Abstract
DNA from the murine pneumotropic virus was extracted from virus in lung tissue of infected mice, and the regulatory region of the genome was amplified by PCR. The regulatory region of individual plasmid cloned DNA molecules appeared to have heterogeneous enhancer segments, whereas the protein-coding part of the genome had a uniform length. Nucleotide sequence analysis revealed that the majority of the DNA molecules had a structure differing from the standard type. A 220-bp insertion at nucleotide position 142 with a concomitant deletion of nucleotides 143 to 148 was prominent. There were two variants of the 220-bp insertion, differing at two nucleotide positions at one of the termini. Other DNA molecules had complete or partial deletions of these structures and surrounding sequences in the viral enhancer. However, the end of the insertion at nucleotide 142 was frequently preserved. The viral early and late promoter activity of the variant regulatory regions was tested in a luciferase reporter assay by using transfected NIH 3T3 cells. In relation to the standard-type DNA, all variants, including a G272T mutant, had much stronger late promoters. In contrast, the early promoter activity was influenced in a positive or negative direction by individual mutations. Also, the activity of the viral origin of DNA replication was affected by the sequence variation of the regulatory region, although the effects were smaller than for the late promoter. Analysis by Southern blotting and quantification using dot blots showed that approximately 103 copies of material related to the 220-bp insert in murine pneumotropic virus DNA was present in mouse and human DNA but not in Escherichia coli DNA. Moreover, analysis by PCR indicated that there were multiple copies in the mouse genome of sequences that were identical or closely related to the 220-bp viral DNA segment. These data together with the nucleotide sequence analysis suggest that the 220-bp insertion is related to a transposable element of a novel type.
Members of the polyomavirus family have DNA genomes with a size of approximately 5 kbp. With this small coding capacity, they depend on host cell functions for their gene expression and DNA replication. The originally discovered mouse polyomavirus is highly tumorigenic in experimental infection, whereas a second mouse polyomavirus, the murine pneumotropic virus (MPtV) discovered by Kilham and Murphy (27)—earlier called Kilham virus—is nontumorigenic in the animal species that have been tested (39). However, MPtV can transform mouse cells in vitro (43).
MPtV, in contrast to other mammalian polyomaviruses, is associated with acute disease. In newborn mice, infection causes a severe interstitial pneumonia (19, 20), whereas exposure of fully immunocompetent mice to MPtV leads to an inapparent infection that becomes persistent. During the acute phase, MPtV can be observed in vascular endothelial cells, mainly in the lung but also in other organs, including liver, spleen, brain, and intestine (19-21, 32). Later in infection, during the persistence phase, virus is mainly present in association with renal tubules (22).
The MPtV genome is a circular, 4,756-bp double-stranded DNA molecule (31; present investigation). As with the other members of this virus family, the early and late regions are encoded by opposite strands of DNA, with the transcriptional promoters and the origin of replication in a ca. 0.5-kb segment located between the two protein-coding regions. However, MPtV is not closely related to the previously characterized mouse polyomavirus (3, 31). Besides the lack of a gene encoding middle T antigen in MPtV DNA, the deduced amino acid sequences of the virally encoded proteins are as distantly related to those of the oncogenic mouse polyomavirus as they are to the proteins encoded by the human polyomaviruses.
A variety of cellular proteins have binding sites in the regulatory region of polyomavirus genomes (25). Most of these sites are located in a segment first identified as an enhancer of the early promoter (10). Later, additional functions of the enhancer in the initiation of viral DNA synthesis and in transcription of the late region have been demonstrated (44). In a given cellular environment, the ability of the enhancer to bind proteins that participate in the assembly of transcription and replication complexes determines whether or not the virus can multiply. Genomes with the enhancer of the standard-type MPtV do not replicate in the presence of large T antigen or express their late genes in a variety of mouse cell lines. However, the substitution of a segment of the MPtV enhancer with a corresponding part of the polyomavirus genome rendered the virus the ability to multiply in mouse fibroblast cells (50).
Whereas coding regions of polyomavirus genomes are genetically stable, a substantial variability of the enhancer has been observed in mouse polyomavirus propagated in cultured cells (17, 26, 30). Corresponding regulatory region variants of simian virus 40 (SV40) were isolated from both infected cells in culture and from immunocompromised monkeys (28), whereas regulatory region variants of the human polyomaviruses BK and JC have been recovered from both immunocompromised and healthy individuals (reviewed in reference 13). The sequence variation in the regulatory region typically consists of duplications, frequently in combination with deletions or other rearrangements. Mutation of the enhancer may alter the host range of mouse polyomavirus in vitro and in vivo (17, 26, 30, 41). In SV40, regulatory region variants isolated from infected animals showed altered growth characteristics in cell culture (29). Studies on JCPyV variants suggested that genomic rearrangements were associated with the persistent state of infection, influencing host cell specificity and virulence, which might lead to human disease (14, 35).
Cellular mobile genetic elements, also called transposons, are distinguished by their ability to insert at new genomic locations. There are two major classes of transposons (15). Class I elements, such as the Alu elements in primates (24) and B1 sequences in rodents (42), are retroelements that use reverse transcriptase to transpose by means of an RNA intermediate. In contrast, class II elements transpose directly from one location in DNA to another. In mammalian genomes, two types of class I retroelements predominate: long and short interspersed nuclear elements (49). Another type of transposable elements, called miniature inverted-repeat transposable elements (MITEs), has been found mainly in plant genomes (46). Various MITEs have common features, including length (74 to 490 bp), terminal inverted repeats of 10 to 15 bp, target site preference, low G+C content, and high copy number (46, 49).
In SV40 a variant was found to have a 157-bp Alu-element-like insertion immediately upstream of the early coding sequences (11). There are additional reports of cellular DNA segments integrated into the regulatory region of polyomavirus genomes (12, 38). However, in these cases, genomes with inserts of cellular DNA were either found as a small minority in a population of normal viral genomes or in a larger proportion of defective viral genomes after repeated high-multiplicity passage of virus in cell culture. In the present investigation, DNA segments apparently derived from repeated sequences in cellular DNA were found integrated in the enhancer of the MPtV genome. Moreover, these genetic elements did change the viral potential of gene expression and DNA replication.
MATERIALS AND METHODS
Animals, cells, and virus.
Pregnant females of C57BL/6 mice were obtained from the Laboratory Animal Resource at Uppsala University, where animals are health monitored and kept behind a barrier. The mouse cell lines NIH 3T3 and FM3A and human HeLa cells were obtained from ECACC (Porton Down, England). The mouse endothelial cell line UAE (1) was kindly provided by T. Ramqvist, Karolinska Institute, Stockholm, Sweden. All cells were cultured in Dulbecco modified Eagle medium (Gibco) supplemented with 10% newborn calf serum. Kilham polyomavirus (MPtV) was obtained from the American Type Culture Collection (Manassas, Va.) as a clarified extract of lung tissue.
Infection and transfection protocols.
Newborn C57BL/6 pups were inoculated intraperitoneally with 2 μl of virus suspension diluted 10-fold in phosphate- buffered saline. At 7 days after infection, livers, spleens, kidneys, and lungs were excised, homogenized, and disintegrated by sonic vibration. For transfection, cultures of NIH 3T3 cells were started at a density of 2.5 × 105 cells per 60-mm-diameter petri dish. The following day the cells were transfected with 4.0 μg of DNA in complex with Lipofectamine according to the manufacturer's instructions (Life Technologies Products).
Cloning, amplification, and sequence analysis of the MPtV and MPtV-related DNA.
The recombinant plasmid pKV19, carrying MPtV DNA in the XbaI site of pUC12 (31), was obtained from Kristina Dörries (Würzburg University, Würzburg, Germany). In this report, pKV19 is called pstMPtV.
Genomic DNA was purified from human HeLa cells; from mouse FM3A, NIH 3T3, and UAE cells; and from Escherichia coli JM109 cells by using a DNeasy Tissue Kit (Qiagen) according to the manufacturer's instructions. Low-molecular-weight DNA prepared (23) from cell extracts of lung tissue of MPtV-infected C3H mice (American Type Culture Collection) and from organs of infected C57BL/6 mice was used as templates for PCR amplification of the regulatory region of the viral genome. Two oligonucleotide primers corresponding to nucleotides (nt) 353 to 337 and nt 4626 to 4643 of MPtV DNA were used. In addition, these primers contained a 5′-terminal SacI recognition sequence for subsequent plasmid cloning. The PCR was performed with the high-fidelity Vent DNA polymerase (New England Biolabs). After SacI cleavage, the PCR product was ligated to the corresponding site of the pGL2-basic plasmid (Promega) and E. coli JM109 cells were transformed. The plasmid carrying the regulatory region of the MPtV genome was called pGL2-basic/MPtVrr.
For amplification of MPtV-related material in mouse genomic DNA, a pair of oligonucleotide primers corresponding to nt 23 to 45 and 178 to 201 of the MPtV “insertion” (see Fig. 5A) were used. Each PCR was carried out by using 250 ng of template DNA, 1× PCR buffer (MBI Fermentas), a 0.3 μM concentration of each primer, 0.2 mM deoxynucleoside triphosphate, MgCl2 at the indicated concentrations, and 1 U of Taq DNA polymerase (MBI Fermentas) in a final volume of 100 μl. The DNA was amplified by 35 reaction cycles (94°C for 45 s, 58°C for 30 s, and 72°C for 30 s). For further analysis, the PCR products were ligated into the pGEM-T vector (Promega), and after transformation of E. coli JM109 cells, the inserts of individual plasmid clones were analyzed by DNA sequencing. The analyses were carried out with an ABI-PRISM Sequenator. In the reactions, commercial oligonucleotide primers (Promega) for sequence analysis of inserts of the pGL2-basic and the pGEM-T vectors were used. All sequence data were proofread visually.
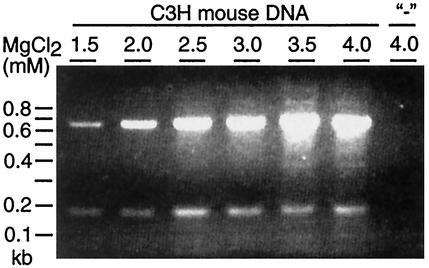
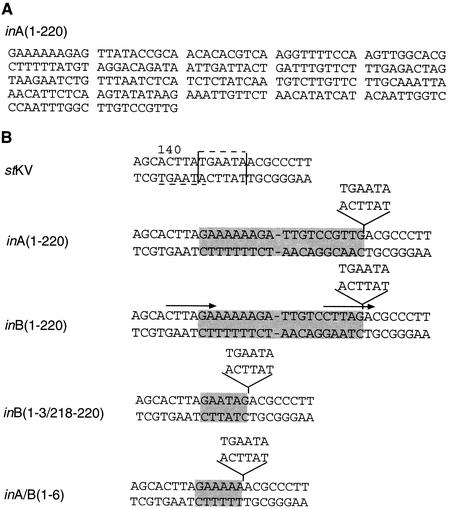
FIG. 5.
Agarose gel electrophoresis of PCR products from amplification of MPtV-related sequences in mouse DNA. DNA was extracted from the spleen of a C3H mouse and then purified. PCR amplification was done for 35 cycles in the presence or absence (−) of template DNA by using a primer pair that would generate a 179-bp product with the inA or inB insert of MPtV DNA as a template. After amplification, DNA was resolved by electrophoresis in a 1.5% agarose gel in the presence of 0.5 μg of ethidium bromide per ml. DNA was visualized in UV light. The final concentration of MgCl2 in the PCR is indicated. The electrophoretic mobility of size markers is displayed to the left.
For Southern blot analysis, genomic DNA was cleaved with BglII and the fragments were resolved by agarose gel electrophoresis and then transferred to a BioTrace polyvinyl difluoride hybridization membrane (Gelman Sciences) by capillary blotting. DNA on the membrane was annealed at 65°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with a 32P-labeled probe prepared by PCR from the insert in MPtV DNA. For estimation of the copy number of MPtV insert-related sequences in genomic DNA, 2.5, 5.0, and 10 μg of each type of genomic DNA were dot blotted to a BioTrace polyvinyl difluoride hybridization membrane (Gelman Sciences). A linearized recombinant plasmid containing a single insert of MPtV DNA was used as standard. The blotted DNA sample was annealed with a 32P-labeled MPtV insert probe as described above.
Reporter assay of gene expression and analysis of viral DNA replication.
The luciferase reporter assay and the analysis of viral DNA replication were performed as described previously (50). pGL2-basic/MPtVrr, carrying the indicated regulatory regions in the early and late promoter orientation relative to the luciferase gene, was used for transfection of NIH 3T3 cells. To obtain the insert in both orientations, it was excised from recombinant plasmids and religated with the vector. Cytoplasmic extracts of the transfected cells were prepared at 42 h posttransfection. For analysis of the activity of the MPtV origin of DNA replication, NIH 3T3 cell cultures were transfected with 2.0 μg of the indicated pGL2-basic/MPtVrr DNAs mixed with 2.0 μg of pcDNA3/MPtV-LT (50). Cells were harvested at 42 to 44 h posttransfection. Low-molecular-weight DNA was extracted from the cells (23) and cleaved with DpnI and a second restriction endonuclease to linearize DpnI-resistant molecules. Thereafter, it was subjected to Southern blot hybridization followed by quantification of signal intensities.
Nucleotide sequence accession number.
Nucleotide sequences of variant nontranslated regions of MPtV DNA have been deposited in the EMBL database: stMPtV (st represents standard type), AJ517507; G272TMPtV, AJ517508; inA/dl143-148MPtV, AJ517509; inB/dl143-148MPtV, AJ517510.
RESULTS
Rearrangements in regulatory region of MPtV DNA.
The observation that gene expression and replication of stMPtV DNA is inefficient in mouse fibroblasts raised the question as to whether virus isolated from infected animals had genomes with a different regulatory region. Virus DNA was extracted from lung tissue of infected C3H mice, and the regulatory region of the genome was amplified by PCR. To identify nucleotide sequence variations, the PCR product was ligated to the plasmid pGL2-basic, and after transformation of E. coli cells, individual plasmid clones were isolated.
Analysis of the PCR products by agarose gel electrophoresis displayed considerable heterogeneity. There was a minor band with a size of 470 bp, corresponding to the size of the standard-type regulatory region, and a major band of 720 bp. In addition, there was a smear of molecules with sizes larger than 720 bp (Fig. 1A).
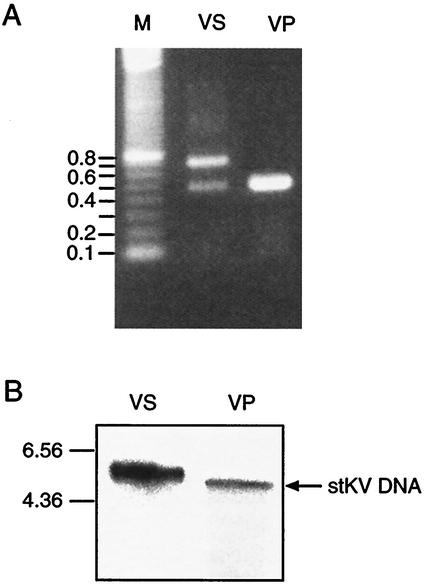
FIG. 1.
Analysis of MPtV genome structure. (A) The regulatory region of MPtV DNA extracted from a crude virus preparation (VS) was amplified by PCR by using primers complementary to the region immediately adjacent to protein coding sequences. As a control DNA from the plasmid pstMPtV (VP) was used. The amplification products were separated by agarose gel electrophoresis. (B) Southern blot analysis of XbaI-digested DNA from lung extracts (VS) and from pstMPtV (VP). Annealing was done with 32P-labeled MPtV DNA isolated from the recombinant plasmid pstMPtV, which was used as a probe. The mobility of DNA size markers is indicated to the left. kb, kilobase pairs.
The high variability of the regulatory region of the MPtV genome led to the question whether most of the DNA molecules were severely rearranged with a corresponding heterogeneity of the coding sequences. Since populations of defective variants have a heterogeneous size, resulting from reiterations of genomic segments (16), viral DNA extracted from virions was analyzed by Southern blotting (Fig. 1B). Low-molecular-weight DNA was extracted from the virus preparation and was linearized by endonuclease XbaI cleavage prior to agarose gel electrophoresis. As a control, pstMPtV DNA cleaved with the same enzyme was used. After blotting, viral DNA was annealed to a 32P-labeled probe prepared from the insert of viral DNA in pstMPtV. DNA extracted from the infected tissue showed a hybridization signal at a position corresponding to a size slightly larger than that of the stMPtV genome (Fig. 1B). This result is consistent with the observation that the majority of the DNA molecules had a larger regulatory region than the standard type. Although the band of viral DNA extracted from virus appeared broader than the corresponding band of plasmid-cloned DNA, the size of the viral DNA molecules indicated that most viral genomes did not have rearrangements outside the regulatory region. Analysis after cleavage with the endonucleases HincII, HindIII, and PvuII (data not shown) led to the same conclusion.
Determination of the nucleotide sequence of the regulatory region in individual plasmid clones revealed a large number of variants. In DNA from 30 plasmid clones, 15 different nucleotide sequences were identified. However, inspection of the sequences revealed that they were all related to the regulatory region of the MPtV genome, as reported by Mayer and Dörries (31), here called stMPtV. In all the plasmid clones, including our stock of the original pstMPtV, there were two extra G residues, at nt 10 and 14. Besides this difference, only 2 of the 30 clones carried the regulatory region of stMPtV DNA. Another two had the standard-type sequence with a G-to-T transversion at nt 272. The remaining plasmid clones all showed more complex alterations of the MPtV regulatory region.
In relation to the structure of the stMPtV enhancer, many of the clones (12 of 30) had an insertion of 220 bp at nt 142, in combination with a deletion of the adjacent 6 bp (nt 143 to 148). Two types of 220-bp insertions were observed that differed in sequence at two positions at one end. The insertion-deletion event apparently occurred in MPtV regulatory regions with either a G or a T residue at nt 272. Besides the isolates with the combination of a 220-bp insertion and a 6-bp deletion, there were 10 types of nucleotide sequences present in the sample that could have arisen by a deletion within the 220-bp insertion or by deletion of the whole insertion and flanking DNA segments. Figure 2 summarizes the basic types of sequences with variation at nt 142 that were observed. In addition to these sequence variations, we found two types of regulatory regions with direct repeats of a segment within the regulatory region of stMPtV DNA. In both cases, the duplication was combined with another genetic event. All the information on the nucleotide sequences of the viral regulatory regions is summarized in Table 1.
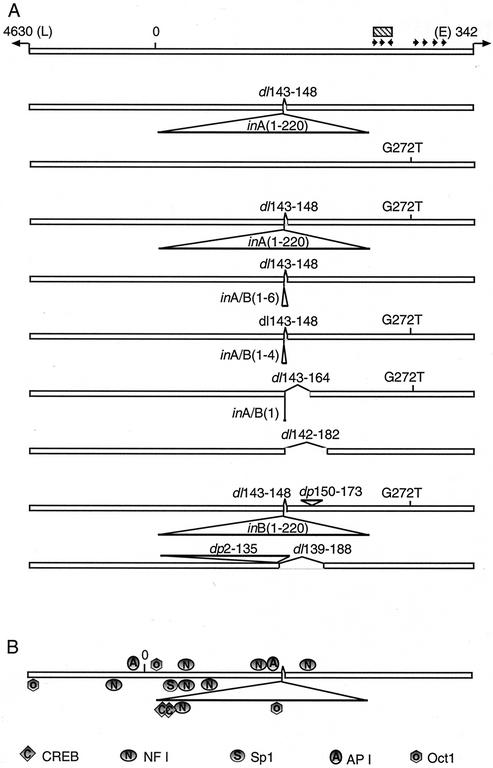
FIG. 2.
Organization of the stMPtV regulatory region and the structure of variant genomes isolated from MPtV in mouse lung tissue. The regulatory region of the viral genome was amplified by PCR and ligated to pGL2-basic DNA. Recombinant plasmids were cloned, and the nucleotide sequence of the regulatory region was determined. The data were related to the published nucleotide sequence of MPtV DNA (31), here called standard type (stMPtV). (A) Short arrows indicate large T antigen recognition pentanucleotides (GPuGGC) in the sense of the early (E→) and late (←L) DNA strands, respectively. The hatched box shows the position of the putative viral replication origin. Abbreviations: inA and inB, insertion type A and type B, respectively (numbers refer to the nucleotide positions of the 220-bp segments); dl, deletion; dp, duplication; G272T, G-to-T base change at nt 272. (B) Predicted recognition sites of DNA-binding transcription factors (40, 47).
TABLE 1.
Nucleotide sequence variation in the regulatory region of genomes from MPtV extracted from infected C3H mice
| No. of clones | nt 272 | Insertion at nt 142 (size [type]) | Deletion | Additional mutation |
|---|---|---|---|---|
| 2 | Ga | |||
| 2 | T | |||
| 4 | G | 220 bpb | 143-148 | |
| 1 | G | 6 bp (GAAAAA) | 143-148 | |
| 2 | G | 142-182 | ||
| 1 | G | 136-185 | ||
| 1 | G | 1 bp (A) | 143-164 | |
| 1 | G | 1 bp (G) | 143-164 | |
| 1 | NDd | 139-188 | dp3-135d | |
| 6 | T | 220 bpb | 143-148 | |
| 2 | T | 4 bp (GAAA) | 143-148 | |
| 3 | T | 143-163 | ||
| 1 | T | 1 bp (G) | 143-164 | |
| 2 | T | 220 bpc | 143-148 | dp150-173d |
| 1 | G | 6 bpc (GAATAG) | 143-148 |
Present in stMPtV.
Insert type A (inA). See legend to Fig. 5.
Insert type B (inB).
dp indicate duplication; ND, not determined.
Since the heterogeneous MPtV genomes were isolated from the lungs of moribund animals, the variability might have arisen in the final stage of the infection and a large proportion of the virus particles might have low viability. To investigate whether the regulatory region genetic variants were infectious, the MPtV prepared from lungs of infected C3H mice was used for infection of newborn C57BL/6 mice. This mouse strain was used because it is derived from Mus musculus, like C3H, and has been shown to be susceptible to MPtV (43). Moreover, we use it routinely for polyomavirus infections. The mice were inoculated intraperitoneally, and at 7 days postinfection, the animals were killed and organs were excised. DNA was extracted from several organs, and the regulatory region of MPtV DNA was amplified by PCR, using primers base pairing to the flanking 5′ termini of the coding regions. Analysis of individual DNA molecules by plasmid cloning and nucleotide sequence determination was done as described above. The result showed (Table 2) that four of the virus variants identified in lung extract were propagated in the infected C57BL/6 mice and that variants with the inB insertion were common. Although only 24 plasmid clones of PCR-amplified DNA were analyzed in this initial study, some conclusions can be drawn. The inA variant combined with the deletion at nt 143 to 148 was found not only in lung but also in kidney, liver, and spleen, whereas the corresponding variant with the inB insertion was identified in kidney, liver, and spleen of the infected C57BL/6 animals. Although this variant was not found in the lungs of infected C3H or C57BL/6 mice, it might, of course, be present—but passed undetected—in both cases. Notably, in the 24 analyzed plasmid clones, there was no regulatory region of the standard type. This observation supports the notion that the insertion-deletion event contributes to the growth efficiency of MPtV.
TABLE 2.
Nucleotide sequence variation in the regulatory region of genomes from MPtV passaged in C57BL/6 micea
| No. of clones | nt 272 | Insertion at nt 142 (size [type]) | Deletion | Additional mutation | Organ distribution
|
|||
|---|---|---|---|---|---|---|---|---|
| Ki | Li | Lu | Sp | |||||
| 1 | G | 143-163 | 1 | |||||
| 8 | T | 220 bp (inA) | 143-148 | 4 | 2 | 2 | ||
| 9 | G | 220 bp (inA) | 143-148 | 2 | 3 | 4 | ||
| 5 | G | 220 bp (inB) | 143-148 | 3 | 1 | 1 | ||
| 1 | G | 220 bp (inB) | 143-148 | dp149-172 | 1 | |||
Organs were taken from six mouse pups at 8 days after intraperitoneal infection. DNA was prepared from pooled organs of each type. Ki, kidney; Li, liver; Lu, lung; Sp, spleen.
Effect of enhancer mutations on promoter activity and on the origin of DNA replication.
The promoters of early and late transcription as well as the origin of DNA replication are located in the regulatory region of the polyomavirus genomes. Moreover, the activity of all three elements depends on specific interactions of the enhancer segment with cellular protein molecules. Thus, the observed variation of the MPtV enhancer might influence the efficiency of both viral gene expression and DNA replication. Although a variety of cell lines and primary cells have been examined for their ability to support MPtV multiplication, none of these was found to be permissive to the virus. However, an enhancer substitution mutant of MPtV was demonstrated to replicate and to form virus after transfection of NIH 3T3 cells (50). Therefore, NIH 3T3 cells were used for the experiments. The activity of the early and late viral promoters was tested, using a luciferase reporter assay, and in addition, the activity of the viral origin of DNA replication was analyzed in the presence of large T antigen expressed from a separate plasmid (pcDNA3/MPtV-LT).
The pGL2-basic vector contains a luciferase reporter gene whose expression depends on the insertion of a DNA segment with promoter activity. For analysis of MPtV early and late promoter activity, the regulatory regions amplified by PCR were cloned in both orientations in relation to the luciferase gene in the pGL2-basic plasmid (pGL2-basic/MPtVrr-E and pGL2-basic/MPtVrr-L). The amount of luciferase activity in cytoplasmic cell extracts prepared at 42 to 44 h posttransfection was taken as a measure of promoter activity. In each experiment, the early and late promoter activities were tested. The stMPtVrr-E and stMPtVrr-L plasmid derivatives were used as standards, and transfection with each plasmid was carried out in triplicate.
In accordance with an earlier report (50), the late promoter of stMPtV DNA had a very low activity in NIH 3T3 cells, whereas the luciferase gene driven by the early promoter was approximately 10-fold more active. The combined data of reporter gene expression are summarized in Table 3. In comparison to the standard-type early promoter, the insertion-deletion decreased the luciferase expression by 65%. In contrast, the G272T mutation increased reporter gene expression from the early promoter by 40%. The two types of mutations had a much larger effect on the activity of the late MPtV promoter. Extracts of NIH 3T3 cells transfected with a plasmid containing a regulatory region with the insertion-deletion at nt 142 contained more than 10-fold more luciferase activity than the cells transfected with the control plasmid of the standard type. The effect of the G272T mutation on the late promoter activity was even larger in relation to the standard-type regulatory region. In this case, the luciferase activity was increased 40-fold. The other regulatory region variants had intermediate effects on the activities of early and late promoters. In individual mutants, the activity of the early promoter was affected positively or negatively in relation to the standard type. In contrast, the viral late promoter uniformly responded to mutation of the regulatory region by increased activity. This positive effect by a variety of nucleotide sequence changes suggests that the late promoter activation was not caused by the creation or disappearance of specific protein binding sites but instead by a more general effect such as remodeling of chromatin.
TABLE 3.
Effect of nucleotide sequence variation on the activity of MPtV early and late promoters and of the origin of DNA replication
| MPtV regulatory region variant | Promoter activitya
|
Viral DNA replicationb | |
|---|---|---|---|
| Early | Late | ||
| stMPtV | 1.00 | 0.08 | 1.0 |
| inA/dl143-148 | 0.35 ± 0.01 | 1.01 ± 0.29 | 1.2 |
| inA/dl143-148/G272T | 0.31 ± 0.06 | 1.54 ± 0.52 | 2.2 |
| G272T | 1.43 ± 0.15 | 3.28 ± 0.28 | 2.1 |
| inA(1-4)/dl143-148/G272T | 0.79 ± 0.02 | 2.30 ± 0.36 | 1.5 |
| dl143-164/G272T | 0.17 ± 0.03 | 1.44 ± 0.08 | 1.0 |
| inA(1-6)/dl143-148 | 1.46 ± 0.28 | 1.94 ± 0.36 | 3.3 |
| dl143-164 | 1.33 ± 0.22 | 1.12 ± 0.24 | 1.5 |
| dl142-182 | 0.69 ± 0.05 | 1.33 ± 0.24 | 2.1 |
| dl136-185 | 0.40 ± 0.13 | 1.92 ± 0.18 | 2.4 |
| inB/dl143-148/dp149-172/ G272T | 0.32 ± 0.04 | 2.32 ± 0.26 | 2.1 |
| dl139-188/dp2-135 | 0.45 ± 0.08 | 2.27 ± 0.33 | 2.4 |
Growing NIH 3T3 cells were transfected with 4.0 μg of plasmid pGL2-basic/MPtVrr with the regulatory region in the early and late orientations relative to the luciferase reporter gene. Transfection was carried out in triplicate, and luciferase activity from duplicate samples of each transfected culture was assayed. Enzyme activity is indicated relative to the level obtained with the early promoter of stMPtV DNA. The variation in luciferase activity is also indicated.
For analysis of the activity of the MPtV origin of DNA replication, cells were transfected with equal amounts of pGL2-basic/MPtVrr and pcDNA3/MPtV-LT. The amount of newly replicated DNA at 42 h after transfection was determined.
The origin of replication in DNA of polyomaviruses has a highly conserved structure. There is an essential core element, consisting of large T antigen binding sites and adjacent sequences. In addition, there are auxiliary elements located in the enhancer, where specific binding of cellular factors cooperates with large T antigen in the initiation of viral DNA synthesis (9).
We showed earlier (50) that MPtV DNA is able to replicate in NIH 3T3 cells when large T antigen is expressed at a sufficiently high level. In the present experiments, NIH 3T3 cells were cotransfected with a pGL2-basic construct, containing the regulatory region of the viral genome, and a second plasmid, pcDNA3/MPtV-LT, expressing the MPtV large T antigen. Thus, the synthesis of reporter plasmid DNA was uncoupled from the early gene expression of the regulatory region being tested.
At ca. 40 h posttransfection low-molecular-weight DNA was selectively extracted, partially purified, and then incubated with the restriction endonuclease DpnI to fragment unreplicated plasmid DNA. Analysis by Southern blotting (Table 3) showed that the activity of the origin of viral DNA replication at a given concentration of large T antigen in NIH 3T3 cells was not significantly increased by insertion-deletion of nucleotides at nt 142. However, the G272T mutation in a genetic background of stMPtV or its deletion-insertion derivatives approximately doubled the amount of replicated viral DNA. Of the remaining variant regulatory regions, the majority was more effective than the standard-type structure, increasing the amount of replicated DNA by a factor of 1.5 to 3.3. The variability of the replication data is not reported because only one experiment included all plasmid constructs. However, the results of several experiments with groups of regulatory region variants were consistent with the replication data reported in Table 3. In general, sequence variations that had the largest effect on the late promoter also increased the activity of the replication origin. However, there was no linear relationship between the effects on the activities of the late promoter and the replication origin.
Relationship of the MPtV enhancer insertion to cellular DNA.
Based on the assumption that stMPtV was a progenitor of the enhancer insertion-deletion mutants, a probable source of the insertion is cellular DNA. In a BLAST search of the mouse DNA database (2) in which the whole 220-bp insertion of inA was used, 64 entries with significant similarity (for smallest-sum P, 0.13 < P < 1.00, 58 to 67% identities) to 202 bp or less of the query sequence were detected (18 November 2002). When the BLAST search was extended to all notated rodent and human sequences, one rat sequence (gb AC079378) with higher similarity (smallest-sum P = 0.072) to inA than the top-scoring mouse sequence (gb AL589735) was revealed. The BLAST search did not reveal more than five sequences with short (20 to 23 bp), perfect, or nearly perfect similarity in the entire nucleic acid database.
The observation that there were 220-bp insertions with two slightly different sequences and that these insertions apparently could be deleted with retention of one endpoint suggested that they might be related to cellular mobile genetic elements. However, analysis of the 220-bp element with the RepeatMasker program (http://genome.washington.edu) did not reveal significant similarity to any known cellular repeated sequence.
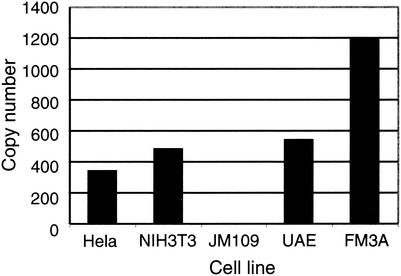
To investigate whether there are nucleotide sequences in cellular genomes related to the 220-bp segment from MPtV DNA, Southern blot analysis was used. DNA was extracted from the mouse cell lines NIH 3T3, UAE, and FM3A; from human HeLa cells; and from E. coli JM109 cells. Purified DNA, 10 μg, was digested with the restriction endonuclease BglII and was then resolved by agarose gel electrophoresis. Ethidium bromide staining of the gel showed that the genomic DNA was cleaved (Fig. 3A and B), and Southern blot analysis, with 32P-labeled 220-bp insert DNA as a probe, showed strong annealing to mouse and human DNA, while no reaction with E. coli DNA was detected (Fig. 3C). The probe did not base pair with a unique DNA fragment. Instead, the annealing pattern was typical for highly repeated sequences. Out of the three mouse cell lines, DNA from FM3A cells, which is derived from C3H mice—the mouse strain which was used for passage of the investigated MPtV—appeared to react more strongly with the probe than did DNA from NIH 3T3 and UAE cells, suggesting that mouse strains differ with respect to representation of the 220-bp-related sequences in their genome. Quantification of probe annealing to dot blot DNA, with MPtV DNA as a standard, indicated that the copy number of nucleotide sequences related to the 220-bp insert in DNA isolated from FM3A cells exceeded 1,000 (Fig. 4).
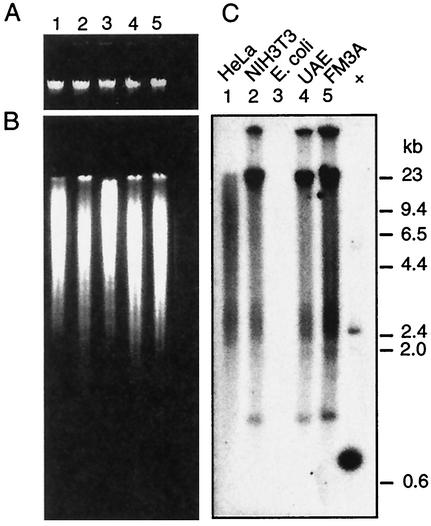
FIG. 3.
Southern blot analysis of MPtV insert-related sequences in cellular DNA. High-molecular-weight DNA was purified from human HeLa cells (lane 1); from mouse NIH 3T3 (lane 2), UAE (lane 4), and FM3A cells (lane 5); and from E. coli JM109 cells (lane 3). (A) Two micrograms of the DNA preparations was subjected to agarose gel electrophoresis and then stained with ethidium bromide. (B) Ten micrograms of the DNA preparations was digested with the restriction endonuclease BglII. The resulting fragments were resolved by agarose gel electrophoresis and then stained with ethidium bromide. (C) The resolved DNA fragments were subjected to Southern blot analysis with 32P-labeled DNA of the 220-bp MPtV insert as a probe. As a positive control, 5 ng of pstMPtV linearized by BglII cleavage was used (+). The positions of size markers (in kilobase pairs) are shown on the right.
FIG. 4.
Estimation of copy number of MPtV insert-related material in genomic DNA. The same DNA preparations and radioactive probe were used as in the experiment described in the Fig. 3 legend. Radioactivity annealing to dot blotted material was quantified in a PhosphorImager. Copy numbers were estimated in relation to the signals obtained with purified pstMPtV DNA.
Although many or even most of the 220-bp related DNA sequences in mouse DNA might be only partially complementary to the viral DNA segment, at least one copy in the mouse genome should be perfectly complementary if the 220-bp segment in MPtV DNA was acquired by transposition. For detection by PCR of such genomic copies, a primer pair was selected that would generate a 179-bp fragment, with inA or inB DNA as a template. In this experiment, DNA isolated from both C3H and C57BL/6 mice was used. From both types of templates, two dominating PCR products were formed. The smaller of these had a size of ca. 180 bp. Figure 5 shows the PCR products from C3H mouse DNA resolved by agarose gel electrophoresis. Similar results were obtained with C57BL/6 DNA (data not shown). Since the specificity of the priming was critical, the PCR was tested at a range of MgCl2 concentrations. For further analysis, DNA was purified from the reactions run at 2.5 mM MgCl2 and was ligated to the plasmid pGEM-T. After transformation of E. coli cells, individual plasmid clones were isolated and then amplified. Nucleotide sequence analysis showed that the smaller PCR product was closely related to the insertion in MPtV DNA, whereas the larger product had little similarity outside the primer positions. The sequence data, summarized in Table 4, show that all 12 plasmid clones that were analyzed contained inserts that were very similar to the 220-bp viral sequence. However, only two of the inserts were identical to the viral DNA.
TABLE 4.
Nucleotide sequence variation of the MPtV-related sequences in mouse DNA amplified by PCRa
| Clone no. | Genome source | Base pair substitutionb | Insertion/deletionb |
|---|---|---|---|
| 1 | C3H | G36A | |
| 2 | C3H | A103G, A193G | in174/dl182 |
| 3 | C3H | A150G | |
| 4 | C3H | T73A, T80A, T136C, A193G | |
| 5 | C3H | T80A, T136C | |
| 6 | C3H | A78G, T87C, A99G | |
| 7 | C3H | T125A, A152G | in58/dl139 |
| 8 | C57BL/6 | T85C, A114G, A145G, T174A | |
| 9 | C57BL/6 | A99G, T158C, A193G | |
| 10 | C57BL/6 | A71G, A106T, A173G, A182C | |
| 11 | C57BL/6 | ||
| 12 | C57BL/6 |
The PCR was done with genomic DNA isolated from C3H and C57BL/6 mice and with primers corresponding to nt 23 to 45 and 178 to 201, respectively of the 220-bp insert in the MPtV genome.
Numbers refers to the position in the 220-bp insert.
DISCUSSION
In comparison to other parts of polyomavirus genomes, their regulatory regions are extremely variable in structure. In various polyomaviruses, a large number of regulatory region variants have been identified (human polyomavirus BK [33, 34], JC [48], and SV40 [29]). Most of the variation occurs in a short segment of the genome first identified in polyomavirus DNA as an enhancer of early transcription (10). Being a target for cellular DNA-binding proteins with regulatory functions in transcription, enhancers in cellular and viral DNA consist of an array of recognition sequences arranged so that their ligands can interact with each other and with the transcription complex (37). Besides the regulatory function in transcription, enhancers influence the activity of adjacent origins of DNA replication (9). The complexity of enhancers relates to need of regulation and cell type specificity of gene expression. For polyomaviruses that have the ability to multiply in several different tissues and at different rates during the acute and persistent phase of the infection, the structure of the enhancer must be a result of intricate selection processes.
Mutation of the enhancer has consequences that are difficult to predict. The cell type specificity of the enhancer function means that the selective effect of the cellular environment varies during the infection of an animal. Secondly, the function of the enhancer is cis acting, and the enhancers that are present in the same cell compete for cognate DNA-binding proteins. Consequently, a genome having an enhancer with superior fitness will suppress the activity of genomes with less fit enhancers (36). This condition is of particular importance in viral DNA replication, leading to the rapid disappearance of genomes with enhancers of inferior fitness. Conversely, a viral genome with an enhancer that has gained in fitness might become predominant in a short period.
In enhancers of polyomavirus genomes, the pattern of variation is quite uniform. Most variants contain either deletions or duplications, compared with the archetype. Although the mutation of enhancers probably is a random process, the genetic alterations observed in individual polyomavirus populations have typical locations that depend on prior selection. Besides deletions, duplications, and point mutations, there are many reported examples of integration of cellular DNA segments. In fact, even the archetype enhancers in polyomavirus genomes might be derived from cellular DNA relatively recently, since their function is not fundamentally different from corresponding elements in the cell.
The present study revealed that MPtV harvested from infected C3H mice had genomes with very heterogeneous enhancers. Out of 30 clonal isolates, only four had identical enhancers and just two were identical to the published standard type of MPtV DNA. Besides deletions and duplications, a large proportion contained an insertion-deletion. None of these genomes was predominant in the population. In addition, the distribution of variants appeared relatively stable. After another passage of MPtV in C57BL/6 mice followed by nucleotide sequence analysis of PCR-amplified regulatory region DNA, several of the previously identified MPtV variant types were still present (Table 2).
The insertions were either 220 bp long (Fig. 1 and 6A) or shorter derivatives of these DNA segments, probably formed by secondary deletion, since one or both ends of the inserted segment were preserved. The interpretation of the data that the standard-type genome was formed by deletion of the 220-bp segment is unlikely, because it does not explain how TGAATA is inserted concomitantly with the deletion. Instead, the sequence data suggested that the 220-bp insert was a cellular transposable element with a target in the enhancer of the MPtV genome. The observation that two types of 220-bp inserts (inA and inB), differing slightly in sequence at one end, were present in the population corroborated our hypothesis. For both types of insertion events, the sequence 5′-TGAATA-3′ was deleted concomitantly with the integration of the cellular DNA segment. The integration might be initiated by cleavage of MPtV DNA 3′ to nt 142 and 5′ to nt 149. There is a sequence with mirror symmetry of the deleted segment in the two strands that has its axis at nt 143. Moreover, in one of the strands, there are TA dinucleotides at both breakpoints, suggesting that an endonuclease with a sequence preference was involved. Since topoisomerase I is participating in MPtV DNA replication, it is a candidate. With this enzyme, the scissile strand has the preferred nucleotides 5′-(A/T)(G/C)(A/T)T-3′ (8) and separate cleavages immediately 3′ to closely spaced recognition sites in opposite strands of DNA would lead to a double-strand break with staggered ends. There are potential cleavage sites in MPtV DNA 3′ to nt 141 in one strand of MPtV DNA and 3′ to nt 148 in the opposite strand. Topoisomerase I cleavage at both these sites would generate a double-strand break with 5′-protruding ends. Such linear molecules of MPtV DNA could be utilized in site-specific recombination.
FIG. 6.
Nucleotide sequences of putative transposon in MPtV DNA and the integration site. The complete nucleotide sequence of the 220-bp inA insertion in the MPtV regulatory region is shown in panel A. The integration site in stMPtV, as well as the junctions between standard-type DNA and selected insertions, is shown in panel B. In the standard-type nucleotide sequence, thin vertical lines indicate the insertion site and dashed horizontal lines indicate a mirror symmetry of the pentanucleotide TGAATA. In the MPtV variant sequences, inserted segments are shaded and the TGAATA deletion is indicated. In inB DNA, the arrows show terminal direct repeats.
Figure 6B depicts the target site and the terminal sequences of the insert. Three base pairs in the 3′ end distinguish inA from inB. As a result of this difference, a 6-bp direct terminal repeat (CTTAGA) is generated by the insertion in the inB variant. Besides the 220-bp insertions, there were variant genomes with shorter inserts, probably formed by secondary deletions. In four of the cloned regulatory regions with nonstandard type (15%), the presumed secondary deletion had removed one end of insertion precisely, as exemplified by inA/B(1-6). This observation suggests the presence of a specific excision mechanism.
In comparison to most transposons of mammalian cells, the 220-bp inserts are short, have no coding potential, and apparently have high target site specificity. These features and the A-T content suggest a relationship with a recently discovered class of elements, MITEs. Although most, but not all, MITEs contain inverted terminal repeats for their transposition (4-7, 45), the MPtV insert and the flanking virus sequence do not contain obvious terminal inverted repeats. Instead, the 220-bp insert has short direct repeats like a retrotransposon. There is an earlier observation (11) of an SV40 mutant with a 175-bp segment related to the Alu family of interspersed repeated sequences in primate DNA. These elements belong to the retrotransposons.
Southern blot analysis of cellular DNA, with the 220-bp segment of MPtV DNA as a probe, confirmed that there were closely related nucleotide sequences in mouse and human DNA but not in DNA from E. coli (Fig. 3). Under hybridization conditions that would result in one signal for a unique DNA sequence, the probe annealed to an array of BglII-cleaved DNA molecules, suggesting that it was complementary to a highly repeated nucleotide sequence. Analysis by dot blot (Fig. 4) showed that the mouse genome might contain as many as 103 copies of the MPtV repeat. A BLAST search of the mouse DNA sequence database confirmed the presence of many sequences related to the 220-bp inA and -B sequence in the mouse genome. Although none of these was identical to the query sequence, PCR amplification with primers complementary to sequences near the end of the 220-bp sequence indicated that there are multiple copies of very closely related sequences in the mouse genome. Although it is difficult to rigorously rule out that the sequence variation observed in these copies was generated during the PCR, the occurrence of single-nucleotide deletions and insertions and the same substitution in more than one clone (T80A, T136C, and A193G) make polymerase errors unlikely. Furthermore, the sequence variation in the larger PCR product was 10-fold less than in the data shown in Table 4. Thus, we conclude that there are a fairly large number of copies closely related to the 220-bp segment in the mouse genome. In addition, some of these copies are probably identical to the DNA segment observed in viral DNA. However, these copies were, apparently, fewer than the number of positive signals in Southern blot and dot blot analysis. An explanation for this discrepancy is that only those copies with nucleotide sequences identical to the primers were detected by PCR.
For cellular genes it has been reported (18) that integration of transposons might alter both the level of expression and the spatial expression pattern of adjacent genes. Transposable elements might also contain tissue-specific enhancers. The individual enhancers isolated from the MPtV genome varied in their activity in gene expression and viral DNA replication (Table 3). These experiments were carried out by transfection of NIH 3T3 cells that are nonpermissive for multiplication of stMPtV (50). However, the sole purpose of the analyses was to demonstrate differences in activity between the standard-type and mutant enhancers. Although the 220-bp inA in combination with the 6-bp deletion added putative CREB, NF1, and Oct1 sites to the enhancer (Fig. 2), it decreased the activity of the early viral promoter in NIH 3T3 cells (Table 3). However, the insertion might be advantageous in other cell types. Moreover, some of the mutant enhancers increased the activity of the viral origin of DNA replication in NIH 3T3 cells.
Besides the insertion-deletions, another frequently occurring variation in the regulatory region of the MPtV genome was the G272T base substitution. This polymorphism, mapping at a large T antigen binding site (unpublished data), had a positive effect on the activity of both the viral early and late promoters and in addition augmented viral DNA replication. This global effect on the regulatory region—expressed in the absence of large T antigen—suggested that the mutation led to a more general consequence, such as modification of chromatin structure. The G272T substitution also provided a genetic marker, based on the assumption that the G↔T mutation occurred at a frequency similar to that of transversions in cellular genomes. Since, both G272 and T272 was observed together with the inA insertion-deletion, as well as in the standard-type background, the insertion events probably occurred much more frequently than the transversion at nt 272. An alternative explanation of the data is that recombination was extraordinarily frequent in MPtV DNA, since the insertion site at nt 142 and the point mutation at nt 272 are very close. Only site-specific recombination might occur at such high frequencies.
The identification of a transposable element in MPtV DNA raises several questions. What family of repeated sequences does the 220-bp MPtV inserts belong to? Does the cellular mobile genetic element have a regular function in the viral life cycle? Can new events of transposition be observed in infection of cultured cells or in infected mice? Is this phenomenon unique to MPtV or ubiquitous in DNA virus evolution? Thus, the MPtV system might provide a useful model for studies of phenomena related to DNA transposition in mammalian cells.
Acknowledgments
The experimental work reported in this paper was supported financially by the Swedish Cancer Society.
REFERENCES
- 1.Bastaki, M., E. E. Nelli, P. Dell'Era, M. Rusnati, M. P. Molinari-Tosatti, S. Parolini, R. Auerbach, L. P. Ruco, L. Possati, and M. Presta. 1997. Basic fibroblast growth factor-induced angiogenic phenotype in mouse endothelium. A study of aortic and microvascular endothelial cell lines. Arterioscler. Thromb. Vasc. Biol. 17:454-464. [DOI] [PubMed] [Google Scholar]
- 2.Blake, J. A., J. E. Richardson, C. J. Bult, J. A. Kadin, and J. T. Eppig. 2002. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res. 30:113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond, S. B., P. M. Howley, and K. K. Takemoto. 1978. Characterization of K virus and its comparison with polyoma virus. J. Virol. 28:337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bureau, T. E., P. C. Ronald, and S. R. Wessler. 1996. A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc. Natl. Acad. Sci. USA 93:8524-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bureau, T. E., and S. R. Wessler. 1994. Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91:1411-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bureau, T. E., and S. R. Wessler. 1994. Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6:907-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bureau, T. E., and S. R. Wessler. 1992. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 9.DePamphilis, M. L. 1993. Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem. 62:29-63. [DOI] [PubMed] [Google Scholar]
- 10.de Villiers, J., and W. Schaffner. 1981. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 9:6251-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhruva, B. R., T. Shenk, and K. N. Subramanian. 1980. Integration in vivo into simian virus 40 DNA of a sequence that resembles a certain family of genomic interspersed repeated sequences. Proc. Natl. Acad. Sci. USA 77:4514-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, D., M. D. Jones, A. Leigh-Brown, and B. E. Griffin. 1982. Mutant din-21, a variant of polyoma virus containing a mouse DNA sequence in the viral genome. EMBO J. 1:461-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorries, K. 1998. Molecular biology and pathogenesis of human polyomavirus infections. Dev. Biol. Stand. 94:71-79. [PubMed] [Google Scholar]
- 14.Elsner, C., and K. Dorries. 1998. Human polyomavirus JC control region variants in persistently infected CNS and kidney tissue. J. Gen. Virol. 79:789-799. [DOI] [PubMed] [Google Scholar]
- 15.Finnegan, D. J. 1992. Transposable elements. Curr. Opin. Genet. Dev. 2:861-867. [DOI] [PubMed] [Google Scholar]
- 16.Fried, M., and B. E. Griffin. 1977. Organization of the genomes of polyoma virus and SV40. Adv. Cancer Res. 24:67-113. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura, F. K., and E. Linney. 1982. Polyoma mutants that productively infect F9 embryonal carcinoma cells do not rescue wild-type polyoma in F9 cells. Proc. Natl. Acad. Sci. USA 79:1479-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard, L., and M. Freeling. 1999. Regulatory changes as a consequence of transposon insertion. Dev. Genet. 25:291-296. [DOI] [PubMed] [Google Scholar]
- 19.Greenlee, J. E. 1981. Effect of host age on experimental K virus infection in mice. Infect. Immun. 33:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenlee, J. E. 1979. Pathogenesis of K virus infection in newborn mice. Infect. Immun. 26:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenlee, J. E., S. H. Clawson, R. C. Phelps, and W. G. Stroop. 1994. Distribution of K-papovavirus in infected newborn mice. J. Comp. Pathol. 111:259-268. [DOI] [PubMed] [Google Scholar]
- 22.Greenlee, J. E., R. C. Phelps, and W. G. Stroop. 1991. The major site of murine K papovavirus persistence and reactivation is the renal tubular epithelium. Microb. Pathog. 11:237-247. [DOI] [PubMed] [Google Scholar]
- 23.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 24.Houck, C. M., F. P. Rinehart, and C. W. Schmid. 1979. A ubiquitous family of repeated DNA sequences in the human genome. J. Mol. Biol. 132:289-306. [DOI] [PubMed] [Google Scholar]
- 25.Jones, N. C., P. W. Rigby, and E. B. Ziff. 1988. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 2:267-281. [DOI] [PubMed] [Google Scholar]
- 26.Katinka, M., M. Yaniv, M. Vasseur, and D. Blangy. 1980. Expression of polyoma early functions in mouse embryonal carcinoma cells depends on sequence rearrangements in the beginning of the late region. Cell 20:393-399. [DOI] [PubMed] [Google Scholar]
- 27.Kilham, L., and H. W. Murphy. 1953. A pneumotropic virus isolated from C3H mice carrying the Bittner milk agent. Proc. Soc. Exp. Biol. Med. 82:133-137. [DOI] [PubMed] [Google Scholar]
- 28.Lednicky, J. A., A. S. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 72:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lednicky, J. A., and J. S. Butel. 2001. Simian virus 40 regulatory region structural diversity and the association of viral archetypal regulatory regions with human brain tumors. Semin. Cancer Biol. 11:39-47. [DOI] [PubMed] [Google Scholar]
- 30.Maione, R., C. Passananti, V. De Simone, P. Delli-Bovi, G. Augusti-Tocco, and P. Amati. 1985. Selection of mouse neuroblastoma cell-specific polyoma virus mutants with stage differentiative advantages of replication. EMBO J. 4:3215-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer, M., and K. Dorries. 1991. Nucleotide sequence and genome organization of the murine polyomavirus, Kilham strain. Virology 181:469-480. [DOI] [PubMed] [Google Scholar]
- 32.Mokhtarian, F., and K. V. Shah. 1980. Role of antibody response in recovery from K-papovavirus infection in mice. Infect. Immun. 29:1169-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monini, P., A. Rotola, D. Di Luca, L. De Lellis, E. Chiari, A. Corallini, and E. Cassai. 1995. DNA rearrangements impairing BK virus productive infection in urinary tract tumors. Virology 214:273-279. [DOI] [PubMed] [Google Scholar]
- 34.Negrini, M., S. Sabbioni, R. R. Arthur, A. Castagnoli, and G. Barbanti-Brodano. 1991. Prevalence of the archetypal regulatory region and sequence polymorphisms in nonpassaged BK virus variants. J. Virol. 65:5092-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman, J. T., and R. J. Frisque. 1999. Identification of JC virus variants in multiple tissues of pediatric and adult PML patients. J. Med. Virol. 58:79-86. [PubMed] [Google Scholar]
- 36.Nilsson, M., M. Osterlund, and G. Magnusson. 1991. Analysis of polyomavirus enhancer-effect on DNA replication and early gene expression. J. Mol. Biol. 218:479-483. [DOI] [PubMed] [Google Scholar]
- 37.Ondek, B., L. Gloss, and W. Herr. 1988. The SV40 enhancer contains two distinct levels of organization. Nature (London) 333:40-45. [DOI] [PubMed] [Google Scholar]
- 38.Oren, M., S. Lavi, and E. Winocour. 1978. The structure of a cloned substituted SV40 genome. Virology 85:404-421. [DOI] [PubMed] [Google Scholar]
- 39.Parsons, D. F. 1963. Morphology of K virus and its relation to the papova group of viruses. Virology 20:385-388. [Google Scholar]
- 40.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rochford, R., B. A. Campbell, and L. P. Villarreal. 1987. A pancreas specificity results from the combination of polyomavirus and Moloney murine leukemia virus enhancer. Proc. Natl. Acad. Sci. USA 84:449-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers, J. H. 1985. The origin and evolution of retroposons. Int. Rev. Cytol. 93:187-279. [DOI] [PubMed] [Google Scholar]
- 43.Takemoto, K. K., and P. Fabisch. 1970. Transformation of mouse cells by K-papovavirus. Virology 40:135-143. [DOI] [PubMed] [Google Scholar]
- 44.Trotot, P., F. Mechali, D. Blangy, and M. Lacasa. 1994. Transcriptional activity in 3T3, F9, and PCC4 embryonal carcinoma cells: a systematic deletion and linker-scanning study of the polyomavirus enhancer. Virology 202:724-734. [DOI] [PubMed] [Google Scholar]
- 45.Tu, Z., and S. P. Orphanidis. 2001. Microuli, a family of miniature subterminal inverted-repeat transposable elements (MSITEs): transposition without terminal inverted repeats. Mol. Biol. Evol. 18:893-895. [DOI] [PubMed] [Google Scholar]
- 46.Wessler, S. R., T. E. Bureau, and S. E. White. 1995. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5:814-821. [DOI] [PubMed] [Google Scholar]
- 47.Wingender, E., X. Chen, R. Hehl, H. Karas, I. Liebich, V. Matys, T. Meinhardt, M. Pruss, I. Reuter, and F. Schacherer. 2000. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yogo, Y., T. Matsushima-Ohno, T. Hayashi, C. Sugimoto, M. Sakurai, and I. Kanazawa. 2001. JC virus regulatory region rearrangements in the brain of a long surviving patient with progressive multifocal leukoencephalopathy. J. Neurol. Neurosurg. Psychiatry 71:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Q., J. Arbuckle, and S. R. Wessler. 2000. Recent, extensive, and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker into genic regions of maize. Proc. Natl. Acad. Sci. USA 97:1160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, S., and G. Magnusson. 2001. Kilham Polyomavirus: activation of gene expression and DNA replication in mouse fibroblast cells by an enhancer substitution. J. Virol. 75:10015-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]