Abstract
Recent studies demonstrated the ability of the recombinant autonomous parvoviruses MVMp (fibrotropic variant of the minute virus of mice) and H-1 to transduce therapeutic genes in tumor cells. However, recombinant vector stocks are contaminated by replication-competent viruses (RCVs) generated during the production procedure. To reduce the levels of RCVs, chimeric recombinant vector genomes were designed by replacing the right-hand region of H-1 virus DNA with that of the closely related MVMp virus DNA and conversely. Recombinant H-1 and MVMp virus pseudotypes were also produced with this aim. In both cases, the levels of RCVs contaminating the virus stocks were considerably reduced (virus was not detected in pseudotyped virus stocks, even after two amplification steps), while the yields of vector viruses produced were not affected. H-1 virus could be distinguished from MVMp virus by its restriction in mouse cells at an early stage of infection prior to detectable viral DNA replication and gene expression. The analysis of the composite viruses showed that this restriction could be assigned to a specific genomic determinant(s). Unlike MVMp virus, H-1 virus capsids were found to be a major determinant of the greater permissiveness of various human cell lines for this virus.
The autonomous replicating parvoviruses can infect a wide variety of animal species, including humans (26). These viruses are able to replicate without assistance of a helper virus, and their DNA does not appear to integrate into the host cell genome. They are strictly dependent on cellular factors for their replication (8), restricting the spectrum of cells that are permissive for parvovirus infection. Parvoviruses are lytic viruses, i.e., they kill the permissive cells which undergo a successful infection. This lytic activity has been ascribed, at least in part, to the nonstructural (NS) viral proteins (4, 23). In vitro transformation of various cells with oncogenes or other treatments was found to correlate with an increase in their permissiveness to parvovirus infection over that of nontransformed cell cultures (6, 25). Furthermore, some of these agents, including the rat parvovirus H-1 and the closely related minute virus of mice (MVM), can protect their host from developing tumors or induce tumor regression in laboratory animals (11, 14). The above-mentioned properties of these viruses, together with their ability to cause clinically asymptomatic infections, point to the potential use of these agents as vectors for the transfer and expression of anticancer genes in tumors (16).
Our laboratory has developed autonomous parvovirus H-1- and MVMp (fibrotropic variant of MVM)-based vectors for cancer therapy in which the early promoter P4 and the sequences encoding the NS proteins have been retained, but the genes coding for the capsid proteins (VP) have been partially deleted and replaced by different transgenes (10, 16, 17, 21, 24, 28). These recombinant viruses express their transgene under the control of the viral P38 promoter that is strongly transactivated by the vector-encoded NS1 protein (18). Consequently, permissive cells accumulate transiently high levels of transgene products upon infection with recombinant parvoviruses (16, 28). The vector genomes are packaged by cotransfection with a helper plasmid expressing both capsid proteins VP1 and VP2 (17). The titers of the virus stocks produced in this way (1 × 107 to 5 × 107 replication units/ml) allowed recent attempts at evaluating the antitumor capacity of recombinant autonomous parvoviruses that carry therapeutic transgenes in recipient mice. H-1 virus vectors expressing human interleukin 2 (16) or the chemokine MCP-3 (28) proved able to inhibit tumor formation or growth under conditions in which the wild-type virus had little or no effect, providing the first evidence of enhanced antitumor activity of recombinant autonomous parvoviruses harboring a therapeutic transgene compared with wild-type viruses. However, parvovirus vector stocks were found to be contaminated at low, but significant levels (0.15 to 1%), with replication-competent viruses (RCVs) probably generated through recombination between overlapping sequences shared by the vector genomes and helper plasmids.
This prompted us to modify the existing vector genomes to minimize the probability of generating RCVs by homologous recombination between the vector backbone and the helper plasmid. To this end, we took advantage of the closely related genomic organization and sequence of the rodent parvoviruses H-1 and MVMp, which show an overall DNA sequence identity of about 80%. We first created chimeric recombinant vector genomes by replacing the right-hand region of the rat H-1 virus DNA with that of the mouse MVMp virus and conversely. A second approach consisted of packaging the genomes of recombinant H-1 and MVMp viruses with capsid proteins from MVMp and H-1 viruses, respectively, i.e., producing pseudotyped viruses. By using both strategies, high-titer virus stocks could be obtained in which the levels of contaminating RCVs dropped dramatically or were even undetectable in a number of cases, without affecting the yields of vector viruses produced. In addition, we analyzed the host range and transducing capacities of the pseudotyped recombinant viruses. The failure of H-1 virus to successfully infect mouse cells that are permissive for MVMp virus was traced back to differences in the genomes but not in the capsids of incoming viruses. In contrast, the determinant of the greater infectivity of H-1 virus compared with MVMp virus for human cells was assigned, to a large extent, to the capsids of the virions.
Construction of chimeric H-1 and MVMp virus genomes and capsid helper plasmids.
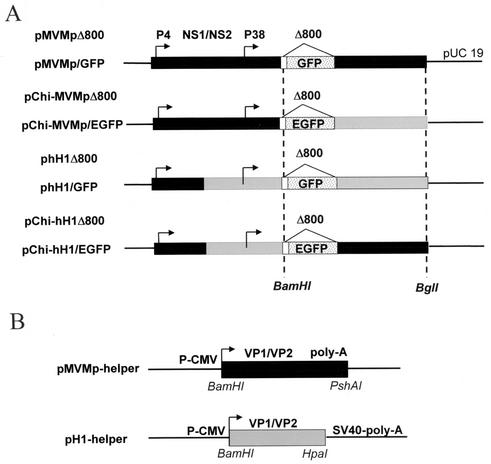
Chimeric viruses were generated by exchanging the region downstream of the multiple cloning site (MCS) between vectors phH1Δ800 and pMVMpΔ800 (17). This region comprises the 3′ ends of the capsid (VP) genes, the internal cis elements that have been reported to be required for viral DNA replication and the right-hand palindrome (17). The exchange was performed by cloning a BamHI-BglI DNA fragment from pMVMpΔ800 (2,433 bp) or phH1Δ800 (2,598 bp) into phH1Δ800 or pMVMpΔ800 (16) that had been digested with the same restriction enzymes, respectively. The resulting chimeric vector DNAs, pChi-hH1Δ800 and pChi-MVMpΔ800, are schematically represented in Fig. 1A.
FIG. 1.
Structures of standard MVMp- and hH1-based vectors, derived chimeras, and helper plasmids. (A) The standard parvoviral vectors were derived from the modified infectious DNA clones (pMVMp and phH1) by deleting about 800 bp from the coding sequence of the structural VP1 and VP2 capsid genes and inserting a MCS (white box) at the VP2 translation initiation codon. Both promoters (P4 and P38) are indicated by arrows. The region encoding the nonstructural proteins (NS1/NS2) is shown. Chimeric MVMp-based vector DNA was constructed by replacing a BamHI-BglI fragment from pMVMpΔ800 with the corresponding region of phH1Δ800, resulting in pChi-MVMpΔ800. Chimeric hH1-based vector DNA was obtained by inserting the BamHI-BglI fragment of MVMpΔ800 into phH1Δ800 cleaved with the same restriction enzymes, resulting in pChi-hH1Δ800. The MVMp (black boxes) and hH1 (grey boxes) DNA sequences are indicated. GFP or EGFP was cloned in standard (pMVMp/GFP and phH1/GFP) and chimeric (pChi-MVMp/EGFP and pChi-hH1/EGFP) DNA vectors by inserting a 731-bp NotI restriction fragment from plasmid pTR/UF2 (30) or a 784-bp SacI-NotI restriction fragment from plasmid pEGFP-1 (Clontech), respectively, into the vectors digested with the same restriction enzymes. (B) DNA helper constructs. The VP1 and VP2 genes from either MVMp or H-1 virus are expressed under the control of the immediate-early promoter of human CMV (P-CMV). Recombinant parvoviruses were produced by cotransfection of human 293T cells with the vector DNA and helper plasmid as indicated in the text. SV40, simian virus 40.
The helper plasmid DNAs were constructed by inserting the entire VP coding region under control of the cytomegalovirus (CMV) immediate-early promoter in expression vectors pBK-CMV (Stratagene) and pcDNA1-1 (Invitrogen) for the production of H-1 and MVMp virus capsid proteins, respectively. These constructs retained the 5′ untranslated sequences of the corresponding mRNAs and the splice donor and acceptor sites for VP1 and VP2. In the helper construct expressing the VP proteins from H-1 virus, most of the 3′-end untranslated sequences of the capsid genes were deleted and replaced by transcription termination and simian virus 40 polyadenylation signals from the expression plasmid, while MVM helper DNA retained its own viral signals. The helper constructs are shown in Fig. 1B.
All the final constructs (Fig. 1) were verified by sequencing. In comparison with the sequences available in the GenBank database, a few mutations were identified in the coding sequence for MVM and H-1 virus capsids, some of which lead to changes in the amino acid composition of the corresponding translated product (Table 1).
TABLE 1.
Nucleotide differences (and predicted amino acid changes) between MVMp or H-1 virus VP-coding sequences from GenBank and the corresponding sequences in pMVMp helper and pH1 helper DNA
| Virus sequence and nucleotide position | Nucleotide change | Amino acid change |
|---|---|---|
| MVM VP | ||
| 3597 | T → C (transition) | Silent |
| 3619 | T → C (transition) | S → P |
| 4111 | G → A (transition) | D → N |
| 4359 | C → T (transition) | Silent |
| H1 VP | ||
| 4486 | T → C (transition) | Silent |
| 3113 | Insertion of ATGa | YD → YDD |
| 3674 | Insertion of G | Frameshift GMPPR → ACLQG |
| 3688 | Deletion of G | Frameshift GMPPR → ACLQG |
| 4310 | A → G (transition) | H → R |
The ATG insertion is split between two codons TAT and GAT GAC (the ATG inserted is underlined).
To construct the recombinant vector DNAs expressing the marker green fluorescent protein (GFP) gene, a humanized GFP cDNA (30) was cloned as a NotI DNA fragment in the MCS of phH1Δ800 (28) or pMVMpΔ800, giving rise to phH1/GFP and pMVMp/GFP, respectively (Fig. 1A). Enhanced GFP (EGFP) (Clontech GmbH, Heidelberg, Germany) was inserted as a SacI-NotI fragment in the MCS of the chimeric vectors, creating pChi-hH1/EGFP and pChi-MVMp/EGFP, respectively (Fig. 1A).
Production of recombinant chimeric and pseudotyped virus stocks devoid of contamination with RCVs.
We examined whether the chimeric recombinant DNAs could be efficiently packaged by the helper constructs after cotransfection of producer cells. To this end, 2 × 106 293T cells/10-cm-diameter dish were cotransfected with 6 μg of vector DNA and 12 μg of the corresponding helper plasmid (28), using the calcium phosphate procedure. Virus stocks were harvested 72 h posttransfection by removing the medium, washing the cultures, and lysing the cells in a solution containing 50 mM Tris-HCl and 0.5 mM EDTA (pH 8.7) by three rounds of freezing and thawing as previously described (16). Cell debris was removed by low-speed centrifugation, and viruses were purified by nonionic iodixanol gradient centrifugation (a modified version of the method in reference 29). Virus stocks were diluted with medium before use.
We also examined whether MVMp and H-1 virus capsids could package phH1/GFP and pMVMp/GFP genomes to form hH1/GFP(MVM) and MVMp/GFP(H1) pseudotype viruses. For this purpose, standard phH1/GFP and pMVMp/GFP DNAs were transfected into producer cells with either MVMp or H-1 virus helper DNA to produce hH1/GFP(MVM) and MVMp/GFP(H1) pseudotyped virus stocks.
Both strategies were tested for their efficiency in recombinant virus production and their suitability to reduce the levels of RCVs in the recombinant virus stocks.
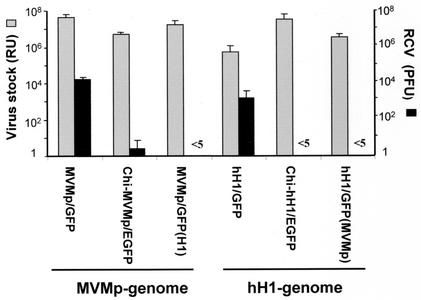
All the virus stocks were produced in parallel, and the titers of the viruses were determined by an infected-cell hybridization assay, using human NB-324K indicator cells, known to be permissive for both H-1 and MVMp viruses. This assay detects viral DNA that became amplified in infected cells, through hybridization to an EcoRV-EcoRI (703-bp) NS-specific 32P-labeled DNA probe. Virus titers are expressed as replication units (RU) per ml (16). Generation of contaminating viruses able to propagate, i.e., to give rise to progeny viruses (usually called RCVs), was revealed and quantified through infection of NB-324K indicator cells, with the recombinant stocks and measurement of either lysis plaques or hybridization using a VP-specific radiolabeled DNA probe. Like wild-type H-1 and MVMp viruses, RCVs from the standard recombinant virus stocks had only slightly (three- to fourfold) higher titers in hybridization versus plaque assays (data not shown), indicating that most of the virus contaminants recovering the VP gene were infectious, i.e., competent for plaque formation. Titers and RCV contamination of recombinant virus stocks produced with the chimeric vectors or through pseudotyping are summarized in Fig. 2. The titers of chimeric and pseudotyped recombinant viruses were similar to, or even higher (Chi-hH1 and hH1 pseudotype), than those of the standard MVMp/GFP and hH1/GFP virus stocks, ranging from 3.5 × 106 to 4.5 × 107 RU/2 × 106 transfected 293T cells. However, the proportion of RCVs was dramatically reduced in chimeric and pseudotyped virus stocks. Indeed, no RCV could be detected without amplification (passages). Plaques formed by RCVs could be detected only after one (chimeric vectors) or (only in one case) after two (pseudotyped vectors) additional rounds of virus amplification, respectively. It was calculated that the recombinant stocks produced from chimeric constructs contained less than 2 × 10−4% RCVs (which means less than 2 PFU/106 RU). This represents a decrease of at least 500-fold in RCV contamination compared with the standard vectors (16). Since at least two amplification steps (passages) were needed to detect RCVs by plaque assay among the pseudotyped recombinants (instead of one for the chimeric viruses), we estimated that the RCV contamination was further reduced at least by a factor of 50 to 100 in the pseudotyped stocks compared with the chimeric ones. However, in most cases, no RCV could be detected even after these amplification steps, suggesting that the recombination rate dropped considerably during the production.
FIG. 2.
Contamination of recombinant virus stocks by RCVs. Standard and chimeric recombinant virus stocks were produced by cotransfecting 293T cells with vector DNA and the corresponding helper plasmid (described in the legend of Fig. 1), while pseudotype virus stocks were produced by cotransfection of standard pMVMp/GFP and phH1/GFP with H1 helper and MVMp helper DNA, respectively, resulting in MVMp/GFP(H1) and hH1/EGFP(MVM). Virus stocks were harvested 72 h posttransfection and purified by centrifugation on iodixanol gradients, and virus titers were determined on NB324K cells by a hybridization assay using a specific 32P-labeled NS DNA probe consisting of a 703-bp EcoRV-EcoRI restriction fragment of either pMVMΔ800 or phH1Δ800. Total levels of virus production are expressed as RU per 2 × 106 transfected cells (grey bars). The proportion of replication-competent viruses present in the recombinant virus stocks was determined by plaque assay and expressed as PFU per 2 × 106 transfected cells (black bars). The results shown are the averages of at least three independent experiments, with standard deviations indicated by the error bars.
To rule out the possibility that these extremely low levels of RCVs detected could be due to lower levels of replication by those viruses rather than to a reduced rate of recombination, the relative rates of replication of the RCVs generated were compared to those of wild-type viruses. To this end, NB-324K cells were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell with either wild-type viruses (H-1 or MVMp) or RCVs from chimeric virus stocks (H-1 or MVMp) and from hH1(MVMp) pseudotype virus, respectively. Three days after infection, the titers of the newly produced viruses on NB-324K cells were determined by hybridization and plaque assays. In all cases, the titers and sizes of the plaques of the progeny viruses were identical (data not shown), indicating that the RCVs were not impaired in their replication ability and behave as wild-type viruses once they are generated. It is worth noting that in a recent study, Dupont et al. (9) modified MVMp virus vector DNA and helper plasmid to reduce recombination and thus the generation of RCVs. Nevertheless, significant levels (0.15%) of contaminating virus were still generated, as determined by hybridization assays. Although these viruses were defective for autonomous propagation, as they could not form plaques, they were able to replicate their DNA after infection and possibly represent defective viruses that arose by nonhomologous recombination (9).
In another approach, split helper plasmids were constructed by mutating or deleting the splice signals for VP1 and VP2 to express these proteins from separate plasmids and thereby reduce the generation of RCVs (3a). hH1 vectors were packaged using the split helpers, resulting in recombinant hH1 virus stocks that were indeed free of infectious RCVs but still comprised VP gene-containing defective viruses able to transfer and replicate their DNA. In this study, we never observed such a discrepancy between the titers of DNA replication-competent and infectious (i.e., propagation-competent) RCVs, as measured by hybridization and plaque assays, respectively (data not shown). The difference in RCV titers between the hybridization and plaque assays was never higher than fourfold and can probably be assigned to the greater sensitivity of the former method. The reason for this discrepancy between our observations and the observations of others is not clear at present. One possibility would be that our vectors, in contrast to those described in the above-mentioned publications, retain and express intact NS2 proteins in addition to NS1. NS2 is known to be involved in various steps of the viral life cycle, including DNA replication, efficient translation of viral mRNA, capsid assembly, and probably generation of viral single-stranded DNA (7, 19, 20). Recently, NS2 was shown to interact with members of the 14-3-3 protein family (3) and to bind to the mouse homologue of the human nuclear export factor Crm1 (2). MVMp virus mutants in which the nuclear export signal site within the NS2 proteins was disrupted showed nuclear retention not only of the viral NS2 products but also of infectious progeny MVMp virus particles, indicating a possible role of NS2 in the efficient release of progeny virions from infected cells (7, 12).
In light of this new function of NS2, we hypothesize that the defective RCVs described previously as contaminants of recombinant virus stocks may represent immature viruses, capable of replicating their DNA upon infection but unable to propagate autonomously. In conclusion, the composite viruses described in this study appear to represent a new generation of autonomous parvoviral vectors devoid of contamination with both DNA replication- and propagation-competent RCVs while retaining the production yields of the original vectors. Interestingly, the use of pseudotyped vectors should help to prevent preexisting neutralizing antibodies against parvoviral capsids from reducing the efficacy of recombinant virus (re)administration in gene therapy protocols.
In vitro cell tropism of chimeric and pseudotyped MVMp and hH1 viruses. (i). Viral DNA replication in mouse and human cells infected with chimeric and pseudotyped viruses.
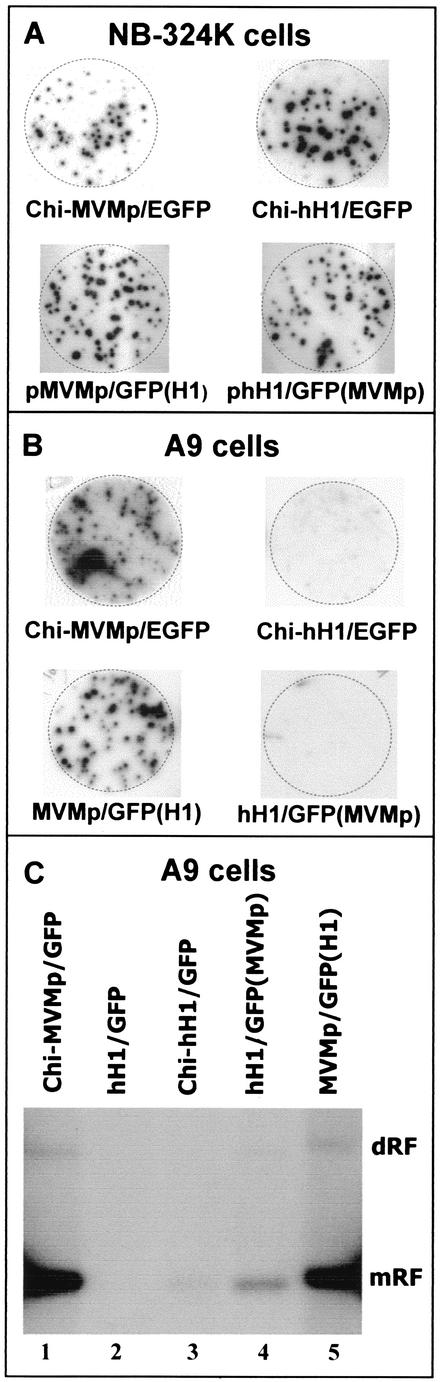
The chimeric viruses (Chi-MVMp and Chi-hH1) and pseudotyped viruses [MVMp(H1) and hH1(MVMp)] were compared to standard viruses (MVMp and hH1) for their ability to infect mouse and human cells. Stocks of standard and composite viruses were produced in parallel in 293T cells, and virus titers were determined by hybridization assay on human NB-324K cells using a 32P-labeled NS-specific DNA probe. All virus preparations had similar titers on the human indicator cells (Fig. 3A). Surprisingly, no replication center could be detected after infection of A9 cells with either Chi-hH1 or hH1(MVMp) virus, even after increasing the MOI up to 100-fold compared to MVMp-based viruses (Fig. 3B). Similarly, the standard hH1 virus failed to replicate in A9 cells (data not shown). This indicated that viruses containing an hH1 genome (standard or chimeric) were unable to amplify their DNA in mouse cells irrespective of whether the capsid was of H-1 or MVMp virus origin. In contrast, MVMp-based viruses were able to amplify their genome in A9 cells, including MVMp/GFP(H1) pseudotyped with H-1 capsids (Fig. 3B). From these observations, we concluded that the origin of the genome and not of the incoming capsid was the major determinant of the difference between MVMp and hH1 viruses in their ability to infect mouse A9 cells. The fact that the MVMp/GFP(H1) virus pseudotype was efficient in replicating its DNA in A9 cells, while Chi-hH1/EGFP virus was not, although both were packaged in H-1 capsids, further suggests that the DNA sequence responsible for this allotropism is located between nucleotides (nt) 991 and 2791, which constitutes the only difference between the two vectors (Fig. 1). Indeed, the hH1 virus genome contains a 995-bp MVMp virus DNA sequence consisting of the left-hand palindrome, the P4 promoter, and part of the NS1/NS2 coding region as described previously (17).
FIG. 3.
Viral DNA amplification after infection of human and mouse cells with chimeric and pseudotyped viruses. (A and B) NB-324K and A9 cells were infected at a MOI of 0.001 RU/cell (A) or 0.03 RU/cell (B) (MVMp-based virus) and 3 RU/cell (B) (hH1-based virus) with the indicated virus stocks and transferred to nitrocellulose membranes 48 h postinfection. Titration of recombinant viruses was performed by hybridization assay and autoradiography (16, 17), using a NS-specific DNA probe as described in the legend to Fig. 2. (C) Southern blotting analysis of low-molecular-weight DNA, isolated by the modified Hirt procedure, 72 h after infection of A9 cells with the indicated virus stocks at a MOI of 3 RU/cell. DNA samples were fractionated by agarose gel electrophoresis, transferred to Hybond N+ nylon membrane, and hybridized with a NS-specific DNA probe. The positions of monomer- and dimer-length RFs are indicated.
We next analyzed by Southern blotting the viral DNA replicative intermediates formed in A9 cells upon infection with the various viruses. Figure 3C shows the DNA bands corresponding to the monomeric and dimeric replicative forms (RFs) from MVMp-based viruses (Fig. 3C, lanes 1 and 5). In contrast, a weak monomeric form and no dimeric RFs were detected in A9 cells infected with hH1-based viruses (Fig. 3C, lanes 2, 3, and 4). This suggested that the hH1-based viruses-whether standard (lane 2), chimeric (lane 3), or pseudotyped (lane 4)-had impaired ability to sustain efficient DNA replication upon infection of A9 cells. This block appeared to occur at an early step of the viral life cycle which prevented the conversion of the virion single-stranded genome into double-stranded DNA (RF), a process known to be independent of the expression of NS proteins (1). This restriction is reminiscent of the interaction of EL4 cells with MVMp3 virus, a variant of the lymphotropic virus strain MVMi (lymphotropic variant of MVM) in which part of the VP coding sequence was replaced by the corresponding region of the fibroblastic virus strain MVMp (22). Indeed, MVMp3 DNA replication was strongly suppressed after infection, while replication of double-stranded RF molecules was unaffected after transfection. It was concluded that the MVMp3 life cycle was arrested most likely at the decapsidation step, prior to conversion of the virion single-stranded genome to RF DNA. Similarly, the entry, transport, and/or decapsidation of the hH1-based viruses might be affected in A9 cells, preventing the delivery of single-stranded DNA into the nucleus. Although it cannot be ruled out, this possibility seems unlikely, since H-1 virus capsids were fully competent for transferring MVMp virus genomes in A9 cells, allowing the MVMp(H1) pseudotype virus DNA to replicate as efficiently as the standard MVMp virus (Fig. 3C, lane 5).
(ii). Protein expression in mouse and human cells infected with chimeric and pseudotyped viruses.
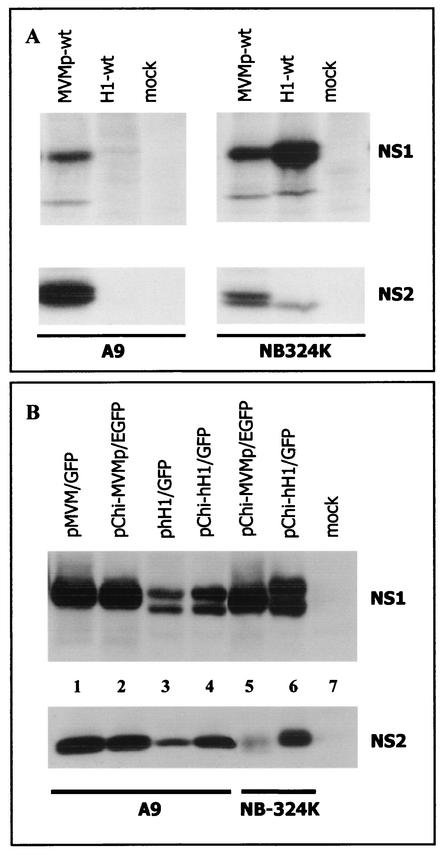
The production of NS1 and NS2 proteins was not detected in A9 cells infected with H-1 virus (Fig. 4A) but could be measured in these cells upon transfection with hH1-based plasmid DNA (Fig. 4B), as determined by Western blotting analysis. However, hH1-based plasmids led A9 cells to accumulate much reduced levels of NS proteins compared with MVMp-based DNAs (Fig. 4B, compare lanes 3 and 4 to lanes 1 and 2). This difference was most pronounced for NS1 polypeptides and could not be accounted for by a weaker binding affinity of the antibodies raised against NS1 from MVMp virus towards NS1 from hH1 virus, since similar NS1 levels were detected in NB-324K cells transfected with either pChi-MVMp or pChi-hH1 plasmid DNA (Fig. 4B, lanes 5 and 6). Altogether, our results showed that irrespective of its mode of delivery (via a MVMp or H-1 virus capsid or as a plasmid) hH1-based DNA underwent restriction(s) to the accumulation of NS products in A9 cells, which may contribute to the impairment of its amplification in these cells. Plasmid DNA transfection experiments indicated that this restriction was still present, at least in part, following conversion of the virion single-stranded genome into double-stranded DNA, arguing for its occurrence at the level of gene expression, transcription template amplification, and/or protein stability. Since no quantitative comparison can be made between the outcomes of transfection and infection experiments, the possibility needs to be considered, however, that an additional, capsid-independent primary barrier takes place in infected A9 cells at or prior to the single- to double-stranded hH1 virus DNA conversion step, limiting the formation of transcription templates and accounting for our failure to detect any NS gene expression in hH1 virus-infected A9 cells. A possible explanation would be that a mouse intracellular factor(s) interacts specifically with the hH1 (or MVMp) virus genome and determines the cell restriction by preventing (or allowing) the conversion of virion single-stranded DNA into double-stranded RF.
FIG. 4.
Expression of NS1 and NS2 proteins in A9 and NB-324K cells. (A) Cells were mock treated or infected at a MOI of 5 PFU/cell with wild-type MVMp (MVMp-wt) or H-1 (H1-wt) virus. At 18 h postinfection, cell cultures were metabolically labeled for 2 h with 200 μCi of Trans35S label (1,000 Ci/mmol; ICN Pharmaceuticals) in Met- and Cys-free Eagle's minimum essential medium supplemented with 5% dialyzed fetal calf serum. Cells were then lysed in modified radioimmunoprecipitation assay (MRIPA) buffer (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate) containing a mixture of protease inhibitors (Complete; Roche Molecular Biochemicals). Equal amounts of labeled protein extracts (107 cpm) were immunoprecipitated with 10 μl of the polyclonal rabbit antiserum SP11 directed against the N terminus of NS1 and NS2 proteins of MVMp virus (3). The immunocomplexes were fractionated by electrophoresis on a sodium dodecyl sulfate-8% polyacrylamide gel and revealed by autoradiography. (B) A9 cells were transfected with 4 μg of plasmid DNA using the Lipofectamine transfection reagent as recommended by the supplier (Gibco BRL), while 25 μg of plasmid DNA was used to transfect NB-324K cells by the calcium phosphate precipitation method. Transfection efficiency was determined by the percentage of (E)GFP-positive cells and was identical for all the constructs. At 48 h after transfection, cells were lysed in MRIPA buffer, and 50-μg portions of protein extracts were analyzed by Western blotting. NS1 and NS2 were revealed by incubating the membrane with rabbit polyclonal antisera directed against the C-terminal part of MVMp NS1 (SP7) (13) and against the major isoform of MVMp NS2 (NS2p), respectively. The NS2p-specific antiserum was producedusing the keyhole limpet hemocyanin-coupled peptide AELGLRPEITWF. The membrane was further incubated with horseradish peroxidase-conjugated goat anti-rabbit antibody, and the immunoreactive proteins were visualized using Western Lightning Chemiluminescence Detection Reagent Plus (Perkin-Elmer Life Sciences).
We suggest that the viral genomic element(s) involved may lie in the above-mentioned DNA sequence from nt 991 to 2791 and be responsible for the recruitment of a cellular factor(s) determining the differential capacity of mouse cells for MVMp versus hH1 virus DNA replication. In agreement with this hypothesis, it was previously reported that the host cell specificity of the fibrotropic and lymphotropic variants of MVM (MVMp and MVMi, respectively) depends not only on determinants located within the coding region of the capsid genes but also on a second sequence overlapping the P38 promoter and the NS coding region (5, 15). It should also be stated that, in contrast with hH1-based genomes (present results), recombinant LuIII genomes pseudotyped with MVMp (but not MVMi) virus capsids were found to be competent for transduction and expression of a marker gene in A9 cells (27). This discrepancy is surprising, given that the H-1 and LuIII parvoviruses are closely related. It is worth noting, however, that, in the latter study, the LuIII genome was deprived not only of the VP but also NS DNA sequences that were replaced by the marker gene. These recombinant LuIII genomes were thus missing the above-mentioned determinant (nt 991 to 2791), which may relieve them from the restriction described in this report.
(iii). The viral DNA region encoding the nonstructural proteins controls the infection of mouse cells, while H-1 virus capsids determine the greater permissiveness of human cells for this virus compared with MVMp virus.
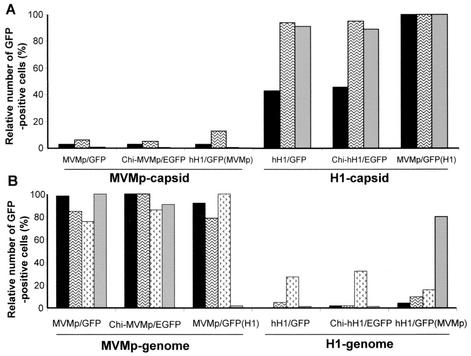
To further analyze the cell tropism exhibited by the chimeric and pseudotyped viruses, we determined their capacity for transducing the reporter GFP gene in three human cell lines of promonocytic (U937), glioma (U373), and cervical carcinoma (HeLa) origin and in four mouse cell lines established from embryo fibroblasts (A9, L929, and C3H10T1/2) and an ascitic tumor (EA). The cell lines were infected at a MOI of 3 RU/cell (human cell lines) or 10 RU/cell (mouse cell lines), and GFP (or EGFP) expression was monitored 48 or 72 h postinfection. While all stocks of viruses could efficiently infect NB-324K cells and yielded similar titers on these indicator cells, the above-mentioned cell lines differed in their relative susceptibility to infection with the various viruses. Therefore, for each cell line, the virus stock that gave the highest number of GFP-positive cells was set at 100%, and GFP transduction by the other virus stocks was expressed relative to this reference value.
As shown in Fig. 5A, human U937 cells showed the highest number of GFP-positive cells when infected with the pseudotype MVMp/GFP(H1) virus, while the chimeric Chi-hH1/EGFP and standard hH1/GFP viruses gave somewhat (2.5-fold) lower scores. In contrast to the H-1-coated viruses, the MVMp-coated viruses yielded 100-fold-less GFP-positive U937 cells, irrespective of the origin of their genome (Fig. 5A). Similarly, HeLa and glioma cells were quite resistant to infection with all viruses packaged with MVMp capsids (Fig. 5A) but were susceptible to the viruses provided with H-1 capsids (Fig. 5A). Thus, in the human cells tested (except for the NB-324K reference line), the H-1 origin of the incoming capsid appeared to constitute a major determinant of virus infectivity, while the hH1 versus MVMp virus origin of the genome made little difference. These data confirm previous observations (6) showing that human cells are usually more susceptible to infection with H-1 virus than with MVMp virus. A parallel can also be drawn between our observations and a report showing that pseudotypes consisting of the genome of the autonomous parvovirus LuIII with the capsid of feline panleukopenia virus or canine parvovirus can be produced but fail to transduce and express a marker gene in a human cell line permissive for LuIII but not for feline panleukopenia virus and canine parvovirus (27).
FIG. 5.
Cell tropism of chimeric and pseudotyped virus MVMp and hH1 vectors. Several human (A) and mouse (B) cell lines were infected with recombinant standard, chimeric, or pseudotyped virus stocks at a MOI of 3 or 10 RU/cell, respectively. GFP (or EGFP) expression was monitored with a fluorescence microscope 48 and 72 h postinfection. The line with the highest number of fluorescent cells after infection with the indicated virus stock was set at 100% for GFP-positive cells. GFP-positive cells from the same cell line infected with all the other virus stocks were expressed relative to this reference value (given as a percentage). A minimum of 200 GFP-positive cells were counted in each cell line. Results are of one representative experiment (of three). (A) Relative number of GFP-positive human cells (U937  , U373
, U373  , and HeLa
, and HeLa  ) after infection with the indicated recombinant virus stocks. (B) Relative numbers of GFP-positive mouse cells (A9
) after infection with the indicated recombinant virus stocks. (B) Relative numbers of GFP-positive mouse cells (A9  , L929
, L929  , Ehrlich ascitic fluid
, Ehrlich ascitic fluid  , and C3H10T1/2
, and C3H10T1/2  ) after infection with the indicated recombinant virus stocks.
) after infection with the indicated recombinant virus stocks.
In contrast to their behavior in human cells, the ability of the composite viruses analyzed in this study to transduce GFP in murine cell lines was strongly dependent on the MVMp virus origin of the genome and not on that of the capsid (Fig. 5B). The C3H10T1/2 line was an exception to this trend, as it yielded a significant proportion of GFP-positive cells upon infection with hH1/GFP(MVMp) virus, a hH1-based genome pseudotyped with MVMp virus capsids (Fig. 5B). Altogether, our data show that in mouse cells, H-1 virus capsids were almost as efficient as MVMp virus capsids for GFP transduction as far as they encapsidated a MVMp-based genome, while these cells were usually resistant to infection with viruses containing a hH1 virus genome, in agreement with the low level of hH1 viral DNA amplification detected in A9 cells (Fig. 3).
Acknowledgments
We are grateful to A. Dege for excellent technical assistance. We are indebted to H. Delius for DNA sequencing, U. Bodendorf for providing NS2p antiserum, and N. Winkelhöfer for help in immunoprecipitation assays.
This work was supported in part by grants from the European Union (Biotech and Quality of Life Programmes).
REFERENCES
- 1.Baldauf, A. Q., K. Willwand, E. Mumtsidu, J. P. Nuesch, and J. Rommelaere. 1997. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J. Virol. 71:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salome. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salome. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Brown, C. S., F. M. DiSumma, J. Rommelaere, A. Y. Dege, J. J. Cornelis, C. Dinsart, and W. J. M. Spaan. 2002. Production of recombinant H1 parvovirus stocks devoid of replication-competent viruses. Hum. Gene Ther. 13:2135-2145. [DOI] [PubMed] [Google Scholar]
- 4.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding nonstructural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colomar, M. C., B. Hirt, and P. Beard. 1998. Two segments in the genome of the immunosuppressive minute virus of mice determine the host-cell specificity, control viral DNA replication and affect viral RNA metabolism. J. Gen. Virol. 79:581-586. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis, J. J., P. Becquart, N. Duponchel, N. Salome, B. L. Avalosse, M. Namba, and J. Rommelaere. 1988. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvoviruses H-1 virus and minute virus of mice. J. Virol. 62:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 8.Deleu, L., A. Pujol, S. Faisst, and J. Rommelaere. 1999. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 73:3877-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont, F., A. Karim, J. C. Dumon, N. Mine, and B. Avalosse. 2001. A novel MVMp-based vector system specifically designed to reduce the risk of replication-competent virus generation by homologous recombination. Gene Ther. 8:921-929. [DOI] [PubMed] [Google Scholar]
- 10.Dupont, F., L. Tenenbaum, L. P. Guo, P. Spegelaere, M. Zeicher, and J. Rommelaere. 1994. Use of an autonomous parvovirus vector for selective transfer of a foreign gene into transformed human cells of different tissue origins and its expression therein. J. Virol. 68:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupressoir, T., J. M. Vanacker, J. J. Cornelis, N. Duponchel, and J. Rommelaere. 1989. Inhibition by parvovirus H-1 of the formation of tumors in nude mice and colonies in vitro by transformed human mammary epithelial cells. Cancer Res. 49:3203-3208. [PubMed] [Google Scholar]
- 12.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salome. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 76:10307-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faisst, S., S. R. Faisst, T. Dupressoir, S. Plaza, A. Pujol, J. C. Jauniaux, S. L. Rhode, and J. Rommelaere. 1995. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J. Virol. 69:4538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faisst, S., D. Guittard, A. Benner, J. Y. Cesbron, J. R. Schlehofer, J. Rommelaere, and T. Dupressoir. 1998. Dose-dependent regression of HeLa cell-derived tumours in SCID mice after parvovirus H-1 infection. Int. J. Cancer 75:584-589. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner, E. M., and P. Tattersall. 1988. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J. Virol. 62:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haag, A., P. Menten, J. Van Damme, C. Dinsart, J. Rommelaere, and J. J. Cornelis. 2000. High efficient transduction and expression of cytokine genes into human tumor cells by means of autonomous parvovirus vectors: generation of antitumor responses in recipient mice. Hum. Gene Ther. 11:597-609. [DOI] [PubMed] [Google Scholar]
- 17.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 18.Lorson, C., J. Pearson, L. Burger, and D. J. Pintel. 1998. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology 240:326-337. [DOI] [PubMed] [Google Scholar]
- 19.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naeger, L. K., N. Salome, and D. J. Pintel. 1993. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J. Virol. 67:1034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olijslagers, S., A. Y. Dege, C. Dinsart, M. Voorhoeve, J. Rommelaere, M. H. Noteborn, and J. J. Cornelis. 2001. Potentiation of a recombinant oncolytic parvovirus by expression of apoptin. Cancer Gene Ther. 8:958-965. [DOI] [PubMed] [Google Scholar]
- 22.Previsani, N., S. Fontana, B. Hirt, and P. Beard. 1997. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J. Virol. 71:7769-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rayet, B., J. A. Lopez-Guerrero, J. Rommelaere, and C. Dinsart. 1998. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J. Virol. 72:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, S. J., A. Brandenburger, C. L. Flemming, M. K. Collins, and J. Rommelaere. 1992. Transformation-dependent expression of interleukin genes delivered by a recombinant parvovirus. J. Virol. 66:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salome, N., B. van Hille, N. Duponchel, G. Meneguzzi, F. Cuzin, J. Rommelaere, and J. J. Cornelis. 1990. Sensitization of transformed rat cells to parvovirus MVMp is restricted to specific oncogenes. Oncogene 5:123-130. [PubMed] [Google Scholar]
- 26.Siegl, G. 1990. Variability, adaptability and epidemiology of autonomous parvoviruses, p. 59-74. In P. Tijssen (ed.), Handbook of parvoviruses. CRC Press, Boca Raton, Fla.
- 27.Spitzer, A. L., F. Maxwell, J. Corsini, and I. H. Maxwell. 1996. Species specificity for transduction of cultured cells by a recombinant LuIII rodent parvovirus genome encapsidated by canine parvovirus or feline panleukopenia virus. J. Gen. Virol. 77:1787-1792. [DOI] [PubMed] [Google Scholar]
- 28.Wetzel, K., P. Menten, G. Opdenakker, J. Van Damme, H. J. Grone, N. Giese, A. Vecchi, S. Sozzani, J. J. Cornelis, J. Rommelaere, and C. Dinsart. 2001. Transduction of human MCP-3 by a parvoviral vector induces leukocyte infiltration and reduces growth of human cervical carcinoma cell xenografts. J. Gene Med. 3:326-337. [DOI] [PubMed] [Google Scholar]
- 29.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]
- 30.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]