Abstract
This report examines the distribution of an RNA polymerase I transcription factor (upstream binding factor; UBF), pre-rRNA processing factors (nucleolin and fibrillarin), and pre-rRNAs throughout mitosis and postmitotic nucleologenesis in HeLa cells. The results demonstrate that nucleolin, fibrillarin, and pre-rRNAs synthesized at G2/M phase of the previous cell cycle are directly recruited to UBF-associated nucleolar organizer regions (NORs) early in telophase before chromosome decondensation. Unlike the fusion of prenucleolar bodies to the nucleoli, this early recruitment of processing factors and pre-rRNAs is independent of RNA polymerase I transcription. In the absence of polymerase I transcription, the initial localization of nucleolin, fibrillarin, and pre-rRNAs to UBF-associated NORs generates segregated mininucleoli that are similar to the larger ones observed in interphase cells grown under the same conditions. Pre-rRNAs are juxtaposed to UBF-nucleolin-fibrillarin caps that may represent the segregated nucleoli observed by electron microscopy. These findings lead to a revised model of nucleologenesis. We propose that nucleolar formation at the end of mitosis results from direct recruitment of processing factors and pre-rRNAs to UBF-associated NORs before or at the onset of rDNA transcription. This is followed by fusion of prepackaged prenucleolar bodies into the nucleolus. Pre-ribosomal ribonucleoproteins synthesized in the previous cell cycle may contribute to postmitotic nucleologenesis.
INTRODUCTION
The nucleus is a complex structure within which various functions and components are spatially and temporally organized and interregulated (reviewed by Lamond and Earnshaw, 1998; Dreyfuss and Struhl, 1999). Nucleoli represent the most prominent example of this organization. The major function of the nucleolus is ribosome biogenesis (reviewed by Busch and Smetana, 1970). rRNAs are synthesized, processed, cleaved, and assembled with ribosomal proteins in the nucleolus before export to the cytoplasm. The nucleolus is composed of three structurally distinguishable constituents: fibrillar centers, dense fibrillar components, and granular components, as observed by electron microscopy (reviewed by Busch and Smetana, 1970; Hadjiolov, 1985). Although the spatial and temporal relationship between rRNA synthesis and the three structural constituents remains to be clarified, it is generally thought that transcription of rDNA takes place in a transient area between the fibrillar centers and the dense fibrillar components and that rRNA processing and preribosomal particle assembly progress from the dense fibrillar components to the surrounding granular components (reviewed by Spector et al., 1993; Shaw and Jordan, 1995; Scheer and Hock, 1999). More recent evidence (reviewed by Pederson, 1998; Garcia and Pillus, 1999) suggests that nucleoli have additional functions related to the cell cycle (Shou et al., 1999; Visintin et al., 1999), cellular aging (Straight et al., 1999), signal recognition particle biosynthesis (Jacobson and Pederson, 1998), small RNA processing, mRNA transport (reviewed by Pederson, 1998), and p53 metabolism (Zhang and Xiong, 1999).
The essential mechanisms of nucleologenesis during postmitotic nucleolar reformation or de novo nucleolar generation in early embryogenesis are not clear. Electron microscopic and immunocytochemical studies show that at least part of nucleologenesis involves fusion between prepackaged prenucleolar bodies (PNBs) and NORs in postmitotic cells (Ochs et al., 1985; Jimenez-Garcia et al., 1994; Fomproix et al., 1998). Such fusion is prevented by treatment with a low concentration of actinomycin D (ActD) that inhibits polymerase I (Pol I) transcription (Ochs et al., 1985). PNBs, spherical bodies of 0.3–2 μm in diameter, are composed of densely packed RNP granules and fibrils (Stevens, 1965). The PNBs generally contain factors involved in pre-rRNA processing, including fibrillarin, nucleolin, B23, argyrophilic proteins, Nop52, PMScl-100 protein, and U3, U8, and U14 snoRNP (Ochs and Busch, 1984; Ochs et al., 1985; Jimenez-Garcia et al., 1994; Fomproix and Hernandez-Verdun, 1999; Savino et al., 1999). Before telophase, these complexes are distributed around the chromosomes and diffusely throughout the mitotic cytoplasm (Gautier et al., 1992; Azum-Gelade et al., 1994). As the other nucleolar components, the Pol I transcription machinery, including the polymerase complex, and the Pol I–specific transcription factors, such as upstream binding factor (UBF) and SL1, bind to NORs throughout mitosis when rDNA transcription is inactivated (Scheer and Rose, 1984; Roussel et al. 1993; Gebrane-Younes et al., 1997). Interestingly, during mitosis the transcription machinery associates only with NORs that have been transcribed during the previous interphase and will be transcriptionally active in the next cell cycle (Roussel et al., 1996). Accordingly, they were named competent NORs.
It has been proposed that the activation of rDNA transcription recruits the processing factors to the sites of transcription, thus facilitating PNB fusion (reviewed by Scheer and Hock, 1999). In this context, several studies have attempted to address the relationship between rDNA transcription and nucleolar assembly. Transcription of rDNA appears to induce nucleolus-like structures. Using a yeast rDNA deletion mutant, Oakes et al. (1998) found that transcription of rDNA by Pol I from a plasmid induced multiple small dense structures, “mininucleoli,” whereas transcription of rDNA by Pol II formed an aggregated larger structure. Insertion of an actively transcribing rDNA repeat into Drosophila polytene chromosomes resulted in dense mininucleoli around the transcription sites (Karpen et al., 1988). These findings demonstrate that transcription of rDNA alone generates dense structures that do not resemble typical nucleoli with well-organized and classically defined structural features. Verheggen et al. (1998) found that during Xenopus laevis development maternal pre-rRNAs are present in nucleolin-concentrated structures with features of transcriptionally competent nucleoli. However, rRNA synthesis was not detected with the use of an in situ run-on method, suggesting that nucleoli form before the activation of Pol I transcription. Another group of studies used ActD (Ochs et al., 1985) or DNA topoisomerase I inhibitor (Weisenberger et al., 1993) or anti-Pol I antibody injection (Benavente et al., 1987) to assess the regeneration of nucleoli in mammalian cells. The results of these studies suggested that typical nucleoli were not assembled when cells progressed into G1 under any of these conditions. Instead, electron micrographs revealed round structures containing dense fibrillar and granular elements, or altered PNBs (Ochs et al., 1985; Benavente et al., 1987). These studies suggest that rDNA transcription may not be necessary or sufficient for the assembly of typical nucleoli. Therefore, nucleolar assembly should be examined as complex integrated events involving rDNA transcription and other not yet defined mechanisms.
In this paper, we examined the temporal distribution of Pol I transcription and pre-rRNA processing factors, including UBF, nucleolin, and fibrillarin, as well as pre-rRNAs throughout mitosis and the beginning of interphase. We report for the first time that nucleologenesis also involves direct recruitment of processing factors and pre-rRNAs to transcription factor–associated NORs very early at the end of mitosis. Unlike the fusion of PNBs, this process is independent of Pol I transcription. We also find that pre-rRNAs synthesized in the previous cell cycle participate in the assembly of daughter cell nucleoli, suggesting that these RNAs may play a role in structuring nucleoli.
MATERIALS AND METHODS
Cell Cultures
HeLa cells were maintained in DMEM supplemented with 10% FBS at 37°C and 5% CO2. Cells were passed twice a week. Mitotic cells were synchronized by treatment with nocodazole at 0.05 μg/ml for 4 h, with colchicine at 0.02 μg/ml for 14 h, or were synchronized by double thymidine block as described (Sirri et al., 1999). In the latter case, 9 h after release from 2′-deoxycytidine, 80% of the cells accumulated in early prophase. ActD was added 1 h before the harvest at a final concentration of 0.04 μg/ml. Mitotic cells were collected through centrifugation and allowed to progress into G1 phase in the presence of the same concentration of ActD. One hour to 4 h after reseeding, cells were fixed in 2% formaldehyde and subjected to immunofluorescence or in situ hybridization studies.
In Situ Hybridization
In situ hybridization was performed according to a modified protocol described previously (Huang and Spector, 1991; Verheggen et al., 1998). Fixed cells were washed three times for 15 min each in PBS and permeabilized in 0.5% Triton X-100 for 5 min at 4°C. For double labeling with immunodetection of UBF, nucleolin, or fibrillarin, cells were incubated with primary and secondary antibodies and subsequently fixed again with 2% formaldehyde for 5 min. Cells were then rinsed in PBS two times for 15 min each and once with 2× SSC followed by hybridization.
The probe corresponding to human rRNA 5′ external transcribed spacer (ETS) core sequences (+693 to +2921) was provided by Dr. J. Sylvester (Sylvester et al., 1986), or a shorter sequence (+933 to +1445) was provided by Dr. M. Olson (Dundr et al., 1998). The probe corresponding to human 28S rRNA (+12,383 to +12,969) was subcloned from p3′ETS that spans the 3′ end of 28S and the 5′ end of the 3′ETS sequence provided by Dr. M. Olson. Probe sequences were excised and gel purified. Probes were labeled by nick translation in the presence of biotin-conjugated dUTP or dCTP (Clontech, Palo Alto, CA). One hundred to 500 ng of labeled probe was dried in a Speed-vac, together with 20 μg of Escherichia coli tRNA and 5 μg of sheared salmon sperm DNA. The pellet was resuspended in 10 μl of deionized formamide, heat-denatured at 75°C for 10 min, and rapidly cooled in ice-water slurry. The final 20-μl hybridization mixture (2× SSC, 10 mM Tris-HCl, pH 7.2, 1 mM EDTA, 5% dextran sulfate, 50% [vol/vol] formamide, denatured probe, tRNA, and sheared salmon sperm DNA) was applied to each coverslip, which was then inverted onto a ribonuclease-free glass slide and sealed with rubber cement. Hybridization was carried out at 37°C in a humidified chamber overnight. After hybridization, cells were washed three times in 2× SSC and once in 1× SSC. Signals were detected by incubation with FITC or Texas Red–conjugated avidin (Jackson ImmunoResearch, West Grove, PA) at a dilution of 1:200 in 4× SSC for 1 h at room temperature or a rabbit anti-biotin antibody (Enzo, Farmingdale, NY) amplified by two-step detection with the use of mouse anti-rabbit and FITC-conjugated goat anti-mouse antibody (Jackson ImmunoResearch). Cells were washed in 4× SSC three times before mounting.
Northern Blot Analysis
RNA was extracted from cells with the use of an RNA Now kit (Biogentex, Seabrook, TX). The RNA extracts were then treated as described previously (Verheggen et al., 1998). RNAs were denatured (50% formamide, 1.84 M formaldehyde, and 10 mM sodium phosphate buffer) for 15 min at 68°C and then run on a 0.8% agarose gel. After transfer onto a nitrocellulose filter, rRNAs were revealed by hybridization with 32P-labeled 5′ETS probes for 20 h at 42°C. Autoradiography was performed with a Phosphorimager (Molecular Dynamics, Sunnyvale, CA). RNA size was determined by comparison with an RNA ladder (GIBCO-BRL, Gaithersburg, MD).
Br-UTP Incorporation Assay
The transcription assay was modified from published studies (Jackson et al., 1993; Wansink et al., 1993). Cells were rinsed once with PBS and once with a Tris buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 25% glycerol, 0.5 mM PMSF, 0.5 mM EGTA). Cells were then permeabilized in the same buffer containing 5 μg/ml digitonin at room temperature for 3 min. Subsequently, cells were incubated in transcription cocktail (100 mM KCl, 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 1 mM PMSF, 2 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.2 mM Br-UTP, 1 U/ml RNAsin) for 5 min at 37°C. At the end of the transcription reaction, cells were gently rinsed three times with PBS and then fixed in 2% paraformaldehyde in PBS for 15 min.
Immunolabeling
For immunolabeling, cells were fixed in 2% freshly prepared formaldehyde in PBS for 15 min. Cells were washed three times for 10 min each in PBS, and coverslips were then incubated with primary antibody. Antibodies used in this study included anti-UBF–specific human serum at a dilution of 1:300 (Chan et al., 1991; Roussel et al., 1993), anti-nucleolin mAb at a dilution of 1:3000 (Pinol-Roma, 1999), anti-fibrillarin human serum at a dilution of 1:100 (Gautier et al., 1994), anti-SC35 at a dilution of 1:1000 (Fu and Maniatis, 1990), anti-hnRNP A1 at a dilution of 1:300 (Pinol-Roma and Dreyfuss, 1992), and anti-Br-UTP mAb at a dilution of 1:500 (Sigma Chemical, St. Louis, MO) for 1 h at room temperature. Cells were rinsed in PBS, incubated with Texas Red–conjugated goat anti-human antiserum or with FITC or Texas Red–conjugated goat anti-mouse antiserum at a dilution of 1:200 for 1 h at room temperature and then washed three times for 10 min each in PBS. The coverslips were mounted onto glass slides with mounting medium containing 90% glycerol in PBS with 1 mg/ml paraphenylenediamine as an antifade agent. The mounting medium was adjusted to pH 8.0 with 0.2 M bicarbonate buffer. Cells were examined with a TCS 4D Leica (Deerfield, IL) confocal microscope equipped with an argon–krypton laser and with a Zeiss (Thornwood, NY) Axiovert microscope equipped with epifluorescence and differential interference contrast optics. Images were captured by a SenSys cooled charge-coupled device camera (Photometrics, Tucson, AZ) with the use of Metamorph Image software (Universal Imaging Corp, West Chester, PA).
Electron Microscopy
Cells were fixed in 4% paraformaldehyde and 2% glutaraldehyde for 20 min and washed in PBS containing 0.3 M glycine and 0.1 M cacodylate buffer. Cells were then postfixed in 1% osmium tetroxide for 1 h, dehydrated by incubation in a series of ascending concentrations of ethanol, and embedded in epon/araldite at 60°C for 48 h. Eighty-nanometer sections were poststained with uranyl acetate–lead citrate and examined at 75 kV with a JEOL (Tokyo, Japan) JEM-1220 transmission electron microscope.
RESULTS
Pre-rRNA Processing Factors Are Directly Recruited to Active NORs at Early Telophase Independently of rDNA Transcription
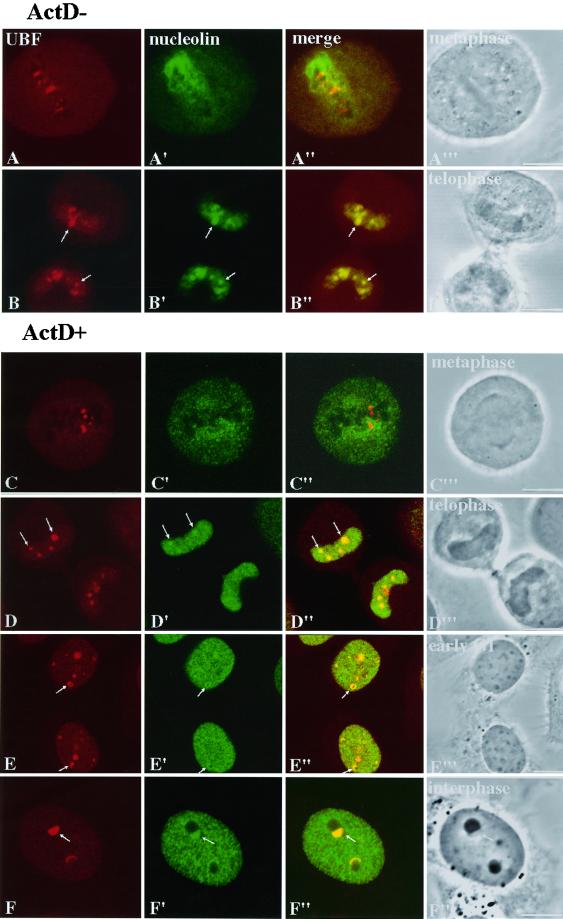
To understand the chronological sequence of nucleologenesis, we examined the distribution of a Pol I transcription factor (UBF), pre-rRNA processing factors (nucleolin and fibrillarin), and the spatial relationship between these two groups of factors throughout mitosis into early G1. UBF was detected by a specific human autoimmune serum (Chan et al., 1991; Roussel et al., 1993), and nucleolin was detected by a mAb, 7G2 (Pinol-Roma, 1999). UBF is associated with competent NORs throughout mitosis (Figure 1, A and B). Nucleolin, in addition to being distributed diffusely throughout the mitotic cytoplasm, appears to sheath the condensed chromosomes during mitosis (Figure 1A′) and forms PNBs at telophase. These observations are similar to the descriptions in the published literature (Scheer and Rose, 1984; Roussel et al., 1996; reviewed by Ginisty et al., 1999). In addition, we find that at early telophase nucleolin is rapidly relocated into daughter cell nuclei and is concentrated in dots in which UBF-associated competent NORs are present (Figure 1, B–B′′) before chromosome decondensation (Figure 1B′′′). Immunolabeling of fibrillarin also shows a similar distribution (our unpublished results). These findings suggest that the recruitment of processing factors to the competent NORs may take place before or at the onset of rDNA transcription and that the initial recruitment may not be in the form of fusion with PNBs. This suggestion is supported by the subsequent observations in cells exiting mitosis in the presence of a low concentration of ActD.
Figure 1.
Nucleolin is recruited directly to UBF-associated NORs early in telophase independent of Pol I transcription. Each row represents the same cells immunolabeled for UBF (first column) and nucleolin (second column), UBF and nucleolin overlay (third column), and differential interference contrast images to identify the cell cycle stages (fourth column). Rows A and B represent cells growing in normal medium (A, metaphase; B, telophase). Rows C–E represent cells at different stages through mitosis to G1 in the presence of 0.04 μg/ml ActD. Nucleolin is recruited before the decondensation of chromosomes (B–B′′′, arrows). The recruitment is not interrupted in the absence of Pol I transcription (D–D′′′, arrows). Toward late telophase, UBF and nucleolin colocalize to cap-like structures that juxtapose or surround dense bodies (Row E, arrows). Row F represents interphase cells treated with 0.04 μg/ml ActD. Cap-like structures (arrows) are associated with segregated nucleoli. All images are 0.8-μm optical sections obtained with the use of confocal laser scanning microscopy. Bars, 10 μm.
ActD has been shown to prevent RNA polymerases from functioning by intercalating into DNA, preferentially targeting G/C-rich sequences (Sobell, 1979). At a very low concentration of 0.04 μg/ml, ActD selectively inhibits RNA Pol I activity as a result of the higher G/C contents of the rDNA (Perry, 1963). Nuclear run-on experiments show that rRNA transcription in HeLa cells is reduced to <1% of its normal level within 1 h of ActD (0.04 μg/ml) treatment (Penman et al., 1968; D. Chen and S. Huang, unpublished results).
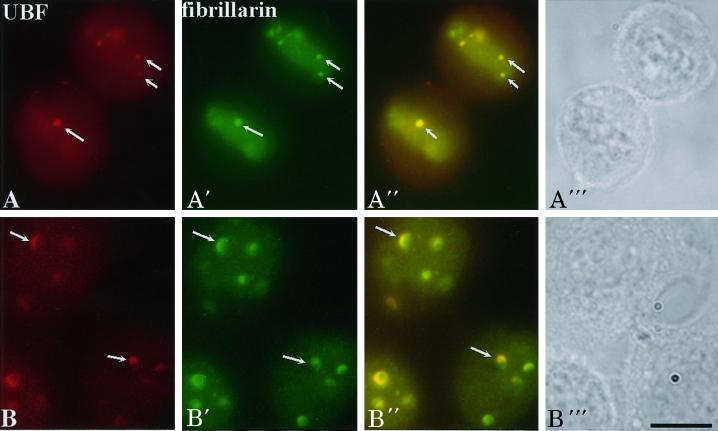
To understand nucleologenesis in the absence of Pol I transcription, we compared the localization of UBF, nucleolin, and fibrillarin throughout mitosis in cells grown in a low concentration of ActD with those grown in drug-free medium. The results demonstrate that the rDNA transcription factor UBF remains associated with competent NORs throughout mitosis and within the newly formed nuclei (Figure 1, C and D) in a similar manner in both treated and nontreated cells (Figure 1, compare A with C and B with D). However, the increase in UBF signal at the active NORs during nucleologenesis no longer takes place in drug-treated cells (Figure 1E), and UBF forms cap-like structures on the newly developed mininucleoli (Figure 1E, arrow). The dynamics of pre-rRNA processing factors throughout mitosis into early G1 is also significantly changed in the presence of ActD. Nucleolin no longer sheathes condensed chromosomes during mitosis (Figure 1C′). Nucleolar assembly in the form of fusion of the PNBs is interrupted (Figure 1E′). However, the initial recruitment of nucleolin (Figure 1D′, arrows) and fibrillarin (Figure 2A′) to UBF-associated NORs continues in drug-treated cells. The recruited nucleolin (Figure 1E′, arrows) and fibrillarin (Figure 2B′) at NORs become colocalized with UBF, forming cap-like structures at early G1 phase. These findings demonstrate that intercalation of ActD into rDNA does not affect the association of rDNA transcription machinery to the NORs during mitosis. Nor does it prevent the initial recruitment of pre-rRNA processing factors to the UBF-associated NORs early in telophase. However, it does disrupt the fusion between PNBs and NORs in a manner similar to the descriptions of Ochs et al. (1985) and Benavente et al. (1987). The findings that the early recruitment of processing factors are Pol I transcription independent support the idea that this process is a novel mechanism of nucleologenesis that takes place in addition to the process of fusion between PNBs and NORs.
Figure 2.
The recruitment of fibrillarin directly to UBF-associated NORs (arrows) in early telophase is not disrupted in the presence of 0.04 μg/ml ActD (A–A′′). When cells progress to early G1, fibrillarin is colocalized with UBF to cap-like structures (B–B′′). Each row represents the same cells immunolabeled for UBF (first column) and fibrillarin (second column), UBF and fibrillarin overlay (third column), and phase contrast to identify the cell cycle stages (fourth column). Bar, 10 μm.
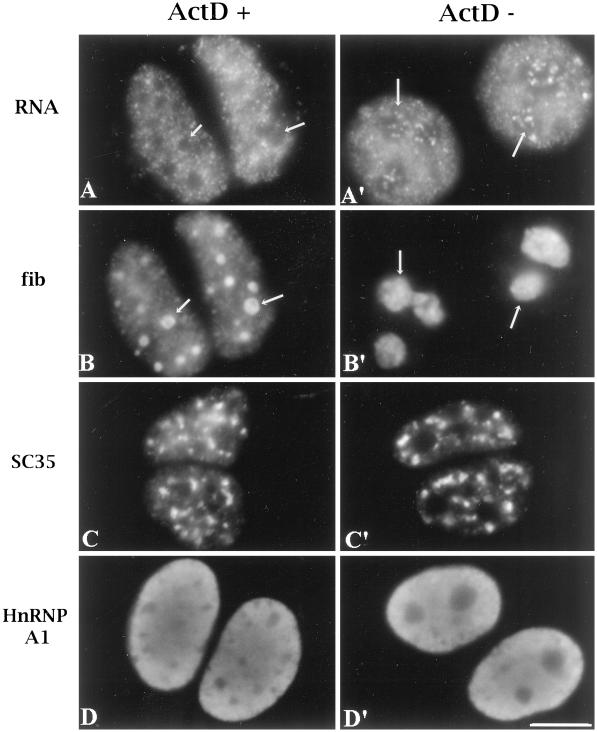
To determine whether treatment of ActD at 0.04 μg/ml may significantly influence other polymerase activities or nuclear structure in general, we monitored several indicators of Pol II transcription and nuclear structure. In situ incorporation of Br-UTP after 5 min of pulse labeling was compared between cells treated and not treated with ActD. Metaphase cells were pretreated with 0.04 μg/ml ActD and reseeded in the presence of the drug. Four hours after replating, the early G1 cells were briefly permeabilized and incubated at 37°C in a transcription cocktail containing Br-UTP for 5 min. The results show that Br-UTP incorporation is not detected in mininucleoli of the G1 cells that exit mitosis in the presence of ActD (Figure 3, A and B, arrows) but is detected in nucleoli of cells grown in drug-free medium (Figure 3, A′ and B′, arrows). In contrast, the nucleoplasmic incorporation of Br-UTP is not obviously changed in cells treated with ActD (Figure 3, A and A′), demonstrating that ActD treatment at this concentration selectively inhibits RNA Pol I transcription with little effect on Pol II and Pol III transcription. Similar observations were also made in interphase cells treated or not treated with the same concentration of ActD (our unpublished results). In addition to Br-UTP incorporation, we also examined the localization of an RNA splicing factor whose distribution is sensitive to RNA Pol II transcription inhibition. Immunolabeling with the use of a mAb specifically recognizing SC35, a member of the SR family (Fu and Maniatis, 1990), does not show significant changes in its speckled labeling pattern in cells treated with the drug (Figure 3C). Furthermore, the distribution of hnRNP proteins that shuttle between the nucleus and the cytoplasm in a transcription-dependent manner, such as hnRNP A1, was also examined. The hnRNP proteins primarily bind pre-mRNA and are implicated in the processing and transport of mRNA (recently reviewed by Krecic and Swanson, 1999). HnRNP A1 becomes cytoplasmically distributed during transcription inhibition with the use of a high concentration of ActD that inhibits all RNA polymerase activities (Pinol-Roma and Dreyfuss, 1992). Presumably, the nuclear localization is dependent on Pol II transcription. When cells were treated with 0.04 μg/ml ActD, the cytoplasmic accumulation of hnRNP A1 was not detected (Figure 3D), suggesting that Pol II transcription is not significantly altered. These results demonstrate that 0.04 μg/ml ActD selectively inhibits RNA Pol I transcription and does not significantly affect the nucleoplasmic transcription and the distribution of other nuclear constituents.
Figure 3.
ActD at a concentration of 0.04 μg/ml selectively inhibits Pol I transcription without significantly affecting other polymerases and general nuclear organization. A and A′ show the in situ incorporation of Br-UTP, and B and B′ show the same cells corresponding to those in A and A′ immunolabeled with anti-fibrillarin antibody. Daughter cells grown in the presence of ActD form mininucleoli that are immunolabeled with anti-fibrillarin antibody (B, arrows). They do not incorporate Br-UTP (A, arrows), whereas untreated nucleoli (B′, arrows) actively incorporate Br-UTP after pulse labeling (A′). Although Br-UTP incorporation in nucleoli declines in treated cells, the nucleoplasmic Br-UTP incorporation from Pol II and Pol III transcription is not altered (A). The speckled distribution of SC35 (C) and the predominantly nuclear localization of hnRNP A1 (D) are also not changed significantly in treated cells. Bar, 10 μm.
Segregated Mininucleoli That Differ from PNBs Are Developed in the Absence of rDNA Transcription
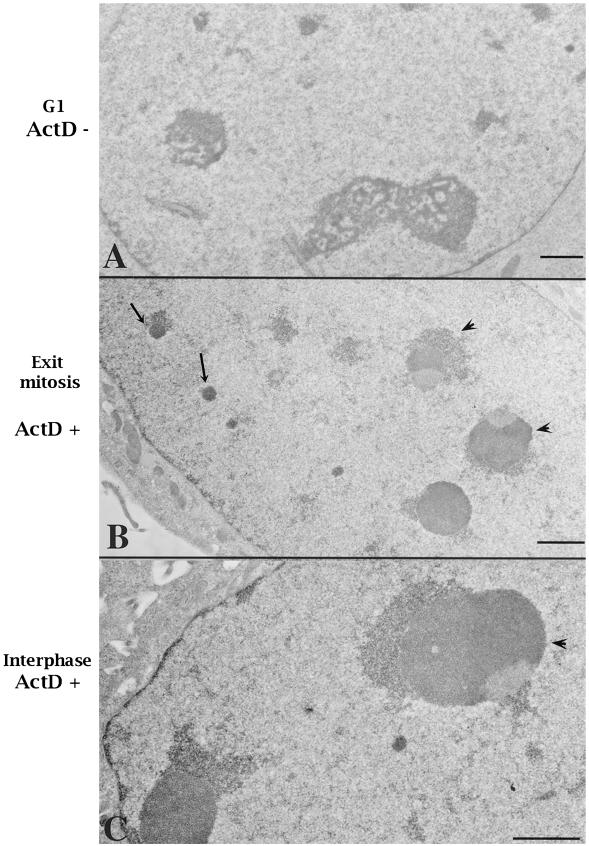
We reexamined nucleolar development in daughter cells progressing into G1 in the absence of Pol I transcription. Mitotic cells that were synchronized with the use of nocodazole were treated with ActD (0.04 μg/ml) 1 h before being released into the cell cycle in the presence of the same concentration of ActD. Four hours after reseeding, the cells were fixed and examined. Compared with untreated cells that form typical nucleoli at early G1, treated cells develop much smaller nucleolus-like structures. These structures are dense and recognizable by Nomarski or phase contrast microscopy (our unpublished results). To analyze them at higher resolution, G1 cells that had exited mitosis in the presence or absence of ActD were optimally fixed and examined by electron microscopy. In cells treated with ActD, two types of nucleolus-related structures were observed. The small, round, and electron-dense structures resemble typical PNBs (Figure 4B, arrows). The PNBs usually are not detectable in G1 cells because most should have fused to form typical nucleoli (Figure 4A). Another type of dense structures (Figure 4B, arrowheads), intermediate in size between PNBs and typical nucleoli, resemble the segregated nucleoli observed in interphase cells grown at the same concentration of ActD (Figure 4C, arrowhead). They contain segregated and densely packed fibrillar and granular components. These structures seem to correspond to those observed at the light microscopic level, in which the UBF-nucleolin-fibrillarin–containing caps juxtapose or surround dense structures (Figures 1E and 2, B and B′). We interpret these structures to be segregated mininucleoli rather than unfused and altered PNBs. These observations suggest that mininucleoli, although significantly changed in structure, are able to assemble in the absence of Pol I transcription.
Figure 4.
Electron microscopic observations demonstrate that cells progressing into G1 phase in the presence of 0.04 μg/ml ActD assemble segregated mininucleoli (B, arrowheads). In addition, unfused PNBs could be visualized in the nucleoplasm of treated cells (B, arrows). (A) A normal G1 cell nucleus. (B) A G1 cell exiting mitosis in the presence of 0.04 μg/ml ActD. (C) An interphase cell treated with the same concentration of ActD. Bars, 1 μm.
Pre-rRNAs Synthesized in the Previous Cell Cycle Are Recruited to Newly Formed Daughter Cell Nuclei
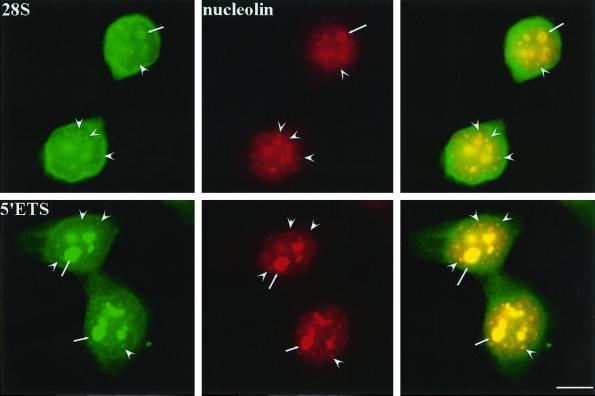
To analyze the distribution of rRNAs through mitosis into early G1, we examined rRNA distribution with the use of in situ hybridization with probes complementary to either 28S rRNAs or 5′ETS core–containing pre-rRNAs. The results demonstrate that 28S rRNAs are present in early G1 nuclei in addition to being distributed in the cytoplasm (Figure 5, top panels, arrows). The nuclear 28S rRNAs along with nucleolin are detected in two types of structures, the growing nucleoli and not-yet-fused PNBs (Figure 5, top panels, arrowheads). To determine whether the rRNAs detected with the use of the 28S probe represent precursor or mature rRNAs, the 5′ETS probe was used to examine the presence of 45S or 30S rRNAs. Double labeling of 5′ETS containing pre-rRNAs and nucleolin (Figure 5, bottom panels, arrows) reveals a similar observation as that observed with the use of the 28S probe. This finding demonstrates that at least part of the rRNAs detected with the use of the 28S probe in the growing nucleoli and PNBs represent 45S or 30S pre-rRNAs. Although the nucleolus-associated pre-rRNAs could be explained as newly synthesized rRNAs upon the activation of rDNA transcription, the PNB-associated pre-rRNAs are most likely derived from the previous cell cycle, because PNBs contain neither rDNA nor Pol I transcription factors and therefore could not be the sites of rDNA synthesis.
Figure 5.
Pre-rRNAs and 28S rRNAs synthesized at the previous cell cycle reenter early G1 daughter cell nuclei. 28S rRNA probe (top) and 5′ETS core probe (bottom) both hybridize to rRNAs that are associated with growing nucleoli (arrows) and nucleolin-containing PNBs (arrowheads). Bar, 10 μm.
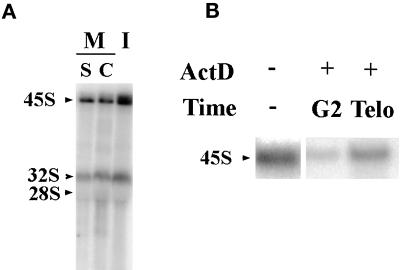
To analyze the stability of pre-rRNAs through mitosis into the next cell cycle, we compared the levels of these RNAs in mitosis and interphase with the use of Northern blotting. Mitotic cells were synchronized with either colchicine or double thymidine blocks. Equal amounts of total RNA from mitotic and interphase cell lysates were hybridized with the 5′ETS core probe. The results demonstrate that both 45S and 30S pre-rRNAs are detectable in mitotic cells with the use of either synchronization protocol (Figure 6A), although the level of the pre-rRNAs is significantly less than that of interphase cells. This finding demonstrates that some of the 5′ETS containing pre-rRNAs made during the previous cell cycle are stable through mitosis. This observation is unlikely to be due to any adverse effects of the drugs used in synchronization because two different methods of synchronization give rise to similar results. To analyze when these pre-rRNAs are synthesized, ActD (0.04 μg/ml) was added at different phases of the cell cycle of synchronized cells. When rDNA transcription is inhibited during G2 through mitosis, the level of the mitotic 45S rRNAs is significantly reduced (Figure 6B). In contrast, when rDNA transcription is inhibited at early telophase, the level of 45S rRNAs is not altered (Figure 6B). These findings suggest that 45S or 30S pre-rRNAs are synthesized predominantly near the G2/M transition and are maintained throughout mitosis into G1 of the next cell cycle.
Figure 6.
Pre-rRNAs (45S and 30S) containing 5′ETS core are detected through mitosis and synthesized during G2/M. (A) Northern blot hybridized with 5′ETS core probe showing that 45S and 30S pre-rRNAs are maintained in mitotic cells (M) synchronized with the use of either colchicine (C) or double thymidine block (S). The levels of these RNAs are less than those in interphase (I). (B) Northern blot hybridized with 5′ETS core probe to RNAs from early G1 cells that were treated with 0.04 μg/ml ActD at G2/M (G2) or telophase (Telo). (−) RNA extracted from control cells that were not treated with ActD. Treatment in G2 significantly reduces the detectable 45S rRNAs, whereas inhibition of Pol I transcription from mitosis to G1 still allows the maintenance of sizable amounts of 45S rRNA.
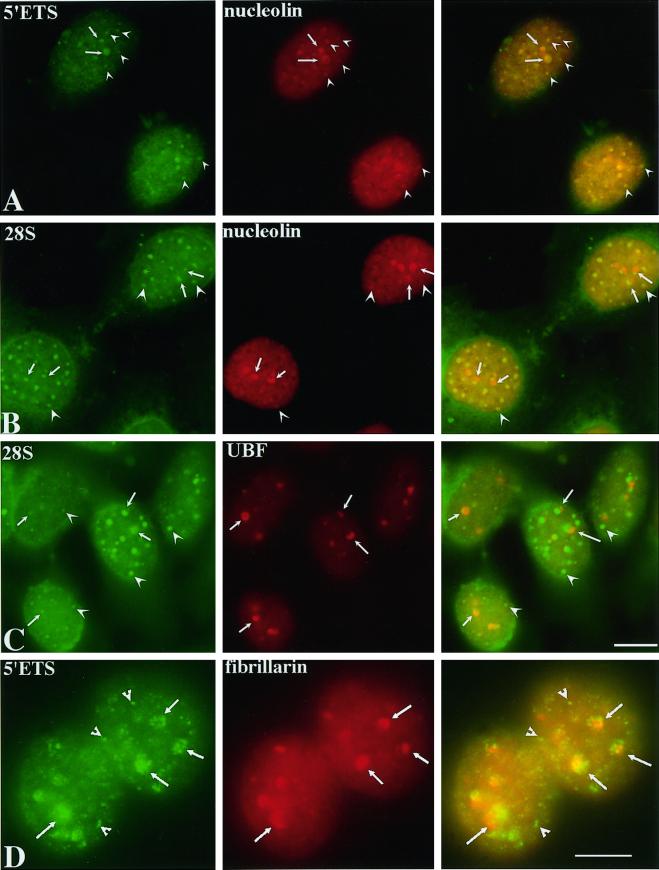
To clarify the origin of pre-rRNAs detected in the growing nucleoli and PNBs at the beginning of the cell cycle, we examined the distribution of these RNAs in cells progressing into G1 in the absence of Pol I transcription (Figure 7). Pre-rRNAs were detected by in situ hybridization with the use of either the 5′ETS core or the 28S probes. The localization of these RNAs was compared with the localization of UBF, fibrillarin, and nucleolin. In the absence of Pol I transcription, pre-rRNAs are observed in 10–20 foci in the nucleoplasm (Figure 7). Some are colocalized with nucleolin-associated PNBs that are unable to fuse with NORs (Figure 7, A and B, arrowheads). Others are associated with the UBF-associated NORs that fail to enlarge into typical nucleoli (Figure 7C, arrows). UBF-nucleolin-fibrillarin cap-like structures surround the pre-rRNA staining (Figure 7, arrows). These caps may represent the dense fibrillar cap-like structures observed by electron microscopy (Figure 4), and pre-rRNAs may represent the granular region (Figure 4). The presence of pre-rRNAs in new daughter nuclei exiting mitosis in the absence of Pol I transcription further supports the notion that the pre-rRNAs detected in PNBs and at NORs early in telophase are derived from the previous cell cycle.
Figure 7.
Pre-rRNAs and possibly 28S rRNAs are recruited to newly formed nuclei in the presence of 0.04 μg/ml ActD. They are colocalized with nucleolin (A and B)-associated PNBs (arrowheads) and are adjacent to UBF (C) and fibrillarin (D)-associated NORs (arrows). Bars, 10 μm.
DISCUSSION
Direct Recruitment of Nucleolar Factors to NORs Is Independent of PNBs and RNA Pol I Transcription
Nucleologenesis involves the assembly of nucleolar components around rDNA-containing chromosomes to establish morphologically well-defined structures at the beginning of interphase. Previous studies have shown that the formation of nucleoli is the consequence of, at least in part, the fusion between preassembled particles containing pre-rRNA processing factors, PNBs, and transcription machinery–associated NORs (Ochs et al., 1985; Jimenez-Garcia et al., 1994; Fomproix et al., 1998). This process is Pol I transcription dependent (Ochs et al., 1985; Benavente et al., 1987). In this report, we provide evidence of a previously undefined mechanism during nucleologenesis. In addition to fusion between NORs and PNBs, pre-rRNA processing factors are directly recruited to NORs early in telophase before chromatin decondensation. These initial recruitments are not disrupted in the absence of Pol I transcription. Because the cell cycle progresses despite Pol I transcription inhibition, our findings suggest that the initial events could be regulated by means other than activation of Pol I transcription. Recently, it was demonstrated that the activation of Pol I transcription is dependent on the level of cdc2–cyclin B activity when cells enter G1 (Sirri et al., 2000). We speculate that the initial recruitment of pre-rRNA processing factors might be regulated by kinases and/or phosphatases involved in the transition from mitosis to interphase. Pre-rRNA processing factors could be recruited to NORs to form a complex with the existing transcription machinery, and the complex could subsequently carry out transcription and processing of rRNA simultaneously. This hypothesis is in agreement with earlier electron microscopic observations of active transcription units that suggest that newly synthesized rRNAs are associated with RNA-binding proteins cotranscriptionally (reviewed by Miller, 1981; Osheim and Beyer 1998). Furthermore, recent studies in the RNA Pol II transcription system show that transcription factors are associated premRNA processing factors and may be recruited to the sites of transcription in the same complex (Gall et al., 1999) and that premRNA processing takes place cotranscriptionally (recently reviewed by Bentley, 1999). We propose that a similar situation may also take place during Pol I transcription.
Structurally Altered Mininucleoli Are Assembled in the Absence of Pol I Transcription
In this report, we reevaluate nucleologenesis in cells treated with a low concentration of ActD. Cells that progress into G1 in the absence of Pol I transcription exhibit two types of nucleolus-related structures, PNBs and segregated nucleoli. The morphological characteristics of these structures are similar to those illustrated by Ochs et al. (1985) and Benavente et al. (1987). They were described as segregated PNBs or round dense fibrogranular structures (Ochs et al., 1985; Benavente et al., 1987). Considering our observations at the light microscopic level, we interpret these structures to be segregated mininucleoli for the following reasons. 1) Morphologically, these structures resemble segregated nucleoli of interphase cells treated with the same concentration of ActD. They are composed of segregated fibrillar and granular components. 2) These structures appear to correspond to those observed at the light microscopic level, in which UBF-nucleolin-fibrillarin cap-like structures juxtapose pre-rRNAs. These segregated structures are composed of both transcription factor–associated NORs and components of pre-rRNA processing factors; thus, they resemble nucleoli more than PNBs, which typically do not contain rDNA and transcription factors. Based on these findings, we propose that nucleologenesis is independent of Pol I transcription. The early recruitment of processing factors and pre-rRNAs to NORs takes place before or at the onset of Pol I transcription. This model is in agreement with the observation that nucleolus-like structures assemble before the onset of rDNA transcription during Xenopus laevis development (Verheggen et al., 1998). However, the development of mature nucleoli by means of PNB fusion does require Pol I transcription, because PNBs fail to fuse with NORs in the presence of a low concentration of ActD, as also described by Ochs et al. (1985), and Benavente et al. (1987).
Pre-rRNAs Synthesized in the Previous Cell Cycle May Contribute to Nucleologenesis
Detection of 5′ETS core–containing pre-rRNAs and 28S rRNAs in cells exiting mitosis reveals that pre-rRNAs (45S and 30S) derived from the previous cell cycle are stable through mitosis and reenter daughter cell nuclei. These RNAs are predominantly synthesized near the G2/M transition of the last cell cycle and are distributed around interchromosomal spaces as well as in the mitotic cytoplasm. At telophase, they are recruited to UBF-associated NORs and are colocalized with nucleolin in PNBs. When Pol I transcription is inhibited, the initial recruitment of pre-rRNAs to the NORs continues. However, the PNB-associated pre-rRNAs no longer fuse with NORs and remain in the nucleoplasm through G1 phase. These findings suggest that the pre-rRNAs may participate in nucleolar assembly. However, it is not clear whether all the rRNAs detected with the use of the 28S probe represent pre-rRNAs. When probes for both 5′ETS-containing and 28S rRNAs of the same length are used, it appears that the hybridization intensity is stronger for 28S rRNAs than for 5′ETS-containing rRNAs, suggesting that 28S rRNAs may also enter daughter cell nuclei. Our findings provide the first evidence that pre-rRNAs synthesized in the previous cell cycle are included in the nucleologenesis in newly formed daughter cell nuclei. These findings are consistent with the biochemical analysis that the 5′ETS-containing 45S, 30S, and the 28S rRNAs are maintained through mitosis (Fan and Penman, 1971; Pinol-Roma, 1999) and that all of them are detected in nucleolin-containing RNPs (Pinol-Roma, 1999). Our observations are also in agreement with the findings described by Dundr et al. (1998) that pre-rRNAs are present in nucleolus-derived foci along with multiple pre-rRNA processing factors through mitosis (Dundr and Olson, 1998). Furthermore, the presence of pre-rRNAs in newly formed mininucleoli in the absence of Pol I transcription is in agreement with the observation that maternal pre-rRNAs are associated with newly formed nucleoli before the onset of rDNA transcription in Xenopus laevis embryos (Verheggen et al., 1998).
What might be the function of pre-rRNAs and their binding proteins during nucleologenesis? Several possible explanations are considered. 1) These rRNAs may form a macromolecular complex with pre-rRNA processing factors, and the complex may contribute to the architecture of newly formed nucleoli. 2) The complex may also represent a way of regulating the availability and the active status of pre-rRNA processing factors. 3) Alternatively, they could be recruited because of their binding to the processing factors. It is also intriguing that the existing pre-rRNAs are segregated from nucleolin, fibrillarin, and UBF in cells that progress into G1 when Pol I transcription is inhibited. The significance and mechanism of the pre-rRNA recruitment to newly formed daughter nuclei will be investigated further.
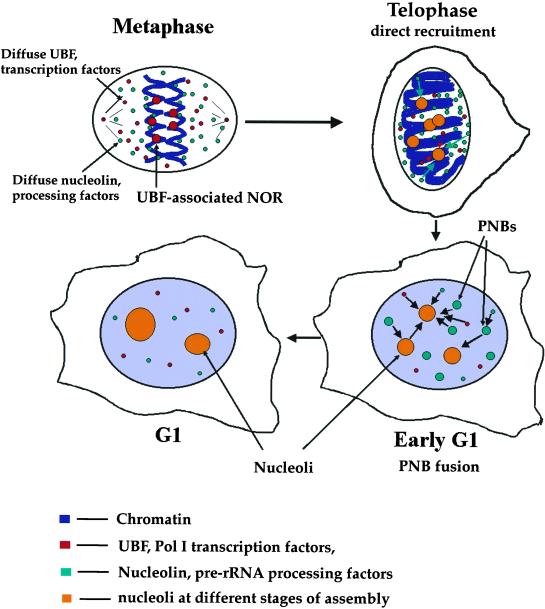
Based on the observations from our laboratory and other laboratories, we propose a revised model of nucleologenesis (Figure 8). At early telophase, competent NORs that are associated with the transcriptional machinery, including UBF, SL1, and RNA Pol I, rapidly and directly recruit pre-rRNA processing factors along with existing pre-rRNAs. This initial recruitment may take place before or at the onset of rDNA transcription. Subsequently, more transcription and processing factors are also recruited in forms of PNBs to small but enlarging nucleoli. The process of PNB fusion requires rDNA transcription. Pre-rRNAs derived from the previous cell cycle may contribute to nucleologenesis.
Figure 8.
Scheme of the revised model of nucleologenesis.
ACKNOWLEDGMENTS
The authors are grateful to J. Sylvester for the rDNA human probes and to G. Géraud for help with confocal microscopy. We acknowledge the Association pour la Recherche sur le Cancer (contract 5304) and the Centre National de la Recherche Scientifique for support. Our thanks also extend to Drs. E.K.L. Chen, S. Pinol-Roma, M. Dundr, and M.O.J. Olson for providing antibodies and rDNA constructs. We would also like to thank Drs. S. Adam, R. Moir, T. Pederson, T. Spann, and D. Spector for helpful comments. This work was partially supported by grants to S.H. from the National Cancer Institute (1 RO1 CA77560-01A1 and 5 K01 CA74988-03).
REFERENCES
- Azum-Gelade MC, Noaillac-Depeyre J, Caizergues-Ferrer M, Gas N. Cell cycle redistribution of U3 snRNA and fibrillarin: presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J Cell Sci. 1994;107:463–475. doi: 10.1242/jcs.107.2.463. [DOI] [PubMed] [Google Scholar]
- Benavente R, Rose KM, Reimer G, Hugle-Dorr B, Scheer U. Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J Cell Biol. 1987;105:1483–1491. doi: 10.1083/jcb.105.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Busch H, Smetana K. The Nucleolus. New York: Academic Press; 1970. [Google Scholar]
- Chan EKL, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF: molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med. 1991;174:1239–1244. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Struhl K. Nucleus and gene expression multiprotein complexes, mechanistic connections and nuclear organization. Curr Opin Cell Biol. 1999;11:303–306. [Google Scholar]
- Dundr M, Olson MOJ. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol Biol Cell. 1998;9:2407–2422. doi: 10.1091/mbc.9.9.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of synthesis and processing of nucleolar components in metaphase-arrested cells. J Mol Biol. 1971;59:27–42. doi: 10.1016/0022-2836(71)90411-6. [DOI] [PubMed] [Google Scholar]
- Fomproix N, Gebrane-Younes J, Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J Cell Sci. 1998;111:359–372. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- Fomproix N, Hernandez-Verdun D. Effects of anti-PM-Scl 100 (Rrp6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp Cell Res. 1999;251:452–464. doi: 10.1006/excr.1999.4578. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- Gautier T, Robert-Nicoud M, Guilly M-N, Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis: a study by confocal laser scanning microscopy. J Cell Sci. 1992;102:729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- Gautier T, Fomproix N, Masson C, Azum-Gelade MC, Gas N, Hernandez-Verdun D. Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol Cell. 1994;82:81–93. doi: 10.1016/s0248-4900(94)80010-3. [DOI] [PubMed] [Google Scholar]
- Gebrane-Younes J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and function of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hadjiolov AA. Cell Biology Monographs. New York: Springer-Verlag; 1985. The nucleolus and ribosome biogenesis; pp. 1–263. [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Miller OL., Jr The nucleolus, chromosomes, and visualization of genetic activity. J Cell Biol. 1981;91:15s–27s. doi: 10.1083/jcb.91.3.15s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Busch H. Further evidence that phosphoprotein C23 (110 kD/pI 5.1) is the nucleolar silver staining protein. Exp Cell Res. 1984;152:260–265. doi: 10.1016/0014-4827(84)90251-9. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Shen E, Carroll RE, Busch H. Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma. 1985;92:330–336. doi: 10.1007/BF00327463. [DOI] [PubMed] [Google Scholar]
- Osheim YN, Beyer AL. EM visualization of transcriptionally active genes after injection into Xenopus oocyte nuclei. Methods Cell Biol. 1998;53:471–496. doi: 10.1016/s0091-679x(08)60891-2. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman S, Vesco C, Penman M. Localization and kinetics of formation of nuclear heterodisperse RNA, cytoplasmic heterodisperse RNA and polyribosome-associated messenger RNA in HeLa cells. J Mol Biol. 1968;34:49–69. doi: 10.1016/0022-2836(68)90234-9. [DOI] [PubMed] [Google Scholar]
- Perry RP. Selective effects of actinomycin D on the intracellular distribution of RNA synthesis in tissue culture cells. Exp Cell Res. 1963;29:400–406. [Google Scholar]
- Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol Biol Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, Andre C, Masson C, Geraud G, Hernandez-Verdun D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci. 1993;104:327–337. doi: 10.1242/jcs.104.2.327. [DOI] [PubMed] [Google Scholar]
- Savino TM, Bastos R, Jansen E, Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Scheer U, Rose KM. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez Verdun D. The mitotically phosphorylated form of the transcription terminator factor TTF-1 is associated with the repressed rDNA transcription machinery. J Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- Sirri V, Roussel P, Hernandez Verdun D. In vivo release of mitotic silencing of rDNA transcription does not give rise to pre-rRNA processing. J Cell Biol. 2000;148:259–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell HM. Drug interactions with DNA. In: Busch H, Crooke ST, Daskal Y, editors. Effects of Drugs on the Cell Nucleus. New York: Academic Press; 1979. [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Stevens BJ. The fine structure of the nucleolus during mitosis in the grasshopper neuroblast cell. J Cell Biol. 1965;24:349–368. doi: 10.1083/jcb.24.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Sylvester JE, Whiteman DA, Podolsky R, Pozsgay JM, Respess J, Schmickel RD. The human ribosomal RNA genes: structure and organization of the complete repeating unit. Hum Genet. 1986;73:193–198. doi: 10.1007/BF00401226. [DOI] [PubMed] [Google Scholar]
- Verheggen C, Le Panse S, Almouzni G, Hernandez-Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis. J Cell Biol. 1998;142:1167–1180. doi: 10.1083/jcb.142.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D, Scheer U, Benavente R. The DNA topoisomerase I inhibitor camptothecin blocks postmitotic reformation of nucleoli in mammalian cells. Eur J Cell Biol. 1993;61:189–192. [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]