Abstract
Proteasome inhibitors reduce the budding of human immunodeficiency virus types 1 (HIV-1) and 2, simian immunodeficiency virus, and Rous sarcoma virus. To investigate this effect further, we examined the budding of other retroviruses from proteasome inhibitor-treated cells. The viruses tested differed in their Gag organization, late (L) domain usage, or assembly site from those previously examined. We found that proteasome inhibition decreased the budding of murine leukemia virus (plasma membrane assembly, PPPY L domain) and Mason-Pfizer monkey virus (cytoplasmic assembly, PPPY L domain), similar to the reduction observed for HIV-1. Thus, proteasome inhibitors can affect the budding of a virus that assembles within the cytoplasm. However, the budding of mouse mammary tumor virus (MMTV; cytoplasmic assembly, unknown L domain) was unaffected by proteasome inhibitors, similar to the proteasome-independent budding previously observed for equine infectious anemia virus (plasma membrane assembly, YPDL L domain). Examination of MMTV particles detected Gag-ubiquitin conjugates, demonstrating that an interaction with the ubiquitination system occurs during assembly, as previously found for other retroviruses. For all of the cell lines tested, the inhibitor treatment effectively inactivated proteasomes, as measured by the accumulation of polyubiquitinated proteins. The ubiquitination system was also inhibited, as evidenced by the loss of monoubiquitinated histones from treated cells. These results and those from other viruses show that proteasome inhibitors reduce the budding of viruses that utilize either a PPPY- or PTAP-based L domain and that this effect does not depend on the assembly site or the presence of monoubiquitinated Gag in the virion.
Proteasome inhibitor treatment reduces virus budding from cells expressing human immunodeficiency virus types 1 (HIV-1) and -2, simian immunodeficiency virus (SIV), and Rous sarcoma virus (RSV). In contrast, the release of equine infectious anemia virus (EIAV) is relatively unaffected by these inhibitors (40, 44, 45, 52, 53). For HIV-1 and -2, the processing of the Gag polyprotein into its mature proteins by the viral protease is also reduced when it is produced from cells with inactivated proteasomes (52). Gag processing induces maturation of the virion: i.e., the structural rearrangement of the virion from the immature to mature form, which is required for infectivity (reviewed in references 54 and 57). HIV-1 produced in the presence of proteasome inhibitors has low infectivity, probably due to this defect in maturation (52).
While retroviruses go through the same replication-cycle and have the same basic genomic organization, they do vary in Gag structure and the cellular location at which the viral structural proteins assemble into particles (7). The Gag polyprotein is the major structural protein of retroviruses and is entirely sufficient for particle formation (reviewed in reference 54). Orthoretroviruses (i.e., those primarily containing RNA in virions as per the International Committee on Taxonomy of Viruses [ICTV] convention [26]) bud from the plasma membrane, yet assemble by two different strategies. Alpha-, gamma-, and deltaviruses and lentiviruses (ICTV nomenclature) assemble on the inner leaflet of the plasma membrane and bud from the cell in a concerted fashion to release immature virions. Alternatively, betaretroviruses first assemble in the cytoplasm to form immature particles that are then transported to the plasma membrane where budding occurs.
Another difference among orthoretroviruses is the sequence used in the late (L) domain of Gag. L domains are centered around short amino acid sequences that are essential for efficient release of virions from the membrane (reviewed in reference 15). Mutations in these sequences cause virions to remain attached to the cell by a membrane tether that has a characteristic lollipop morphology (19) or appears as a series of particles trapped in a distended beads-on-a-string morphology (66). The three types of sequences that have been found to be required for L domain function are as follows: PPPY for RSV (62, 63), Mason-Pizer monkey virus (MPMV) (64), and murine leukemia virus (MuLV) (67); PTAP for HIV-1 (12, 19, 25); and YPDL for EIAV (5, 47). L domains are located primarily in two different regions of the Gag polyprotein, usually in the C terminus for lentiviruses or between the matrix (MA) and capsid (CA) proteins for non-lentiviruses. Both plasma membrane and cytoplasmic assembly schemes appear to require L domains (64). Interestingly, these L domains seem to be interchangeable and function independent of their position in Gag (43, 47, 63, 66).
Mechanistically, it is not clear how retroviruses are released from cells, although it seems clear that this requiresinteractions between L domains and cellular proteins (reviewed in references 4, 15, and 16). Several cellular proteins have been shown to interact with L domain sequences. Prime candidates for cellular proteins involved in budding are Tsg101, which binds PTAP (17, 32, 56); class I W-W domain-containing proteins, including the LDI-1 member of the Nedd4 family, which bind PPPY (16, 27); and the AP-50 subunit of the AP-2 complex, which binds YPDL (48). Interestingly, these proteins appear to be involved in protein trafficking and the endocytic pathway. The AP-2 complex functions in clathrin-mediated endocytosis (28), while Tsg101 is a ubiquitin-conjugating enzyme-like protein that, although unable to ubiquitinate proteins, functions in many cellular processes, including ubiquitin-mediated sorting of proteins into the multivesicular body budding pathway (14). While the cellular function of LDI-1 is unknown, Nedd4 is a ubiquitin ligase required for the ubiquitination and endocytosis of membrane proteins from the cell surface (49). Expression of fragments of LDI-1 and Tsg101 can specifically inhibit the budding of their respective viruses, suggesting a functional link between these proteins and viral budding (11, 27). Additionally, depletion of Tsg101 levels in HIV-1-producing cells reduces budding, further supporting a role for this protein in virus release.
The ubiquitination system attaches ubiquitin to lysines in targeted proteins by forming an isopeptide bond between the C terminus of ubiquitin and the ɛ-amino group of lysines (reviewed in references 50, 59, and 61). This process generally involves three steps: the E1 ubiquitin-activating enzyme attaches ubiquitin to an E2 ubiquitin-conjugating enzyme, which interacts with an E3 ubiquitin ligase to specifically recognize and ubiquitinate the target protein. While there is only one functional E1 enzyme, there are many E2 enzymes and probably even more E3 enzymes (50). Ubiquitin itself can be ubiquitinated on one of its internal lysines to form extended chains. The most commonly found modification on proteins is polyubiquitination (i.e., the addition of a ubiquitin chain with four or more proteins), which functions to target proteins for degradation by 26S proteasomes (6, 59, 61). In contrast, the addition of chains of less than four ubiquitin molecules to a lysine, most commonly the attachment of a single ubiquitin (monoubiquitination), is not normally a signal for proteasomal degradation; rather, this modification functions in various cellular processes, including endocytosis (24, 55). Proteins can be monoubiquitinated at several sites and not serve as proteasome degradation signals.
Retroviruses interact with the ubiquitination system during assembly, as evidenced by the finding that small amounts of monoubiquitinated HIV-1, SIV, MuLV, and EIAV Gag proteins are present inside virions (39, 40). Furthermore, ubiquitination of a minimal Gag construct requires a PPPY L domain, demonstrating an interaction with a ubiquitin ligase activity (53). Interestingly, the ubiquitinated Gag found in viruses is modified near their L domains (39, 40). Together, these results demonstrate that retroviruses interact with the ubiquitination system during assembly and budding, apparently in an L domain-dependent manner. Interestingly, ubiquitin has been shown to be part of the budding machinery for RSV (45).
The ubiquitination of Gag and the proteasome activity requirement for viral budding suggest that other retroviruses might also interact with the ubiquitin-proteasome pathway. In this report, we have investigated whether retroviruses that differ in their L domain usage or assembly strategy are sensitive to proteasome inhibitors. These results revealed that proteasome inhibitors can reduce the budding of viruses that assemble in the cytoplasm. However, even though Gag-ubiquitin conjugates were found inside mouse mammary tumor virus (MMTV) virions, the budding of this virus did not require proteasome activity, indicating that interaction with the ubiquitination system does not correlate with sensitivity to proteasome inhibitors.
MATERIALS AND METHODS
Cell culture.
The following chronically infected cell lines and culture media were used: HIV-1MN-infected H9 cell line Clone 4 in RPMI 1640; Moloney MuLV-infected NIH 3T3, EIAV-infected Cft2h, M-PMV-infected A204 cells; and MMTV-producing Mm5MT cells in Dulbecco's modified Eagle's medium. All media were supplemented with 10% (vol/vol) fetal bovine serum (except the medium used with NIH 3T3 cells, which was supplemented with 10% [vol/vol] calf serum), 2 mM glutamine, and 100 U (each) of penicillin and streptomycin per ml. All media and medium additives were obtained from Invitrogen (Carlsbad, Calif.). The M-M cell line was produced by infecting Mm5MT cells with Mo-10A1, a Moloney MuLV that contains the 10A1 envelope (41), and culturing them for 2 weeks.
Steady-state immunoblot analysis.
Duplicate infected cell cultures (either a ∼90% confluent monolayer in a 75-cm2 flask or 5 × 106 cells in suspension) were washed three times in phosphate-buffered saline (PBS [without Ca2+ and Mg2+]), and then 1 ml of medium was added with or without proteasome inhibitors for 10 min to allow for the release of preformed virions. The medium was then removed, and 9 ml of fresh medium with or without the respective inhibitors was added. The inhibitors used in this study were either a combination of 10 μM carbobenzoxyl-l-leucinyl-l-leucinyl-leucinal (zLLL) and lactacystin (both obtained from Biomol, Plymouth Meeting, Pa.) or 1 μM PS-341 (Millennium Pharmaceuticals, Cambridge, Mass.). At various time points from the initial exposure to the inhibitors, supernatants were removed, and the virions present were isolated by being pelleted through 20% sucrose as previously described (37). Cells were lysed in a mixture of 0.1% (vol/vol) Triton X-100, 10 mM Tris (pH 7.5), and 100 mM NaCl, and the nuclei were removed by centrifugation at 2,500 × g for 5 min at 4°C to produce cytoplasmic and nuclear fractions. Immunoblot analysis of virions and cytoplasmic and nuclear fractions was carried out as previously described with Immobilon-P filters (Millipore, Inc., Watertown, Mass.) and Enhanced ChemiLuminescent detection (23). Antibody against ubiquitin clone, 2C5, was obtained from MLB International (Watertown, Mass.); goat serum against HIV p24CA (goat 81) and rabbit sera against HIV-1 p6Gag (DJ-30552), MuLV p15MA (DJ-39529), and p30CA (DJ-39461) were produced by the AIDS Vaccine Program (NCI-Frederick, Frederick, Md.); goat sera against MPMV p27CA and MMTV p27CA were obtained from ViroMed Biosafety Products (Camden, N.J.). Scanning densitometry was performed with the Scion Image program (Scion Corp., Frederick, Md.) and an Epson Expression 1600 scanner (Epson USA, Long Beach, Calif.). Signal densities were measured and compared by using sampling boxes of similar sizes.
Protein analysis.
Double-sucrose-density-purified MMTV (prepared as previously described in reference 3) was used for preparative reversed-phase high-pressure liquid chromatography (HPLC) under conditions described in reference 22. Briefly, a virion preparation of 204 mg of total protein, as assayed by the Lowry method (31), was applied to an RCM μBondapak C18 column (25 by 100 mm) (Waters Corp., Milford, Mass.) by using a continuous water-acetonitrile gradient from 18 to 60% (vol/vol) with 0.1% trifluoroacetic acid at 50°C. Samples from the fractions containing protein peaks were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie brilliant blue staining and by ubiquitin immunoblotting. Fractions containing proteins to be analyzed were blotted onto Immobilon-P filters and detected with Ponceau S stain, and the bands of interest were excised and subjected to Edman degradation protein sequence analysis on an Applied Biosystems (Foster City, Calif.) Procise model 494 microsequencer as previously described (22).
Sequence comparison.
A local homology search was performed with the BestFit utility in the SeqWeb 2.0 software package (GCG Corporation, Madison Wis.).
RESULTS
We have previously used a sensitive pulse-chase technique to demonstrate that proteasome inhibitors reduce both the release of HIV-1 and -2 virions from cells and the processing of Gag in those virions by approximately fourfold (52). To extend this result, we wanted to study other retroviruses that differ in L domain type, Gag organization, or assembly site. However, our pulse-chase analysis technically requires high levels of viral protein expression and sensitive immune reagents, two requirements that did not allow for the study of the viruses that we wished to examine (data not shown). As an alternate method, we treated chronically infected cell lines with proteasome inhibitors and analyzed samples of virions, cytoplasm, and nuclei prepared at various times by immunoblotting. We have previously used this time course immunoblot technique to study the effect of proteasome inhibitors on EIAV budding, which is relatively resistant to these compounds (40).
Validation of the time course immunoblot technique.
To demonstrate that this method can adequately detect the reduction in budding observed with proteasome inhibitor treatment, we used immunoblotting to analyze the HIV-1 produced from chronically infected cells. Clone 4 cells, which produce constitutively high levels of HIV-1MN (42), were first washed before being placed in fresh medium either without or with a combination of proteasome inhibitors, 10 μM reversibly acting peptide aldehyde zLLL (also called MG-132), and the lactone derivative lactacystin, which irreversibly alkylates proteasome enzymes (9, 30). These inhibitor concentrations completely block proteasome activity with little effect on translation or cellular integrity during the treatment period (38, 51, 52). Unlike pulse-chase experiments, in which the fate of newly synthesized Gag can be observed, this immunoblot-based method can only detect the steady-state levels of Gag. Thus upon proteasome inhibition, some Gag will already be in the final stages of budding, potentially producing particles that can confound this analysis and mask the true effect of proteasome inactivation on viral budding. To reduce the number of particles that bud immediately after the washing steps, the medium on the cells was removed after a postwash incubation for 10 min at 37°C and replaced with the corresponding fresh medium.
Virion samples were prepared from treated or untreated cultures at various time points for 8 h from the initial exposure to the inhibitors and analyzed by immunoblotting, first with p6Gag antiserum, followed by stripping and detection with anti-p24CA serum (Fig. 1A). All of the treated and untreated samples were processed and analyzed in parallel under the same conditions. For both exposures, the bands in the treated samples were less intense than those in the untreated samples, revealing that proteasome inhibitor treatment of these chronically infected T cells decreased HIV-1 budding, similar to the pulse-chase results with acutely infected T cells (52). Unlike the very specific proteasome inhibitor lactacystin, the zLLL peptide aldehyde can indiscriminately form hemiacetal adducts with hydroxyl side groups in some enzymes and inhibit their activity, specifically that of proteases (30). This has led to artifactual results in studies of EIAV Gag processing (40, 44). To confirm the time-course results, another proteasome inhibitor, PS-341, was used in this assay. This reversible dipeptide boronic acid inhibitor is highly specific for the proteasome, tightly interacting with the hydroxyl group in threonine 1 of the proteasome's catalytic β-subunit (Ki = 0.6 nM) (2). Cells were washed and treated as described above with medium without inhibitors, the 10 μM zLLL-lactacystin combination, or 1 μM PS-341, and the virions released into the supernatant over an 8-h period were isolated from the cultures and examined by p24CA immunoblotting (Fig. 1B). The results showed that both of these inhibitors decreased virus budding to nearly the same extent as the untreated culture. The signal densities of the bands were measured by Scion Image densitometry software. The results, presented in Table 1, show that the 8 h of treatment with the 10 μM zLLL-lactacystin combination or 1 μM PS-341 reduced the level of HIV-1 budding by approximately fourfold. These results are similar to our previously reported pulse-chase findings for HIV-1 (52). Therefore, this time course immunoblot method can be used to study the short-term effects of virus release.
FIG. 1.
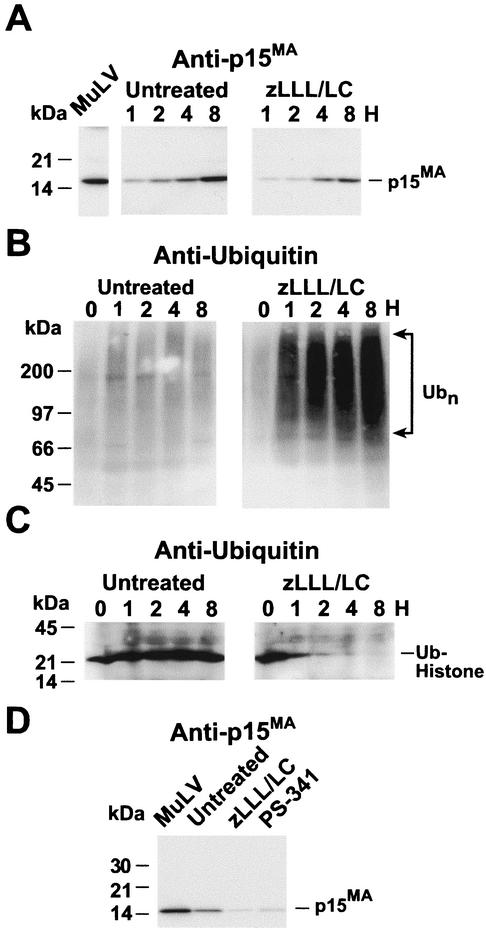
Immunoblot analysis of treated HIV-1-infected cultures. (A) Time course immunoblots of virion samples produced from cells in the presence or absence of a 10 μM zLLL-lactacystin (zLLL/LC) combination. The number of hours posttreatment is indicated above each lane. (B) Immunoblot of virion samples produced from cells during 8 h of treatment with the indicated inhibitors. (C) Ubiquitin (Ub) immunoblots of cytoplasmic extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. Ubn, polyubiquitinated proteins. (D) Ubiquitin immunoblots of nuclear extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. The antiserum used is indicated above each blot, and each sample is labeled above its respective lane. “HIV-1” indicates virus control, a sample of a 24-h harvest of virions prior to treatment. Molecular mass markers are indicated on the left, and the detected bands are identified on the right.
TABLE 1.
Density of immunoblot samples
| Virus and treatment | Densitya | Relative density of treated vs. untreated |
|---|---|---|
| HIV-1 | ||
| None | 123 | 1 |
| zLLL-lactacystin | 22 | 0.18 |
| PS-341 | 29 | 0.24 |
| MuLV | ||
| None | 87 | 1 |
| zLLL-lactacystin | 11 | 0.13 |
| PS-341 | 19 | 0.22 |
| M-PMV | ||
| None | 43 | 1 |
| zLLL-lactacystin | 11 | 0.26 |
| PS-341 | 11 | 0.26 |
| MMTV | ||
| None | 167 | 1 |
| zLLL-lactacystin | 145 | 0.87 |
| PS-341 | 143 | 0.86 |
Mean pixel gray value of band − mean pixel gray value of background.
To demonstrate that the inhibitor treatment effectively inactivated the proteasomes, cytoplasmic fractions of the zLLL-lactacystin-treated cells from the time course experiment were examined by ubiquitin immunoblotting and compared to the corresponding untreated samples (Fig. 1C). The results clearly showed that proteasome inhibitor treatment induced an intense and rapid accumulation of polyubiquitinated protein conjugates within 1 h of treatment, indicative of an effective block in proteasome function.
To determine if the ubiquitinating machinery in general is affected by this proteasome inhibitor treatment, we examined the nuclei of zLLL-lactacystin-treated cells from the time course experiment for the presence of a 21-kDa histone-ubiquitin conjugate that has been previously used as a measure of monoubiquitination activity (8, 13, 29, 33). Histones 2A and 2B are reversibly monoubiquitinated in a dynamic process that appears to regulate transcription (18, 34, 35, 46, 60). Therefore, interruption of the monoubiquitination of histones causes a rapid decrease in the steady-state levels of ubiquitinated histones (8, 13, 29, 33). Ubiquitin immunoblot analysis of nuclear extracts revealed that the amount of monoubiquitinated histone began to decrease after 1 h of inhibitor treatment, with no histone conjugates detectable after 8 h (Fig. 1D). These results show that this proteasome inhibitor treatment reduced the ability of the cell to monoubiquitinate histones, and presumably other proteins, in addition to inducing the accumulation of polyubiquitinated proteins.
MuLV budding is sensitive to proteasome inhibitors.
Unlike HIV-1, MuLV possesses a PPPY-based L domain in p12Gag, located between MA and CA in Gag (66), similar in sequence and position to the RSV L domain. To determine if MuLV budding is reduced by proteasome inhibitors, NIH 3T3 cells that were chronically infected with MuLV were treated with the 10 μM zLLL-lactacystin combination by the same procedure used in the HIV-1 experiments described above. Immunoblotting of virion samples isolated at different times during the 8-h treatment with p15MA antiserum showed that, compared to the untreated samples, less virus was released in the presence of the inhibitors (Fig. 2A). Therefore, similar to our results with HIV-1 presented above, MuLV budding is sensitive to proteasome inhibitors and thus is dependent on proteasome function. As was the case with HIV-1, examination of the cytoplasmic fraction samples from this time course experiment by ubiquitin immunoblotting confirmed that polyubiquitinated proteins accumulated rapidly, within 1 h of treatment, demonstrating the efficacy of the proteasome block in the NIH 3T3 cells (Fig. 2B). Ubiquitin immunoblots of nuclear extracts also demonstrated that proteasome inhibitor treatment decreased ubiquitinated histone levels after 1 h posttreatment (Fig. 2C).
FIG. 2.
Immunoblot analysis of treated MuLV-infected cultures. (A) Time course immunoblots of virion samples produced from cells in the presence or absence of 10 μM zLLL-lactacystin (zLLL/LC) combination. The number of hours posttreatment is indicated above each lane. (B) Ubiquitin immunoblots of cytoplasmic extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. Ubn, polyubiquitinated proteins. (C) Ubiquitin immunoblots of nuclear extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. (D) Immunoblot of virion samples after 8 h of treatment with the indicated inhibitors. The antiserum used is indicated above each blot, and each sample is labeled above its respective lane. “MuLV” indicates virus control, a sample of a 24-h harvest of virions prior to treatment. Molecular mass markers are indicated by arrows on the left, and the bands are identified by arrows on the right.
To confirm the time course result, the MuLV-infected cells were exposed to the highly specific PS-341 inhibitor. A p15MA immunoblot of the virions produced during 8 h by cells treated with PS-341 or the zLLL-lactacystin combination produced results similar to those for HIV-1 (Fig. 2D): the signals from the treated samples, in comparison to that in the untreated sample, were reduced. Densitometry analysis showed that there was approximately four- to fivefold less signal in the treated samples than in the untreated control (Table 1). Together, these results demonstrate that MuLV budding is reduced by proteasome inhibitors to a similar extent as HIV-1.
M-PMV budding is decreased by proteasome inhibitors.
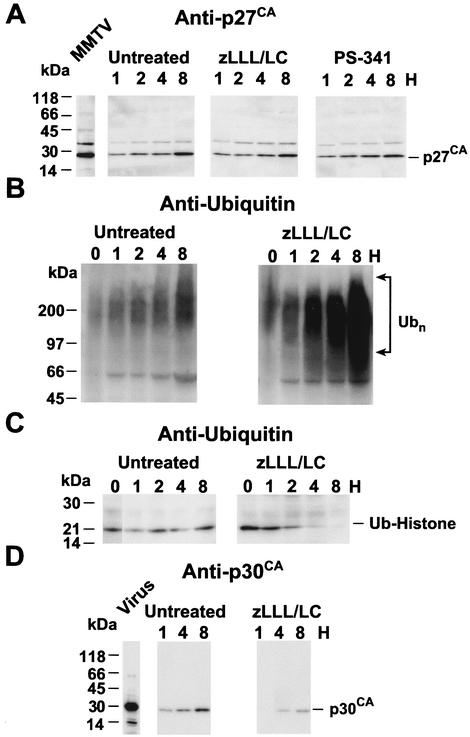
To date, the importance of the ubiquitin-proteasome pathway has only been investigated for retroviruses that assemble at the plasma membrane. To examine the proteasome inhibitory effect on a betaretrovirus (i.e., one that assembles in the cytoplasm), A204 rhabdomyosarcoma cells chronically infected with M-PMV were analyzed by the time course immunoblot procedure with the 10 μM zLLL-lactacystin combination. Immunoblot analysis with p27CA antiserum revealed that proteasome inhibitor treatment decreased the amount of virions released from the cell (Fig. 3A). Examination of the cytoplasmic fractions by ubiquitin immunoblotting showed that, similarly to the other cell lines tested, polyubiquitinated proteins accumulated within 1 h, indicating that the inhibitors blocked the proteasome (Fig. 3B). Consistent with the data presented above, the ubiquitinating activity of the cell was inhibited, as revealed by the loss of monoubiquitinated histones in the nuclei of the cells after 8 h of treatment (Fig. 3C). Analysis of virions produced from cells after an 8-h treatment of virus-producing cells with the zLLL-lactacystin combination or PS-341 showed that the levels of viral budding were decreased for both these inhibitors to a comparable extent (Fig. 3D): the scanned densities of the both bands were approximately fourfold lower for the inhibitor-treated samples than those that were untreated (Table 1). Based on these immunoblot data, proteasome function is required for efficient budding of this betaretrovirus.
FIG. 3.
Immunoblot analysis of treated M-PMV-infected cultures. (A) Time course immunoblots of virion samples produced from cells in the presence or absence of a 10 μM zLLL-lactacystin (zLLL/LC) combination. The number of hours posttreatment is indicated above each lane. (B) Ubiquitin (Ub) immunoblots of cytoplasmic extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. Ubn, polyubiquitinated proteins. (C) Ubiquitin immunoblots of nuclear extracts from the 8-h untreated (8U) and treated (8PI) cells. (D) Immunoblot of virion samples after 8 h of treatment with the indicated inhibitors. The antiserum used is indicated above each blot, and each sample is labeled above its respective lane. “M-PMV” indicates virus control, a sample of a 24-h harvest of virions prior to treatment. Molecular mass markers are indicated by arrows on the left, and the bands are identified by arrows on the right.
MMTV budding does not require proteasome activity.
Like M-PMV, MMTV also assembles in the cytoplasm. While M-PMV uses a PPPY-type L domain, the L domain for MMTV has not been characterized. The MMTV Gag protein sequence does not contain either a PTAP or PPPY sequence in the region between MA and CA (pp21, p3, or p8) where the RSV, MuLV, and M-PMV Gag proteins contain their L domains. However, MMTV Gag does contain a PSAP sequence in p27CA and two YXXL motifs in p10MA and pp21. Viral budding from Mm5MT cells, a chronically MMTV-infected mouse mammary gland cell line, was examined by the time course immunoblot procedure in the presence of the 10 μM zLLL-lactacystin combination or 1 μM PS-341. Immunoblot analysis of the virions harvested and isolated at various time points revealed that the budding of MMTV was not detectably reduced by the proteasome inhibitors: the amounts of virions present in the supernatants from the untreated and the treated samples were essentially indistinguishable from one another (Fig. 4A and Table 1). The faint band above the p27CA band in the immunoblot is probably due to the presence of a small amount of an incompletely processed p8Gag-p27CA Gag polyprotein fragment. The failure to reduce virus budding was not due to ineffective proteasome inhibition in the Mm5MT cells, because, consistent with the other experiments, the zLLL-lactacystin combination (Fig. 4B) or PS-341 (data not shown) treatment produced a rapid and intense accumulation of polyubiquitinated proteins. Similarly, the level of monoubiquitinated histones declined, and these modified histones were no longer present in the nuclei from the treated cells after 8 h, confirming the interruption of the ubiquitination system in these cells (Fig. 4C).
FIG. 4.
Immunoblot analysis of treated MMTV-infected cultures. (A) Time course immunoblots of virion samples produced from cells with a 10 μM zLLL-lactacystin (zLLL/LC) combination, 1 μM PS-341, or no treatment. The number of hours posttreatment is indicated above each lane. (B) Ubiquitin (Ub) immunoblots of cytoplasmic extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. Ubn, polyubiquitinated proteins. (C) Ubiquitin immunoblots of nuclear extracts from untreated and treated cells. The number of hours posttreatment is indicated above each lane. (D) Time course immunoblots to detect MuLV p30CA in virion samples produced from M-M cells in the presence or absence of a 10 μM zLLL-lactacystin combination. The antiserum used is indicated above each blot, and each sample is labeled above its respective lane. “MMTV” indicates virus control, a sample of a 24-h harvest of virions prior to treatment. Molecular mass markers are indicated on the left, and the detected bands are identified on the right.
It is possible that despite the clear interruption of the ubiquitin-proteasome system, our results could be an artifact of the cell line used. To address this question, Mm5MT cells were infected with MuLV, an inhibitor-sensitive virus. This cell line, M-M, was then treated with a 10 μM zLLL-lactacystin combination to see if the budding of MuLV was reduced as expected. Immunoblot analysis of the virions harvested and isolated at various time-points revealed that the budding of MuLV was decreased in the presence of inhibitors compared to that in the untreated controls (Fig. 4D). These data demonstrate that inhibition of proteasomes in Mm5MT cells can reduce retroviral budding. Thus, the insensitivity of MMTV budding to proteasome inhibitors is due to a property of this virus rather than an artifact of the Mm5MT cell line.
MMTV Gag is ubiquitinated.
Ubiquitinated Gag proteins have been found in several viruses (HIV-1, SIV, and MuLV) that are sensitive to proteasome inhibitors. Therefore, these viruses appear to interact with both the ubiquitination and proteasome systems. However, the p9Gag protein of EIAV Pr55Gag is monoubiquitinated, yet the budding of EIAV is relatively unaffected by proteasome inhibitors (40), suggesting that these two phenomena are not necessarily linked. To determine if MMTV Gag interacts with the ubiquitination system, we searched for Gag-ubiquitin conjugates in virions. A 204-mg (total protein) portion of a purified MMTV stock was separated by preparative reversed-phase HPLC. Screening of 5% of the selected fractions by ubiquitin immunoblotting suggested that, in addition to free ubiquitin (present as a 5-kDa band), some 20- to 30-kDa ubiquitin-reacting bands were present in fractions 92 to 98 (Fig. 5). The proteins in this size range in fractions 94 and 96 were separated by SDS-PAGE and blotted onto filters, and their respective bands were excised and subjected to N-terminal protein sequence analysis. The results detected two ubiquitin-MMTV Gag conjugates (Fig. 5). Fraction 94 contained a 30-kDa band that produced both ubiquitin and p14NC sequences at a 2-to-1 molar ratio. Therefore, this protein is p14NC conjugated to two ubiquitin molecules. From these data, it is not possible to determine whether this protein is monoubiquitinated twice or simply diubiquitinated on one lysine. Fraction 96 contained a 21-kDa band that yielded a sequence of both p3 and ubiquitin at a 1:1 ratio. Considering that the p3 protein itself does not contain lysines and given the apparent 21-kDa size of this conjugate, this protein is probably a p3-p8 intermediate processing product that is conjugated to ubiquitin. Thus, two Gag proteins are ubiquitinated in MMTV, unlike those in our previous studies, which found only one Gag protein modified in other viruses (39, 40). It has not been determined if these two modifications are found on the same Gag polyprotein or are present even in the same virion. The total amount of Gag-ubiquitin conjugates present in MMTV could not be estimated, although the amounts found here are not more than 1% of the total amount of Gag applied to the HPLC column. Thus, similarly to the Gag proteins of several other retroviruses we have previously characterized, MMTV interacts with the ubiquitinating system, even though its budding is insensitive to proteasome inhibitors.
FIG. 5.
HPLC analysis of MMTV. The HPLC A206 chromatogram of the entire MMTV separation is presented at the top, with the area analyzed denoted in a box. The chromatogram of the region examined is presented with the sequence data above and ubiquitin (Ub) immunoblots below the respective fractions. Sequences are presented in the single-amino-acid code, with “X” denoting an ambiguous amino acid determination.
Ubiquitin levels and inhibitors.
In the absence of proteasome activity, free ubiquitin levels have been observed to decline over time, apparently because of the inability of the cell to recycle the ubiquitin sequestered in the polyubiquitinated proteins that accumulate under these conditions (33). However, another study found that free ubiquitin levels were relatively stable after proteasome inhibitor treatment (36). It has been proposed that decreased free ubiquitin levels caused by proteasome inhibition would block the ubiquitination of Gag (45, 53). This, in turn, could prevent a ubiquitination of Gag that might be required for virus release, resulting in the observed budding defect. To determine the levels of free ubiquitin during the inhibitor treatment of cells, we compared the signal intensities of the ubiquitin bands present in immunoblots of the same 8-h cytoplasmic lysates from the zLLL-lactacystin-treated and untreated HIV-1-, MuLV-, and MMTV-infected cells analyzed as described above (Fig. 1C, 2C, and 4C, respectively). Lanes containing treated samples were compared to a series of lanes, each with decreasing amounts of untreated sample (Fig. 6). The immunoblots of the HIV-1-infected Clone 4 cells showed that the intensity of 10 μl of the treated cell lysate (from 4 × 104 cells) was between that of the 10- and 5-μl samples of untreated lysate (from 4 × 104 to 2 × 104 cells, respectively). Therefore, blocking proteasomes only produced a small difference (less than a twofold effect) in ubiquitin levels after 8 h of treatment. For MuLV-infected cells, there was less of a difference: the intensities of the 10-μl lysates (from approximately 2 × 104 cells) were essentially equal for both samples, indicating little, if any decrease in free ubiquitin levels. The results with the MMTV-infected cell lysate samples were similar to those from the HIV-1 infected cells: the intensity of the 10-μl treated sample (from approximately 2 × 104 cells) was similar to that of the 5-μl untreated sample. Corresponding results were found in a study of EIAV-infected cells (data not shown). These results show that despite the accumulation of polyubiquitinated proteins, the level of free ubiquitin did not drastically decrease immediately following proteasome inhibition.
FIG. 6.
Ubiquitin levels in proteasome inhibitor-treated cells. Ubiquitin (Ub) immunoblots of the 8-h cell lysate samples from protease inhibitor (PI)-treated culture are compared to a dilution series of the corresponding untreated cell lysate samples. The infected cells examined and their treatment are indicated above the respective blots, and the amount of cytoplasmic preparation loaded is indicated above each lane.
DISCUSSION
Our data show that proteasome inhibitors reduce the budding of MuLV and M-PMV. Given that proteasome inhibitors also reduce the budding of HIV-1, HIV-2, SIV, and RSV, most retroviruses require active proteasomes for efficient budding. The reduction in M-PMV budding in the presence of proteasome inhibitors demonstrates that retroviruses that assemble in the cytoplasm can be affected by this treatment as well. However, we found that the budding of another betaretrovirus, MMTV, was unaffected by proteasome inhibitors, similar to the recent findings for EIAV (40, 44). It has been proposed that the insensitivity of EIAV budding to proteasome inhibitors might be due to a ubiquitin-like motif (NVKEKDQ) in the p9Gag region of EAIV Gag that could function to promote virus release in the presence of these inhibitors (44). A comparison of MMTV Gag and ubiquitin protein sequences (using the BestFit program) showed no such homology. Therefore, it is unlikely that the results with MMTV can be explained by a ubiquitin homology signal. Understanding the differences between MMTV, EIAV, and retroviruses that are sensitive to proteasome inhibitors should help our understanding of the retroviral budding processes.
One of the current proposed mechanisms for the proteasome inhibitor effect postulates that ubiquitination of Gag is required for virus budding (reviewed in references 15 and 58). In this model, the loss of proteasome activity causes free ubiquitin to be depleted, resulting in a loss of the ubiquitination activity that modifies Gag. We have previously shown that over long periods of treatment (36 h), ubiquitin does decline sixfold (52). However, the data presented here show that free ubiquitin levels did not appear to change dramatically during the relatively short time frame (8 h) of these experiments, even though virus budding was clearly reduced. Therefore, a simple decrease in free ubiquitin is not likely to be the cause of the reduced virus budding in our experiments. Despite the relative stability of free ubiquitin levels, the monoubiquitination of histones was clearly inhibited after 1 to 2 h of proteasome inhibitor treatment in all of the cell lines used in this study, suggesting that the ubiquitination of Gag would also be rapidly affected.
The link between the ubiquitination of Gag and retroviral budding is not clear. While experiments with RSV show that ubiquitin is part of the budding machinery (45), it is not known whether the ubiquitination of Gag plays a role in the budding of other retroviruses. We show here that a small amount of MMTV Gag is monoubiquitinated (or in the case of p14NC potentially diubiquitinated), similar to our previous findings for HIV-1, MuLV, and EIAV. For EIAV and MMTV, the observed Gag ubiquitination events do not correlate with a requirement for proteasome activity in budding. Additionally, the elimination in HIV-1 or MuLV Gag of the lysines that are known acceptors for monoubiquitination does not have any detectable effect on virus budding (12, 38, 67), although unidentified ubiquitination sites in Gag could function in budding. Overall, these data do not support a model of Gag ubiquitination being required for virus budding, although they most certainly do not exclude it. As proposed previously, the observed ubiquitination of Gag could be simply a consequence of Gag interacting with a cellular ubiquitinating activity (38, 40).
An alternative mechanism to the direct ubiquitination of Gag for the proteasome effect is centered around the accumulation of Gag proteins as defective ribosomal products (DRiPs) upon proteasome inhibition (51, 52). DRiPs are misfolded or improperly modified proteins that result from errors in protein synthesis and folding (65). These dead-end proteins are normally removed from the cytoplasm by polyubiquitination, which targets them for degradation by the 26S proteasome. However, without active proteasomes, these defective proteins accumulate. In the case of a highly cooperative process such as virion assembly, defective Gag proteins could interfere with the Gag-Gag interactions during particle formation. Thus, proteasome inactivation increases the levels of Gag DRiPs that, in turn, might act in a dominant-negative fashion, interfering with assembly, decreasing virus budding, and reducing processing by the viral protease. The insensitivity of MMTV shown here as well as that of EIAV, while not compatible with this model, may be due to other factors—perhaps L domain-mediated effects—that cause these proteins to be either segregated from or otherwise unaffected by the accumulation of Gag-DRiPs. Comparisons of Gag-DRiP formation between HIV-1, EIAV, and MMTV might help clarify these points.
While an L domain for MMTV has not been identified, the regions of MMTV Gag that are ubiquitinated may provide a clue to its location. MMTV Gag does not contain a PPPY or PTAP sequence. There is a PSAP sequence in p27CA, although the presence of a functional L domain in capsid would be unprecedented. Since HIV-1, MuLV, and EIAV Gag are ubiquitinated near their respective L domains and L domains are functionally linked to ubiquitination (53), MMTV Gag might contain L domains in p8Gag and p14NC that use unique sequences.
To date, all of the retroviruses that are sensitive to proteasome inhibitors use or appear to use PTAP- or PPPY-based L domains. Since the viruses that are resistant to proteasome inhibitors, EIAV and MMTV, use either YPDL or possibly another type of L domain, respectively, proteasome inhibitors appear to specifically act on viruses that use the PTAP and PPPY domains. Recent reports have suggested that PPPY serves as an L domain for the matrix protein of vesicular stomatitis virus (10, 21) and that the PPXY sequence is required for an L domain function in the Ebola virus VP40 matrix protein (20, 32). Perhaps proteasome inhibitors might also affect the budding of these unrelated viruses.
While the mechanism by which proteasome inhibitors decrease HIV-1 budding and infectivity is not yet clear, they have been proposed as antiviral therapeutics for AIDS (52). Even though proteasome function is essential, certain specific inhibitors have yielded promising results as anticancer agents (1), thereby demonstrating that this cellular target can be safely interrupted in humans. Further work on the mechanism(s) of action for the proteasome inhibitor effect on retroviral budding should allow us a better understanding of this important portion of the viral life cycle and potentially allow for new therapies, either as proteasome inhibitor-based pharmaceuticals or aids to the development of small molecule inhibitors to inhibit viral budding and Gag processing.
Acknowledgments
We acknowledge Julian Bess for providing the MMTV preparation and Robert Gorelick for critical reading of the manuscript.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400. U.S. was supported by grant Schu11/2-1, a Heisenberg grant from the Deutsche Forschungsgemeinschaft, and a grant from the German Human Genome Research Project.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Adams, J. 2002. Preclinical and clinical evaluation of proteasome inhibitor PS-341 for the treatment of cancer. Curr. Opin. Chem. Biol. 6:493-500. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J., M. Behnke, S. Chen, A. A. Cruickshank, L. R. Dick, L. Grenier, J. M. Klunder, Y. T. Ma, L. Plamondon, and R. L. Stein. 1998. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8:333-338. [DOI] [PubMed] [Google Scholar]
- 3.Bess, J. W., Jr., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type 1, and other retroviruses. J. Virol. 66:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, C. A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203-205. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J., S. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 8.Cox, J. H., P. Galardy, J. R. Bennink, and J. W. Yewdell. 1995. Presentation of endogenous and exogenous antigens is not affected by inactivation of E1 ubiquitin-activating enzyme in temperature-sensitive cell lines. J. Immunol. 154:511-519. [PubMed] [Google Scholar]
- 9.Craiu, A., M. Gaczynska, T. Akopian, C. F. Gramm, G. Fenteany, A. L. Goldberg, and K. L. Rock. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 272:13437-13445. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deveraux, Q., R. Wells, and M. Rechsteiner. 1990. Ubiquitin metabolism in ts85 cells, a mouse carcinoma line that contains a thermolabile ubiquitin activating enzyme. J. Biol. Chem. 265:6323-6329. [PubMed] [Google Scholar]
- 14.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11:R932-R934. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature (London) 381:744-745. [DOI] [PubMed] [Google Scholar]
- 17.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 18.Goldknoff, I. L., and H. Busch. 1977. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. USA 74:864-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson, L. E., R. C. Sowder II, G. Smythers, and S. Oroszlan. 1987. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J. Virol. 61:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hull, R. 2001. Classifying reverse transcribing elements: a proposal and a challenge to the ICTV. International Committee on Taxonomy of Viruses. Arch. Virol. 146:2255-2261. [DOI] [PubMed] [Google Scholar]
- 27.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchhausen, T. 2002. Clathrin adaptors really adapt. Cell 109:413-416. [DOI] [PubMed] [Google Scholar]
- 29.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 30.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 31.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 255:265-275. [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 33.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 34.Mueller, R. D., H. Yasuda, C. L. Hatch, W. M. Bonner, and E. M. Bradbury. 1985. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. J. Biol. Chem. 260:5147-5153. [PubMed] [Google Scholar]
- 35.Ohe, Y., H. Hayashi, and K. Iwai. 1979. Human spleen histone H2B. Isolation and amino acid sequence. J. Biochem. (Tokyo) 85:615-624. [DOI] [PubMed] [Google Scholar]
- 36.Ohtani-Kaneko, R., K. Takada, M. Iigo, M. Hara, H. Yokosawa, S. Kawashima, K. Ohkawa, and K. Hirata. 1998. Proteasome inhibitors which induce neurite outgrowth from PC12h cells cause different subcellular accumulations of multi-ubiquitin chains. Neurochem. Res. 23:1435-1443. [DOI] [PubMed] [Google Scholar]
- 37.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J. Virol. 73:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 39.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D. E., J. Keller, K. Sill, and A. Rein. 1992. Phenotypes of murine leukemia virus-induced tumors: influence of 3′ viral coding sequences. J. Virol. 66:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott, D. E., S. M. Nigida, Jr., L. E. Henderson, and L. O. Arthur. 1995. The majority of cells are superinfected in a cloned cell line that produces high levels of human immunodeficiency virus type 1 strain MN. J. Virol. 69:2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 47.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 50.Scheffner, M., S. Smith, and S. Jentsch. 1998. The ubiquitin-conjugation system, p. 65-98. In J.-M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum Press, New York, N.Y.
- 51.Schubert, U., L. C. Anton, A. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 52.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 55.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogt, V. 1997. Retroviral virions and genomes, p. 27-70. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 58.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 60.West, M. H. P., and W. M. Bonner. 1980. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 8:4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 62.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yewdell, J. W., L. C. Anton, and J. R. Bennink. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157:1823-1826. [PubMed] [Google Scholar]
- 66.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]