with phosphate-buffered saline (PBS), trypsinized, and mixed with the nonadherent cells. Cells were pelleted, washed in PBS, and homogenized in 50 μl of passive lysis buffer (Promega). Luminescence was measured with 5- and 10-μl samples of extract. For cotransfection experiments, 1.5 μg of reporter plasmid and 1.0 μg of unr expression vector (or empty vector) were mixed with 9 μl of Fugene 6 reagent. For dicistronic RNA analysis, 3 × 106 cells were transfected with 10 μg of plasmids and 30 μl of Fugene 6 reagent. Twenty-four hours following transfection, RNA was isolated and subjected to Northern blot analysis.
Luciferase and CAT analysis.
Firefly and Renilla luciferase activities were determined by using the dual-luciferase reporter system (Promega). Luciferase activities were measured with a Lumat LB9501 luminometer (Berthold), and the IRES activity was determined by calculating the Rluc/Fluc ratios. CAT activity in cell extracts was determined by the liquid scintillation method as described previously (7).
RESULTS
Generation of unr−/− ES cell lines.
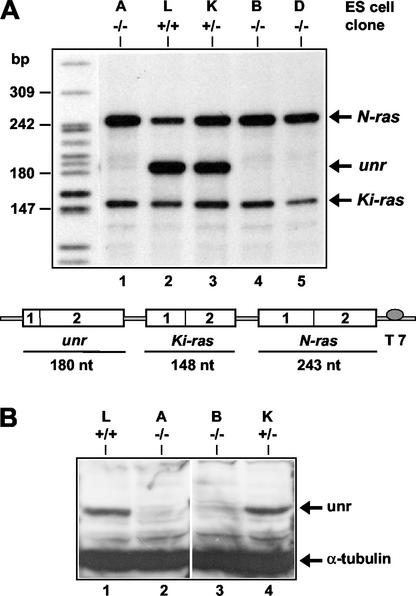
We previously reported the targeted disruption of the unr gene in mouse ES cells and the generation of mice heterozygous for the unr locus (6). Briefly, the targeting vector was designated to encompass the promoter and 5′ end of the unr gene, leading to inactivation of unr transcription. To generate unr−/− ES cell lines, we intercrossed unr+/− mice and isolated individual blastocysts (39). Eight ES cell lines were generated and genotyped by Southern blot analysis. The probe, located in the 5′ end of the unr gene, allowed us to distinguish the wild-type from the mutated unr alleles (Fig. 1A). Five ES clones were unr−/−, two were unr+/−, and one was unr+/+ (Fig. 1B). The absence of unr expression in three unr−/− clones, designated A, B, and D, was confirmed by an RNase protection assay. The probe (Fig. 2A) was designed to simultaneously detect the unr, N-ras and Ki-ras messages (the amount of Ki-ras mRNA was used for normalization of the results) (6). As expected, no unr transcripts were detected in the homozygous unr−/− ES clones A, B, and D (Fig. 2A), whereas in the heterozygous unr+/− clone K, unr expression was about 60% of the unr+/+ clone L (after normalization). N-ras expression was increased two- to three-fold in unr−/− clones compared with the unr+/+ clone L (Fig. 2A), in agreement with the observation of an increased expression of N-ras in tissues of heterozygous unr+/− mice (6). The absence of unr expression in two unr−/− ES clones (A and B) was further confirmed by Western blotting (Fig. 2B) with anti-unr rabbit polyclonal antibody (23).
FIG. 1.
Genotype of ES cell clones mutated at the unr locus. (A) Schematic representation of the 5′ end of the wild-type unr allele and of the mutated allele generated by homologous recombination; BamHI sites are indicated by the letter B. (B) Southern blot analysis of ES cell clones. Genomic DNA from eight independent ES cell clones (A, B, C, D, E, G, L, and K [lanes 1 to 8, respectively]) was digested with BamHI and hybridized with the unr probe P5 indicated in panel A. This probe detects a 4.3-kb fragment and a 2.2-kb fragment from the wild-type (wt) and mutated (mt) alleles, respectively.
FIG. 2.
Expression of unr in ES cell clones. (A) RNase protection analysis of RNA isolated from wild-type, unr+/−, and unr−/− ES cells. Probe UNK comprises unr, N-ras and Ki-ras sequences. Ten micrograms of total RNA from five ES clones of each genotype as shown in Fig. 1 was analyzed with probe UNK. The molecular size marker on the left was generated with end-labeled MspI fragments of pBR322. The protected fragments are indicated on the right. (B) Immunoblot of wild-type, heterozygous unr+/−, and homozygous unr−/− ES cells. Lysates (25 μg) prepared from genotyped ES cell clones were analyzed by Western blotting with anti-unr and anti-α-tubulin antibodies. Arrows indicate unr (85 kDa) and α-tubulin (55 kDa).
When cultured on mouse embryonic fibroblast (MEF) feeder cells, unr−/− cells were morphologically indistinguishable from wild-type ES cells. In addition, unr−/− cells grew normally on MEF feeders (with doubling times of 12 h), suggesting that unr is not necessary for the growth of undifferentiated ES cells.
The rhinovirus and poliovirus IRESs work efficiently in wild-type ES cells.
The dicistronic plasmids pCRPolL, pHRVL and pCREL contain the poliovirus, HRV, and EMCV IRESs, respectively, inserted between Rluc and Fluc. Translation of RLuc, the first cistron, is cap dependent, whereas expression of the second cistron, Fluc, depends on the activity of the IRES located between the two reporter genes. As a negative control, plasmid pCRHL, which contains a hairpin but no IRES between the two cistrons, was used. The dicistronic plasmid pD128 contains the FMDV IRES inserted between CAT (upstream cistron) and Fluc (downstream cistron). For this plasmid, translation of CAT is cap dependent, whereas Fluc expression depends on FMDV IRES activity. As a control, plasmid pDmt, which contains a hairpin between CAT and Fluc, was used.
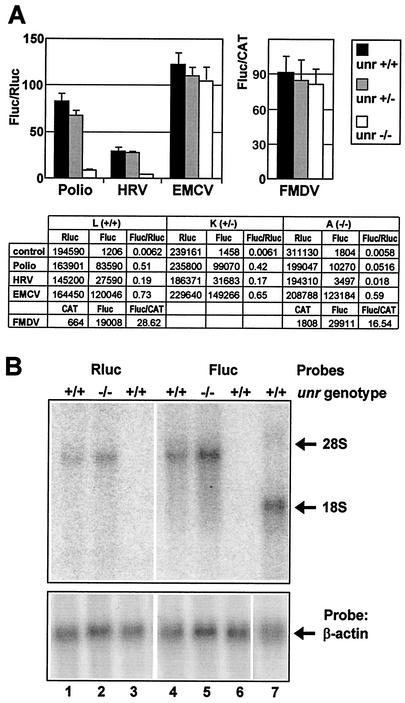
These plasmids were first transfected into wild-type ES cells that had been depleted of feeders prior to transfection. We determined that the contribution of residual feeders to the Rluc and Fluc activities was less than 1% and therefore negligible. We then determined the poliovirus, HRV, EMCV, and FMDV IRES activities in unr+/+ (clone L) and unr+/− (clone K) ES cells. In unr+/+ cells (Fig. 3A, black bars), the relative IRES activities were 80-fold for poliovirus, 30-fold for HRV, 120-fold for EMCV, and 90-fold for FMDV those of the negative controls. For comparison, the relative EMCV IRES activity in Cos-7 cells transiently transfected with the same dicistronic construct was 14.5 (19).
FIG. 3.
Effect of unr expression on the activity of picornaviral IRES elements. (A) The dicistronic constructs pCRpolL, pHRVL, pCREL, and pD128 containing the poliovirus, HRV, EMCV, and FMDV IRESs, respectively, and the control construct pCRHL were transfected in unr+/+, unr+/−, and unr−/− ES cells. Twenty-four hours after transfection, cells were analyzed for Fluc, Rluc, and CAT activities. The ratio of the enzymatic activities of the second cistron to the first cistron are presented by histograms for unr+/+ (clone L, black bars), unr+/− (clone K, gray bars), and unr−/− (clone A, light gray bars) cells. Fluc/Rluc ratios (left) or Fluc/CAT ratios (right) were normalized to the values obtained with control plasmids (pCRHL or pDmt, respectively). Three independent experiments were performed, each time in triplicate. Rluc and Fluc activities (relative light units) and CAT (cpm) activities are presented in the lower panel of the figure. (B) Northern blot analysis of total RNA extracted from transfected ES cells. Total cellular RNA was isolated from unr+/+ and unr−/− ES cells transfected with pHRVL (lanes 1, 2, 4, and 5) or pSGluc (lane 7), or from untransfected ES cells (lanes 3 and 6). Fifteen micrograms of total RNA was analyzed by Northern blotting with the Rluc and Fluc probes (top) and with a β-actin probe as a loading control (bottom). The migration of 28S and 18S rRNA is indicated on the right.
These results show that ES cells have the capacity to sustain efficient picornaviral IRES-directed initiation of translation. In the heterozygous unr+/− cells (clone K, dark gray bars) the four viral IRES elements were as efficient as in unr+/+ cells, indicating that a twofold decrease in unr expression had no effect on these IRES activities.
Rhinovirus and poliovirus IRES-directed translation is impaired in unr−/− ES cells.
We next examined the effect of unr disruption on the internal initiation of translation directed by picornaviral IRES elements. In unr−/− cells (clone A, Fig. 3A, upper panel, light gray bars), both the HRV and poliovirus IRES activities were severely impaired, their activities being about 10-fold lower than those in unr+/+ or unr+/− cells. In contrast, the EMCV and FMDV IRES activities did not significantly differ between the unr+/+, unr+/−, and unr−/− cells. These results were confirmed after transfection in another unr−/− ES clone (clone B, data not shown). A decrease in IRES activity (Fluc/Rluc ratio) could result from a decrease in Fluc (second cistron) translation, an increase in Rluc (first cistron) translation, or a combination of both. As shown in Fig. 3A (lower panel), the Rluc activity did not vary significantly in unr+/+, unr+/−, and unr−/− cells, independently of the transfected plasmid. In contrast, the Fluc activity directed by the HRV and poliovirus IRES elements was specifically decreased in unr−/− cells. Therefore, the reduction in HRV and poliovirus IRES activities in unr−/− cells was the consequence of a decrease in Fluc activity and not of an increase in Rluc activity. Thus, in ES cells, unr is required for efficient translation directed by the HRV and poliovirus IRES elements, but not for the internal initiation of translation directed by the EMCV and FMDV IRESs.
To exclude that the results obtained above could be due to differential RNA degradation of the two cistrons, we checked the integrity of the dicistronic HRV mRNA in unr+/+ and unr−/− cell lines. Total RNA was extracted from transfected ES cells and analyzed by Northern blotting with Rluc and Fluc probes. In both in unr+/+ and unr−/− cells, a single band of the size expected (∼3.5 kb) for the dicistronic mRNAs was detected (Fig. 3B). As controls, RNA isolated from nontransfected cells was analyzed and showed no specific hybridization to the Rluc and Fluc probes (Fig. 3B, lanes 3 and 6), while after transfection of the monocistronic pSGluc plasmid, a single band of greater mobility (1.7 kb) was detected (Fig. 3B, lane 7). Thus, the integrity of the dicistronic HRV mRNA was not affected in transfected unr−/− cells. Moreover, quantification of the Rluc, Fluc, and β-actin (used for loading control) signals revealed that the Fluc/β-actin and Fluc/β-actin ratios differed by ∼1.5-fold in the unr+/+ and unr−/− cells, excluding a major change of the dicistronic mRNA metabolism in the absence of Unr.
Altogether, these results show that unr is required in vivo for internal initiation of translation directed by the picornaviral type I IRES elements of both HRV and poliovirus, but is dispensable for the activity of the type II IRES elements of EMCV and FMDV.
Rhinovirus and poliovirus IRES activities are complemented by Unr cDNA.
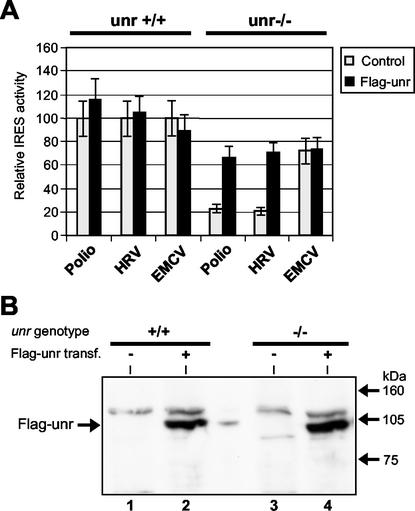
We then tested whether reintroduction of Unr in trans into Unr-deficient cells would restore HRV and poliovirus IRES activities. In a calibration experiment, dicistronic plasmids containing the poliovirus or the EMCV IRES were cotransfected into unr−/− ES cells with increasing amounts of a unr-expressing vector (pSGFlag-unr) or an empty vector (pSG5). We observed a specific stimulation of poliovirus IRES activity when 0.3 to 1 μg of PSGFlag-unr was added, while a larger amount of plasmid (3 μg) led to an inhibition of cap-dependent and IRES-dependent translation initiation for both dicistronic plasmids (data not shown).
We then performed cotransfections by using 1 μg of pSG5 or pSGFlag-unr, respectively, along with 1.5 μg of dicistronic plasmids (Fig. 4A). pSFlag-unr protein expression was confirmed by Western blotting with an anti-Flag monoclonal antibody (Fig. 4B). In unr+/+ cells (Fig. 4A, left panel), the activities of the poliovirus, HRV, and EMCV IRESs were similar when the control vector pSG5 and the unr-expressing vector pSGFlag-unr were cotransfected. In contrast, in unr−/− cells (Fig. 4A, right panel), the activities of the HRV and poliovirus IRESs were markedly reduced. Transfection of pSGFlag-unr then rescued poliovirus as well as HRV IRES activities to about 66 and 69%, respectively, of their activities in unr+/+ cells. In contrast, EMCV IRES activity was not markedly reduced in unr−/− cells and was not significantly affected by transfection of pSG5 or pSGFlag-unr. These results thus confirmed that the defect in HRV and poliovirus IRES activities resulted from the absence of Unr expression.
FIG. 4.
Rescue of HRV and poliovirus IRES activity by unr. unr+/+ and unr−/− ES cells were transfected with 1.5 μg of dicistronic plasmids containing the poliovirus, HRV, and EMCV IRESs, respectively. Each of the above plasmids was transfected together with 1 μg of either pSG5 (control) or pSG Flag-unr plasmids, respectively. Twenty-four hours after transfection, cells were analyzed for Fluc and Rluc activity (A) or Flag-unr protein expression (B). (A) The IRES activities in unr+/+ and unr−/− cells were determined as described in the legend to Fig. 3; the values were taken as 100 for each dicistronic plasmid cotransfected with pSG5 in unr+/+ cells. (B) Immunoblot analysis with anti-Flag antibody to detect the expression of Flag-unr protein. Lysates were prepared from unr+/+ (lanes 1 and 2) or unr−/− (lanes 3 and 4) ES cells transfected with 1 μg of pSG5 (lanes 1 and 3) or 1 μg of pSG Flag-unr (lanes 2 and 4) plasmids.
DISCUSSION
Previous in vitro evidence indicated that the cellular RNA-binding protein Unr is required for efficient translation of HRV RNA (20). To analyze in vivo the role of Unr in the internal initiation of translation driven by picornaviral and cellular IRESs, we have generated unr−/− ES cells.
Generating isogenic cell lines deficient in specific genes is a powerful tool for analysis of the biochemical functions of the encoded protein of interest in vivo. The level of unr expression in wild-type ES cells was similar to that observed in other cell lines (31), making these cell lines suitable to assess the consequences of unr inactivation on internal initiation of translation. ES cell lines were established from eight blastocysts, five of which were homozygous for the unr mutation. Within the limits of detection, the homozygous unr−/− cells were defective in unr expression, reflecting an essentially null mutation. Once established, the mutant cells grew with the same doubling time as wild-type cells, indicating that unr does not control cell growth of these undifferentiated cells, whereas development of mouse embryos was seriously affected (6; O. Boussadia and H. Jacquemin-Sablon, unpublished observations).
Here we report that HRV and poliovirus IRES-dependent translation is about 10-fold less efficient in unr−/− ES cells than in unr+/+ or unr+/− cells. Reintroduction of a unr expression vector into unr−/− ES cells efficiently restores HRV and poliovirus IRES activities. In contrast, the IRES elements from EMCV and FMDV are as effective or decreased by less than twofold in unr−/− ES cells compared with unr+/+ cells. Altogether, our results demonstrate that unr functions as a specific regulator of translation initiation directed by the HRV and poliovirus elements in vivo.
The observed decrease in HRV and poliovirus IRES activity (Fluc/Rluc ratio) in unr−/− ES cells reflects the specific inhibition of translation of the second, IRES-dependent cistron (Fluc). This conclusion is based on the following observations. (i) An 8- to 12-fold decrease in Fluc activity is observed specifically after transfection of dicistronic plasmids containing HRV and poliovirus elements. (ii) Repeated transfections of all dicistronic plamids showed that in unr−/− cells, Renilla luciferase activity (the first cistron) did not change significantly compared with that in unr+/+ cells. (iii) Northern blot analysis of the HRV dicistronic mRNA from transfected cells indicates that IRES integrity was maintained in unr−/− cells. Moreover, the limited variations in the amount of the Fluc/Rluc mRNA in the unr−/− and unr+/+ cells indicate that unr expression did not significantly affect the steady-state level of the dicistronic mRNA. Thus, in agreement with previously reported in vitro data (20), our results indicate that physiological unr concentrations do not significantly modulate cap-dependent translation and that the relative decrease in the HRV and poliovirus IRES activities in the unr-knockout ES cells was not a secondary consequence of competing translation of the first cistron.
On the basis of their primary sequences and secondary structures as well as their requirements for efficient translation initiation in reticulocyte lysates, picornaviral IRES elements have been assigned to three groups: the type I IRES elements of the enterovirus and rhinovirus group, the type II IRES elements of the cardiovirus and aphthovirus group, and the type III IRES of hepatitis A virus (1). Our results establish that Unr functions in vivo as a specific regulator of translation directed by picornaviral type I, but not type II IRES elements. The direct involvement of Unr in HRV IRES-directed translation has been investigated in detail in vitro. (i) In HeLa cell extracts, Unr could be cross-linked to the HRV 5′ UTR, but not to the EMCV 5′ UTR (4). (ii) In rabbit reticulocyte lysates, Unr acts synergistically with PTB to stimulate HRV IRES activity (20), and this effect depends on the interaction of Unr with the HRV IRES, since mutations introduced in the Unr cold shock domains that knock out the RNA-binding activity of these domains impaired the Unr activity. PCBP2 enhances the activity of Unr and PTB on the HRV IRES, while PCBP2 alone has no effect on HRV IRES activity. In contrast, the stimulation of poliovirus IRES activity by Unr was uncertain, although purified Unr protein interacts both with HRV and poliovirus IRES elements. Recombinant Unr protein was unable to stimulate poliovirus IRES activity in rabbit reticulocyte lysate, whether Unr was added separately or in combination with PTB and PCBP2. Stimulation of the poliovirus IRES was only obtained with PTB, while PCBP2 had no effect. In contrast, purified native Unr protein (the so-called B-type activity) purified from HeLa cells stimulated poliovirus translation by a factor of 1.8 (20). Thus, the requirements of the HRV and the poliovirus IRES for added recombinant Unr, PTB, and PCBP2 appear to be different in reticulocyte lysates (which contain various endogenous levels of these factors). We can only speculate whether the combination of RNA-binding proteins that promotes internal initiation directed by a given IRES differs from one tissue to another or according to the proliferation or differentiation status of the cells. Moreover, factors other than those tested in vitro, including Unr, PTB, and PCBP2 (20), that possibly contribute to the poliovirus IRES activity may be limiting in rabbit reticulocyte lysate, but not in ES cells. Nevertheless, our findings that Unr strongly regulates both HRV and poliovirus IRES activities in vivo indicate that Unr is one of the key regulators of type I IRES activity.
The mechanism by which ITAFs facilitate the recruitment of ribosomal subunits is yet unknown. One hypothesis is that ITAFs have a chaperone activity (17, 22) and help to fold the IRES into the conformation required for maximal IRES activity. This hypothesis is based mainly on the structural properties of these RNA-binding proteins. All of them have multiple-RNA-binding domains, namely cold shock domains for Unr, RNA recognition motif (RRM) domains for La, and PTB and KH domains in PCBP2. Moreover, most of them may dimerize in solution, like PTB, La, or PCBP2. Accordingly, these proteins may make several contacts with the IRES, like PTB (25). Understanding the role of Unr in IRES-mediated translation will require a detailed mapping of Unr binding sites on the HRV and poliovirus elements. The strong effect of Unr on HRV and poliovirus IRES-directed translation that we observed in ES cells should facilitate the determination of cis RNA sequences within the HRV and poliovirus elements required for regulation by Unr. Our system also provides a powerful experimental approach to analyze Unr domains required for stimulation of IRES activity. This could be investigated by cotransfection of constructs that express wild-type or mutant Unr proteins into unr−/− ES cells and analysis of their ability to restore HRV and poliovirus IRES activities.
In contrast to the large number of in vitro studies, there are only a few reports concerning the role of ITAFs in picornaviral IRES-directed translation in vivo. Transient overexpression of PTB in BSC-1 or HeLa cells was shown to stimulate cap-independent translation directed by picornaviral type I (poliovirus) and type III (hepatitis A virus) IRES elements as well as by the flaviviral hepatitis C virus IRES (14). PCBP proteins appear to be required for poliovirus IRES-directed translation, since addition of antibodies against PCBP1 and PCBP2 inhibited viral translation in Xenopus oocytes (12).
The results presented here show that ES cells sustain efficient internal initiation of translation directed by the four picornavirus IRES elements that we analyzed. Interestingly, it was recently reported that two cellular IRES activities, c-myc and FGF-2, are regulated during mouse embryonic development; while these IRESs are very active in 11-day-old embryos, they are inactive or poorly active in adult organs. In contrast, the EMCV IRES was found to be active in both embryonic and adult tissues (9, 10). It would be interesting to examine whether the HRV and poliovirus IRES activities depend on the differentiation state of ES cells.
Our results demonstrate that Unr is required in vivo for HRV and poliovirus IRES activity. Our approach using Unr-deficient cells implies that IRES activities were analyzed at physiological Unr concentrations. It was recently reported that PTB and nPTB, a neural-specific homologue of PTB, promote translation initiation directed by the IRES of the GVII strain of Theiler's murine encephalomyelitis virus and are critical for viral pathogenicity (35). It is tempting to speculate that Unr could affect production of rhinovirus and of poliovirus, since at least in the latter case, a correlation between translational efficiency and neurovirulence has been established (2, 35).
Acknowledgments
We thank A. Jacquemin-Sablon for critical reading of the manuscript, A. Dias for technical assistance, and D. Lepesteur for advice on ES cell culture and excellent technical assistance. We are grateful to F. Sainteny and W. Vainchenker for receiving us in their laboratory and for helpful discussions.
This work was supported by a grant from the Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A., M. T. Howell, J. G. Patton, and R. J. Jackson. 1993. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 74:1775-1788. [DOI] [PubMed] [Google Scholar]
- 5.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiations of translation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boussadia, O., F. Amiot, S. Cases, G. Triqueneaux, H. Jacquemin-Sablon, and F. Dautry. 1997. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 420:20-24. [DOI] [PubMed] [Google Scholar]
- 7.Cassinotti, P., and M. Weitz. 1994. Increasing the sensitivity of a common CAT assay. BioTechniques 17:36-40. [PubMed] [Google Scholar]
- 8.Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5:597-605. [DOI] [PubMed] [Google Scholar]
- 9.Créancier, L., P. Mercier, A.-C. Prats, and D. Morello. 2001. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol. 21:1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Créancier, L., D. Morello, P. Mercier, and A. C. Prats. 2000. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell. Biol. 150:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doniger, J., D. Landsman, M. A. Gonda, and G. Wistow. 1992. The product of unr, the highly conserved gene upstream of N-ras, contains multiple repeats of the cold shock domain (CSD) a putative DNA-binding motif. New Biol. 4:389-395. [PubMed] [Google Scholar]
- 12.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 14.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergenic poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 17.Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270:20871-20874. [DOI] [PubMed] [Google Scholar]
- 18.Hershey, J. W. B., and W. C. Merrick. 2000. Pathways and mechanism of protein synthesis. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Huez, I., L. Créancier, S. Audigier, M.-C. Gensac, A.-C. Prats, and H. Prats. 1998. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 18:6178-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt, S. L., and R. J. J. Jackson. 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5:344-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquemin-Sablon, H., G. Triqueneaux, S. Deschamps, M. Le Maire, J. Doniger, and F. Dautry. 1994. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucleic Acids Res. 22:2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. J. Jackson. 1995. Direct evidence that polypirimidine tract binding protein (PTB) is essential for internal ribosomal entry site of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 25.Kolupaeva, V. G., C. U. Hellen, and I. N. Shatsky. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of the encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199-1212. [PMC free article] [PubMed] [Google Scholar]
- 26.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landsman, D. 1992. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 20:2861-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomakin, I. B., C. U. T. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, M. B., N. Sonenberg, and J. W. B. Hershey. 2000. Origins and principles of translational control. Translational control of gene expression, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [DOI] [PMC free article] [PubMed]
- 30.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. L. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicolaiew, N., G. Triqueneaux, and F. Dautry. 1991. Organization of the human N-ras locus: characterization of a gene located immediately upstream of N-ras. Oncogene 6:721-730. [PubMed] [Google Scholar]
- 32.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvment of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. T. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 35.Pilipenko, E. V., E. G. Viktorova, S. T. Guest, V. I. Agol, and R. P. Roos. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20:6899-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyronnet, S., J. Dostie, and N. Sonenberg. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 38.Raught, B., A. C. Gingras, and N. Sonenberg. 2000. Regulation of ribosomal recruitment in eukaryotes. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Robertson, E. J. 1987. Embryo-derived stem cells, p. 71-112. In E. J. Robertson (ed.), Teratocarcinomas and embryonic stem cells: a practical approach. IRL Press, Oxford, United Kingdom.
- 40.Sachs, A. B., and S. Buratowski. 1997. Common themes in translational and transcriptional regulation. Trends Biochem. Sci. 22:189-192. [DOI] [PubMed] [Google Scholar]
- 41.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting at the Beginning, Middle, and End: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Triqueneaux, G., M. Velten, P. Franzon, F. Dautry, and H. Jacquemin-Sablon. 1999. RNA binding specificity of UNR, a protein with five cold shock domains. Nucleic Acids Res. 27:1926-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, B. L., J. H. C. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weil, D., S. Brosset, and F. Dautry. 1990. RNA processing is a limiting step for murine tumor necrosis factor β expression in response to interleukin-2. Mol. Cell. Biol. 10:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, X., P. Fong, N. Iizuka, D. Choate, and D. R. Cavener. 1997. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 17:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]