Abstract
Finger insertion mutations of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) (T69S mutations followed by various dipeptide insertions) have a multinucleoside resistance phenotype that can be explained by decreased sensitivity to deoxynucleoside triphosphate (dNTP) inhibition of the nucleotide-dependent unblocking activity of RT. We show that RTs with SG or AG (but not SS) insertions have three- to fourfold-increased unblocking activity and that all three finger insertion mutations have threefold-decreased sensitivity to dNTP inhibition. The additional presence of M41L and T215Y mutations increased unblocking activity for all three insertions, greatly reduced the sensitivity to dNTP inhibition, and resulted in defects in in vitro DNA chain elongation. The DNA chain elongation defects were partially repaired by additional mutations at positions 210, 211, and 214. These results suggest that structural communication between the regions of RT defined by these mutations plays a role in the multinucleoside resistance phenotype.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is responsible for replication of the HIV-1 genome and is therefore an important target for antiretroviral therapy. Many inhibitors of HIV-1 RT are nucleoside analogues that are converted to their active triphosphate forms by host cell kinases and are incorporated into the viral genome by HIV-1 RT. Because nucleoside analogues lack a 3′-OH group, their incorporation prevents further extension of the DNA chain. HIV-1 RT becomes resistant to nucleoside analogues primarily through two mechanisms (8, 12): (i) increased discrimination against the compounds (9, 13, 35) leading to decreased incorporation and less chain termination and (ii) increased ability to remove chain terminators from blocked DNA chains, allowing DNA synthesis to resume (1, 3, 19, 20, 22, 26). The removal occurs through excision of the 3′-terminal nucleotide by transfer to one of several potential acceptor substrates, including pyrophosphate (PPi) or ATP, generating the triphosphate form of the chain terminator or a dinucleoside polyphosphate, respectively. Most zidovudine (AZT) resistance mutations, including M41L, D67N, K70R, T215F/Y, and K219Q (29), confer increased removal, compared to wild-type (WT) RT (3, 19, 22, 25, 26). The effect of these mutations is greatest when a nucleoside triphosphate, such as ATP, is used as the acceptor substrate, whereas there is little if any effect of AZT resistance mutations on removal when a nucleoside diphosphate or PPi is used as the substrate (3, 16, 20, 22, 25).
The efficiency of the removal reaction under physiological conditions will depend on the rate of the reaction and the ability of deoxynucleoside triphosphates (dNTPs) to inhibit it. HIV-1 RT bound to a chain-terminated primer-template will still bind the dNTP complementary to the next nucleotide on the template strand (34) to form a stable complex. Since chemical bond formation is prevented due to lack of a 3′-OH, this complex has been named a dead-end complex (DEC). While in DEC, excision of the chain-terminated primer-template is precluded (23). The balance between removal versus DEC formation depends on several factors, including the nature of the chain-terminating nucleotide analogue present at the 3′ end of the DNA chain, the HIV-1 RT used in the removal reaction, and the concentration of dNTPs. Primer-templates chain terminated with 3′-azido-3′-deoxythymidine-5′-monophosphate (AZTMP) are excellent substrates for nucleotide-dependent removal and poor substrates for DEC formation (KddNTP for DEC formation, 40 to 220 μM) (22, 23). Isel et al. (12) have also reported that DEC formed with AZTMP-terminated primer-template are less stable than those formed with 2′,3′-dideoxyadenosine-5′ monophosphate (ddAMP)- or 2′,3′-didehydro-3′-deoxythymidine-5′-monophosphate (d4TMP)-terminated primer-templates. Therefore, removal of AZTMP can occur efficiently in the presence of physiological concentrations of dNTPs. Primer-templates terminated with nucleotide analogues such as ddAMP or d4TMP that contain smaller 3′ substituents are also removed efficiently but form DEC at low dNTP concentrations (KddNTP, 3 to 20 μM) (12, 22, 23, 25, 26). Therefore, removal of these nucleotide analogues under physiological conditions will depend on the intracellular concentration of dNTPs, which have been reported to range from 0.14 to 5.6 μM in resting lymphocytes (10, 11, 28, 33), 2.4 to 26 μM in mitogen-stimulated lymphocytes (10, 11), and 15 to 170 μM in CEM lymphoblasts (28, 33). Although AZT resistance mutations confer an increase in removal of most chain terminators and likely confer some resistance to many types of chain terminators in HIV-infected individuals, the impact of these mutations on phenotypic resistance assays is likely underestimated due to very high dNTP levels present in the cells used in these assays. Nevertheless, it is now recognized that these mutations play a role in resistance to a variety of nucleoside inhibitors (8).
While AZT resistance mutations are specific for resistance to AZT in phenotypic assays (15), there are other classes of mutations that confer phenotypic resistance to multiple nucleoside analogues (14, 21, 37). One such class contains a threonine-to-serine mutation at codon 69 together with a dipeptide insertion between amino acids 69 and 70 in HIV-1 RT. These so-called finger insertion mutations are present in approximately 0.5 to 2.3% of nucleoside RT inhibitor-experienced individuals (30, 31, 36) and usually appear after exposure to AZT in combination with other nucleoside analogues. The finger insertion mutations are generally present in combination with AZT resistance mutations (7, 14, 21, 31, 37). The inserted dipeptide is most often SS or SG but can also consist of many other combinations of small amino acids (14, 21, 31, 37).
The finger insertion mutations confer little, if any, ability to discriminate against incorporation of chain terminators (17, 19) but have been shown to increase ATP-dependent removal of chain terminators (4, 16, 19, 20) and to decrease inhibition of the removal reaction by dNTPs (4, 19, 20). However, there is still little known about differences, if any, between various dipeptide insertions or the combined effects of these mutations with AZT resistance mutations on the biochemical properties of HIV-1 RT. We therefore conducted a detailed study on the effects of three sets of finger insertion mutations, 69S-SS, 69S-SG, and 69S-AG by themselves or in the background of two different sets of AZT resistance mutations, either M41L/T215Y or M41L/L210W/R211K/L214F/T215Y, on nucleotide-dependent removal, sensitivity of the removal reaction to dNTPs, and effects of the mutations on DNA-dependent DNA polymerization.
Removal of ddAMP from blocked primer-templates by WT and mutant RT.
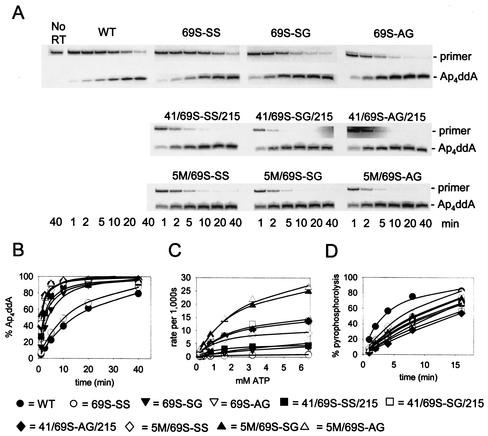
The enzymes used in our studies were WT RT, RT containing the AZT resistance mutations M41L/T215Y (41L/215Y), or RT containing one of three sets of finger insertion mutations, 69S-SS, 69S-SG, or 69S-AG, present in either WT background or together with the AZT resistance mutations 41L/215Y or 41L/210W/211K/214F/215Y (5M) (Table 1). Removal of ddAMP was detected by the transfer of [32P]ddAMP from a labeled primer terminus to ATP to form [32P]dinucleoside tetraphosphate (Ap4ddA). DNA primer L32 was annealed with DNA template WL50 and was chain terminated by extending with [32P]ddATP as previously reported (22). The [32P]ddAMP-terminated L32 primer-WL50 template was reisolated and incubated with excess WT or mutant RT in the presence of ATP. Products were separated by polyacrylamide gel electrophoresis, and [32P]Ap4ddA was quantitated by phosphorimaging as a function of time of incubation with 3.2 mM ATP (Fig. 1A and B) and as a function of ATP concentration (Fig. 1C and Table 2). There was no increase in Ap4ddA synthesis conferred by the 69S-SS mutation, in agreement with published data on ATP-dependent removal of AZTMP (19). The 69S-SG and 69S-AG mutations, however, did confer modest (three- or fourfold) increases over results for WT RT. The difference between the 69S-SG/AG and the 69S-SS mutations was also observed in RT-containing AZT resistance mutations. The 41L/69S-SS/215Y mutations conferred about a 10-fold increase in Ap4ddA synthesis over WT RT, consistent with an additive effect of the 10-fold increase conferred by the 41L/215Y mutations (25) and minimal increase due to the 69S-SS mutations. The 41L/69S-SG/215Y and the 41L/69S-AG/215Y mutations resulted in 21-fold increases in Ap4ddA synthesis relative to WT RT, also consistent with an additive effect of the finger insertion mutations and the AZT resistance mutations. Addition of the mutations 210W, 211K, and 214F to the 41L/215Y mutations in HIV-1 RT containing the finger insertion mutations further increased the Ap4ddA synthesis activity by approximately 50%. Two enzymes, 5M + 69S-SG RT and 5M + 69S-AG, formed Ap4ddA 32 times more efficiently than did WT RT. Similar methods were used to measure the pyrophosphorolysis reaction catalyzed by WT and mutant enzymes. The [32P]ddAMP-terminated primer-template was incubated with WT or mutant RTs in the presence of PPi, and [32P]ddATP was quantitated by phosphorimaging (Fig. 1D). The mutant enzymes had pyrophosphorolysis activities that were at most 60 to 70% of the WT activity.
TABLE 1.
Nomenclature of HIV-1 RT mutants useda
| Name | Mutations |
|---|---|
| WT | None |
| 69S-SS | T69S and an insertion of SS after 69 |
| 69S-SG | T69S and an insertion of SG after 69 |
| 69S-AG | T69S and an insertion of AG after 69 |
| 41L/215Y | M41L, T215Y |
| 41L/69S-SS/215Y | M41L, T69S, an insertion of SS after 69, and T215Y |
| 41L/69S-SG/215Y | M41L, T69S, an insertion of SG after 69, and T215Y |
| 41L/69S-AG/215Y | M41L, T69S, an insertion of AG after 69, and T215Y |
| 5M+69S-SS | M41L, T69S, an insertion of SS after 69, L210W, R211K, L214F, and T215Y |
| 5M+69S-SG | M41L, T69S, an insertion of SG after 69, L210W, R211K, L214F, and T215Y |
| 5M+69S-AG | M41L, T69S, an insertion of AG after 69, L210W, R211K, L214F, and T215Y |
His-tagged HIV-1 RT was prepared as previously described (24). The RNA-dependent DNA polymerase activities of the WT and mutant RTs were as follows: WT, 35,000 U/mg; 69S-SS, 40,000 U/mg; 69S-SG, 23,000 U/mg; 69S-AG, 41,000 U/mg; 41L/215Y, 27,000 U/mg; 41L/69S-SS/215Y, 33,000 U/mg; 41L/69S-SG/215Y, 38,000 U/mg; 41L/69S-AG/215Y, 33,000 U/mg; 5M + 69S-SS, 25,000 U/mg; 5M + 69S-SG, 29,000 U/mg; and 5M + 69S-AG, 27,000 U/mg, where 1 U is equal to the amount of enzyme required for incorporation of 1.0 nmol of [3H]dTMP in 10 min when using poly(rA)/oligo(dT) as the substrate (32).
FIG. 1.
Effect of finger insertion mutations and AZT resistance mutations on removal of [32P]ddAMP from blocked primer-template through transfer to ATP or PPi. (A) [32P]-ddAMP-terminated L32 primer-WL50 template (5 nM) was incubated with the indicated RT and 3.2 mM ATP for 1, 2, 5, 10, 20, or 40 min in reaction buffer (40 mM HEPES, pH 7.5, 20 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol, 2.5% glycerol, and 80 μg of bovine serum albumin/ml) at 37°C. Products were separated on a 20% denaturing polyacrylamide gel. (B) The radioactivity in primer and Ap4ddA was quantitated by phosphorimaging, and the percent Ap4ddA formed was plotted versus time. (C) Ap4ddA synthesis as a function of ATP concentration. Reactions were carried out as for panel A but with 0.2 to 6.4 mM ATP and for 20 s to 10 min, depending on the enzyme, to allow a maximum of 35% Ap4ddA formation. (D) PPi-dependent formation of [32P]ddATP (pyrophosphorolysis). Experiments were performed as described in Fig. 1A legend but with 5 μM PPi substituted for ATP, and incubation times were 1, 2, 4, 8, and 16 min. The amount of radioactivity in primer and ddATP was quantitated by phosphorimaging, and the percentage of label in ddATP was plotted versus time. For panels B, C, and D, the symbols represent data points obtained with the RTs indicated at the bottom of the figure, and the lines represent the best fit to hyperbolae when Sigmaplot 4.0 was used. L32 primer, 5′-CTACTAGTTTTCTCCATCTAGACGATACCAGA-3′; and WL50 template, 5′-GAGTGCTGAGGTCTTCATTCTGGTATCGTCTAGATGGAGAAAACTAGTAG-3′.
TABLE 2.
Kinetic parameters for Ap4ddA formation by WT and mutant RTs
| HIV-1 RT used | Values for formation of Ap4ddAa
|
|
|---|---|---|
| kcat/Km, ATP (M−1 S−1) | Increase (n-fold) versus WT RT | |
| WT | 0.38 ± 0.02 | 1.0 |
| 69S-SS | 0.38 ± 0.03 | 1.0 |
| 69S-SG | 1.2 ± 0.1 | 3.2 |
| 69S-AG | 1.6 ± 0.2 | 4.2 |
| 41L/215Y | 1.9 ± 0.4 | 9.0∗ |
| 41L/69S-SS/215Y | 4.0 ± 0.1 | 10 |
| 41L/69S-SG/215Y | 8.0 ± 0.9 | 21 |
| 41L/69S-AG/215Y | 7.8 ± 0.8 | 21 |
| 5M+69S-SS | 7.0 ± 1.8 | 18 |
| 5M+69S-SG | 12 ± 2.0 | 32 |
| 5M+69S-AG | 12 ± 2.0 | 32 |
Inhibition of ATP-dependent removal of ddAMP by the next complementary dNTP.
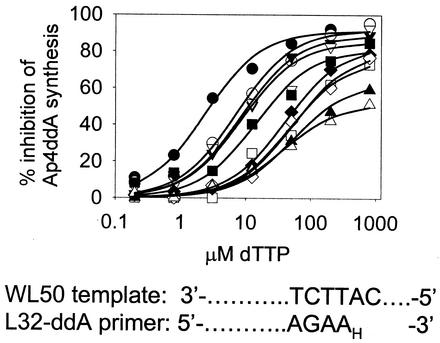
The large increase in ATP-dependent removal of ddAMP conferred by the combination of AZT resistance mutations and finger insertion mutations suggests a mechanism for resistance for the mutant HIV-1 to dideoxyinosine (ddI) (which is converted to its active form, ddATP, by host enzymes). However, removal of ddAMP by either WT or AZT-resistant RTs is very sensitive to inhibition by dNTPs (50% inhibitory concentrations [IC50s] approximately 3 to 20 μM [22, 24, 25]). We therefore investigated the effects of the finger insertion mutations on the sensitivity of the removal reaction to dTTP, the dNTP complementary to the next nucleotide position on the template. Briefly, the synthesis of Ap4ddA was determined as described above in the presence of increasing concentrations of dTTP. The percent inhibition of Ap4ddA synthesis is shown as a function of dTTP concentration (Fig. 2), and the IC50 for dTTP inhibition was determined for each RT (Table 3). WT RT was the most sensitive to inhibition by dTTP with an IC50 of about 3 μM. Each of the finger insertion mutations decreased sensitivity to dTTP inhibition of Ap4ddA synthesis by about threefold. Together with the 41L/215Y mutations, the finger insertion mutations decreased sensitivity to dTTP inhibition by 5- to 18-fold. Sensitivity to dTTP inhibition was decreased only about 25% by the 41L/215Y mutations alone (Table 3) (25). Therefore, the effect of the combination of finger insertion mutations with 41L/215Y mutations on dTTP inhibition was synergistic. When the 41L/210W/211K/214F/215Y mutations were present as well as the finger insertion mutations, dTTP inhibition was decreased by 16- to 23-fold relative to WT RT. The decreased sensitivity to dTTP inhibition of Ap4ddA synthesis by the mutant RTs is likely related to decreased formation or decreased stability of DEC formed by the mutant enzymes with ddAMP-terminated primer-template and dTTP (data not shown).
FIG. 2.
Ability of the complementary dNTP to inhibit dinucleoside polyphosphate synthesis. Inhibition of Ap4ddA synthesis by dTTP. The ATP-dependent removal of ddAMP was performed as described in the Fig. 1 legend in the presence of 0.2 to 800 μM dTTP. The amount of Ap4ddA was measured by phosphorimaging, and the percent inhibition of Ap4ddA formation was plotted versus dTTP concentration. Symbols are identified in the legend to Fig. 1. Lines represent the theoretical curves obtained by fitting the data to hyperbolae by using Sigmaplot 4.0 and were used to determine IC50s (Table 3). A portion of the L32-ddAMP-WL50 sequence is shown at the bottom of the figure.
TABLE 3.
Ability of dTTP, the dNTP complementary to the next nucleotide position on the template, to inhibit Ap4ddA synthesis by HIV-1 RT and ddAMP-terminated primer-template
| HIV-1 RT used | Values for inhibition of Ap4ddA synthesisa
|
|
|---|---|---|
| IC50 (μM) of dTTP | IC50 versus WT | |
| WT | 2.8 ± 0.5 | 1.0 |
| 69S-SS | 8.6 ± 2.0 | 3.1 |
| 69S-SG | 7.3 ± 1.7 | 2.6 |
| 69S-AG | 8.8 ± 0.4 | 3.1 |
| 41L/215Y | 4.9 ± 2.3 | 1.2b |
| 41L/69S-SS/215Y | 14 ± 1 | 5.0 |
| 41L/69S-SG/215Y | 42 ± 17 | 15 |
| 41/69S-AG/215Y | 50 ± 25 | 18 |
| 5M+69S-SS | 64 ± 14 | 23 |
| 5M+69S-SG | 48 ± 9 | 17 |
| 5M+69S-AG | 44 ± 11 | 16 |
Effects of the finger insertion mutations on HIV-1 RT DNA-dependent polymerization.
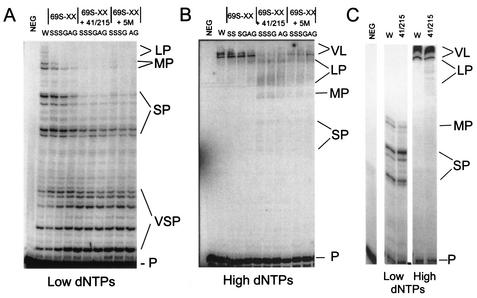
The decreased sensitivity of mutant RTs to dNTP inhibition and decreased ability to form stable ternary complexes with dNTP and primer-template suggested that dNTP incorporation might also be altered by these mutations. We therefore measured the ability of each enzyme to carry out DNA-dependent polymerization on M13 template-primer in the presence of low, 625 nM (Fig. 3A) or high, 200 μM (Fig. 3B) concentrations of dNTPs. A comparison of DNA chain elongation with 41L/215Y RT and isogenic WT RT at each dNTP concentration is shown in Fig. 3C.
FIG. 3.
DNA-dependent polymerization by WT and mutant RTs. 5′-[32P]-labeled 35-mer DNA primer (5′-AAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGAC) annealed to M13 template (2.5 nM) was incubated with excess RT (100 nM) at 37°C for 5 min followed by incubation with either a 625 nM (Low dNTPs [A and C]) or 200 μM (High dNTPs [B and C]) concentration of each dNTP for 2.5 min at 37°C. Reactions were stopped by heating at 90°C for 5 min. Samples were separated by electrophoresis through a 10% denaturing polyacrylamide gel. Extension products are indicated at the right of each panel as VL (very long products), LP (long products), MP (intermediate products), SP (short products), VSP (very short products), and P (unextended primer). The enzyme used for the extension is indicated above each lane: NEG, no enzyme; W, WT RT; 69S-XX, 69S-SS, SG, or AG RT as indicated; 41/215, 41L/215Y RT with 69S-XX mutations as indicated; 5M, 41L/210W/211K/214F/215Y RT with 69S-XX mutations as indicated.
At low levels of dNTPs, WT and 41L/215Y RTs had the greatest DNA chain elongation activity, but short products still predominated (Fig. 3A and C). The three RTs containing only the 69S-XX mutations had reduced DNA polymerization activities compared to WT RT. Of the three finger insertion mutations, the 69S-SS mutant was slightly more active than the 69S-SG and AG mutants, with more products in the SP and intermediate-length size ranges and fewer very short products than the other two enzymes. RTs containing the finger insertion mutations in combination with the 41L/215Y mutations showed a dramatic reduction in DNA-dependent polymerization activity, with products in the very short range predominating and no difference between the three finger insertion mutations. The 41L/69S-XX/215Y RTs were much more defective in primer extension than either 41L/215Y RT or the 69S-XX RTs, and it is clear that the deficiency in chain elongation is due to interaction between these mutations. Enzymes containing the finger insertion mutations in combination with the 41L/210W/211K/214F/215Y mutations also had greatly reduced polymerization activity compared to WT RT.
At high dNTP concentrations (200 μM), WT RT and 41L/215Y RT extended almost all chains into very long products (Fig. 3B and C). Each of the three enzymes containing the finger insertion mutations in the absence of AZT resistance mutations could also extend the majority of DNA to very long products. Even at these high dNTP levels, however, the enzymes containing the finger insertion mutations in combination with the 41L/215Y mutations produced much shorter products. With the addition of mutations at 210W, 211K, and 214F (5M), longer products were observed (Fig. 3B), although the DNA chains did not reach the lengths seen with WT or 41L/215Y RT. In summary, the DNA-dependent DNA polymerase activity was greatest for WT RT with WT ∼ 41L/215Y > 69S-XX > 69S-XX + 5M > 41L/69S-XX/215Y.
Conclusions.
AZT resistance mutations confer an increase in ATP-dependent removal of many chain terminators from blocked DNA ends (3, 4, 16, 17, 19, 22, 23, 25, 26). The concentration of dNTPs in infected cells, however, should inhibit removal of most chain terminators other than AZTMP under physiological conditions. Our results suggest a mechanism for multidrug resistance conferred by the combination of finger insertion and AZT resistance mutations. In addition to a large increase in ATP-dependent removal of ddAMP from blocked DNA chains (up to 32-fold increase over WT RT, mostly conferred by the AZT resistance mutations), the combination of finger insertion and AZT resistance mutations conferred a greatly reduced sensitivity of the removal reaction to dNTPs (up to 23-fold-decreased sensitivity relative to WT RT), which would enable the mutant RT to remove nucleotide analogues such as ddAMP and d4TMP, in addition to AZTMP, more efficiently than WT RT under physiological conditions.
The 69S-XX mutations confer several different properties on HIV-1 RT; some of these depend on the nature of the amino acids inserted, while others do not. While the 69S-SS mutations conferred no increase in removal compared to WT RT, the 69S-SG and 69S-AG mutations increased removal by three- or fourfold. In combination with the AZT resistance mutations, 41L/215Y, the 69S-SG and 69S-AG mutations conferred up to a 21-fold increase in ATP-dependent removal. Since removal of ddAMP by WT RT or by several enzymes containing AZT resistance mutations is sensitive to inhibition by physiological concentrations of dNTPs (12, 25), an increase in removal might not by itself be expected to confer significant levels of resistance to ddI in vivo. However, the most dramatic in vitro effect of the 69S-XX finger insertion mutations, especially when present in combination with the 41L and 215Y mutations, was to decrease the sensitivity of the removal reaction to inhibition by dNTPs. The addition of mutations at 210W, 211K, and 214F had little additional effect, except in the case of the 41L/69S-SS/215Y combination, where the addition of the mutations at 210, 211, and 214 resulted in a fivefold decrease in dNTP inhibition. The decrease in sensitivity to dNTP inhibition was due to a reduced tendency of the mutant RTs to get trapped as a DEC together with primer-template and dNTP. The combination of finger insertion mutations and AZT resistance mutations would therefore confer both an increase in ATP-dependent removal of chain terminators from blocked DNA chains and decreased sensitivity of the removal reaction to dNTP inhibition. These effects could explain the cross-resistance to many chain terminators, such as AZT, stavudine (d4T), ddI, dideoxycytosine (ddC), and abacavir (14).
The severe defect in DNA chain elongation that we observed for the combination of 69S-XX mutations with the 41L/215Y mutations is consistent with impaired dNTP binding suggested by the decreased sensitivity to dNTP inhibition of the unblocking reaction. Previous studies (17, 19; P. R. Meyer et al., unpublished data) have shown small decreases in binding affinity between mutant RTs and primer-template and in affinity for dNTP substrates; however, the defect in DNA synthesis is likely to depend on the precise genetic makeup of the mutant enzymes. A detailed investigation of the catalytic parameters of DNA synthesis by these enzymes is not yet available.
The combination of 41L/215Y mutations and 69S-XX mutations is usually observed only in patients receiving multiple nucleoside analogue therapy (14, 37), and Lukashov et al. (18) have reported that a virus containing a 69S-XX/215Y combination was completely replaced by WT virus within 1 month after termination of therapy with d4T and 3′ thiacytidine (lamivudine). These results strongly suggest that viruses with these combinations of mutations are at a selective disadvantage in the absence of therapy, as would be predicted from the DNA chain elongation defects that we have observed. We have shown that addition of mutations 210W, 211K, and 214F to the 41L/69S-XX/215Y combination partially repairs the defect in DNA chain elongation, suggesting that these mutations should improve viral replication fitness. Quiñones-Mateu et al. (27) have recently reported that the removal of the 69S-SS mutations from the genome of a virus containing 69S-SS mutations in a background containing a variety of mutations, including AZT resistance mutations, produced a virus with reduced fitness. This result suggests that the contribution of the 69S-SS mutations to fitness depends on the mutational environment in the rest of the RT coding region.
How are the 69S-XX mutations generated? One possibility is that the 69S mutation increases the likelihood of the RT slipping during DNA synthesis after it has replicated the 69S codon. This could lead to rereplication of the 69S codon. If this occurred twice, an insertion of 6 bases would result. Since the codon at position 68 also encodes a serine, another possibility is that the inserted codons arise from a rereplication of both the 68 and 69 codons, as suggested by Boyer et al. (2). Published reports support the latter hypothesis. The sequence of the first and second inserted serine codons of six different HIV-1 viruses containing the 69S-SS insertion was identical in sequence to the codons at positions 68 and 69, respectively (5, 37). If the 69S-XX insertion mutations were indeed created by rereplication of codons 68 and 69, all finger insertion mutations should initially be 69S-SS. The two inserted serines would then be under selective pressure to change to any amino acids that would confer a growth advantage. The 69S-SS mutations conferred no detectable increase in ATP-dependent removal of chain terminators, while both the 69S-SG and the 69S-AG mutations did confer such an increase. The 69S-SS did confer reduced sensitivity to dNTP inhibition of the removal reaction, which could explain its selection; however, removal activity could be improved by subsequent mutation of the SS codons to codons for other amino acids. The in vitro results are consistent with viral drug susceptibility assays, since the 69S-SG mutations are associated with two- to sevenfold-greater resistance to AZT, ddI, ddC, and d4T than the 69S-SS mutations (17, 37). Taken together, these results lead to the prediction that the 69S-SS mutations may be replaced by 69S-SG or other amino acid insertions over time due to selective pressure. The predicted evolution has, in fact, been reported for several HIV-1-infected individuals (6, 14, 31).
Our results reinforce the importance of ATP-dependent removal of chain terminators in resistance to nucleoside analogues. Initially, it was shown that AZT resistance mutations confer an increase in ATP-dependent removal of AZTMP from blocked DNA chains (3, 16, 22), which would be a likely mechanism for AZT resistance. Since then, a role of ATP-dependent removal has also been suggested as a mechanism of resistance to d4T (17, 20, 23) and as a mechanism of multinucleoside resistance (4, 18; this report). Therefore, chain-terminating compounds that are refractory to removal and drugs that block the removal reaction could greatly increase the antiviral potency of nucleoside analogues and are important candidates for future drug research.
As this paper was about to be submitted, a paper by Boyer et al. (4) appeared showing that excision of various chain-terminating nucleotides was enhanced in RT containing 69S-XX insertion mutations even in the presence of relatively high levels of dNTPs. These authors showed that ternary complex stability was diminished in the mutant RT and proposed that enhanced excision of these chain terminators could be explained by destabilization of the ternary complex of RT with dNTP resulting from the amino acid insertions in the finger domain.
Acknowledgments
This work was supported by amFAR fellowship 70567-31-RF (to P.R.M.), a scholarship from Wenner-Gren Stiftelsen, a grant from the Swedish Research Council (to J.L.), and NIH grant AI39973 (to W.A.S.).
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. L., J. Lisziewicz, F. Lori, and S. H. Hughes. 1999. Analysis of amino insertion mutations in the fingers subdomain of HIV-1 reverse transcriptase. J. Mol. Biol. 286:995-1008. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 76:9143-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briones, C., A. Mas, G. Gómez-Mariano, C. Altisent, L. Menéndez-Arias, V. Soriano, and E. Domingo. 2000. Dynamics of dominance of a dipeptide insertion in reverse transcriptase of HIV-1 from patients subjected to prolonged therapy. Virus Res. 66:13-26. [DOI] [PubMed] [Google Scholar]
- 6.Briones, C., A. Mas, M. Pérez-Olmeda, C. Altisent, E. Domingo, and V. Soriano. 2001. Prevalence and genetic heterogeneity of the reverse transcriptase T69S-S-X insertion in pretreated HIV-infected patients. Intervirology 44:339-343. [DOI] [PubMed] [Google Scholar]
- 7.de Jong, J. J., J. Goudsmit, V. V. Lukashov, M. E. Hillebrand, E. Baan, R. Huismans, S. A. Danner, J. H. ten Veen, F. de Wolf, and S. Jurriaans. 1999. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 13:75-80. [DOI] [PubMed] [Google Scholar]
- 8.de Mendoza, C., O. Gallego, and V. Soriano. 2002. Mechanisms of resistance to antiretroviral drugs—clinical implications. AIDS Rev. 4:64-82. [PubMed] [Google Scholar]
- 9.Feng, J. Y., and K. S. Anderson. 1999. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry 38:9440-9448. [DOI] [PubMed] [Google Scholar]
- 10.Gao, W.-Y., R. Agbaria, J. S. Driscoll, and H. Mitsuya. 1994. Divergent anti-human immunodeficiency virus activity and anabolic phosphorylation of 2′,3′-dideoxynucleoside analogs in resting and activated human cells. J. Biol. Chem. 269:12633-12638. [PubMed] [Google Scholar]
- 11.Gao, W.-Y., A. Cara, R. C. Gallo, and F. Lori. 1993. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc. Natl. Acad. Sci. USA 90:8925-8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isel, C., C. Ehresmann, P. Walter, B. Ehresmann, and R. Marquet. 2001. The emergence of different resistance mechanisms toward nucleoside inhibitors is explained by the properties of the wild type HIV-1 reverse transcriptase. J. Biol. Chem. 276:48725-48732. [DOI] [PubMed] [Google Scholar]
- 13.Krebs, R., U. Immendörfer, S. H. Thrall, B. M. Wöhrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 14.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Sturmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob. Agents Chemother. 43:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larder, B. A., B. Chesebro, and D. D. Richman. 1990. Susceptibilities of zidovudine-susceptible and -resistant human immunodeficiency virus isolates to antiviral agents determined by using a quantitative plaque reduction assay. Antimicrob. Agents Chemother. 34:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennerstrand, J., D. K. Stammers, and B. A. Larder. 2001. Biochemical mechanism of human immunodeficiency virus type 1 reverse transcriptase resistance to stavudine. Antimicrob. Agents Chemother. 45:2144-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukashov, V. V., R. Huismans, M. F. Jebbink, S. A. Danner, R. J. de Boer, and J. Goudsmit. 2001. Selection by AZT and rapid replacement in the absence of drugs of HIV type 1 resistant to multiple nucleoside analogs. AIDS Res. Hum. Retrovir. 17:807-818. [DOI] [PubMed] [Google Scholar]
- 19.Mas, A., M. Parera, C. Briones, V. Soriano, M. A. Martínez, E. Domingo, and L. Menéndez-Arias. 2000. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 19:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mas, A., B. M. Vázquez-Álvarez, E. Domingo, and L. Menéndez-Arias. 2002. Multi-drug resistant HIV-1 reverse transcriptase: involvement of ribonucleotide-dependent phosphorolysis in cross-resistance to nucleoside analogue inhibitors. J. Mol. Biol. 323:181-197. [DOI] [PubMed] [Google Scholar]
- 21.Masquelier, B., E. Race, C. Tamalet, D. Descamps, J. Izopet, C. Buffet-Janvresse, A. Ruffault, A. S. Mohammed, J. Cottalorda, A. Schmuck, V. Calvez, E. Dam, H. Fleury, F. Brun-Vézinet, and The ANRS AC11 Resistance Study Group. 2001. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrob. Agents Chemother. 45:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, P. R., S. E. Matsuura, A. A. Tolun, I. Pfeifer, A. G. So, J. W. Mellors, and W. A. Scott. 2002. Effects of specific zidovudine resistance mutations and substrate structure on nucleotide-dependent primer unblocking by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naeger, L. K., N. A. Margot, and M. D. Miller. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 46:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quiñones-Mateu, M. E., M. Tadele, M. Parera, A. Mas, J. Weber, H. R. Rangel, B. Chakraborty, B. Clotet, E. Domingo, L. Menéndez-Arias, and M. A. Martínez. 2002. Insertions in the reverse transcriptase increase both drug resistance and viral fitness in a human immunodeficiency virus type 1 isolate harboring the multi-nucleoside reverse transcriptase inhibitor resistance 69 insertion complex mutation. J. Virol. 76:10546-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy, B., C. Beuneu, P. Roux, H. Buc, G. Lemaire, and M. Lepoivre. 1999. Simultaneous determination of pyrimidine and purine deoxyribonucleoside triphosphates using a polymerase assay. Anal. Biochem. 269:403-409. [DOI] [PubMed] [Google Scholar]
- 29.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 30.Sugiura, W., M. Matsuda, Z. Matsuda, H. Abumi, A. Okano, T. Oishi, K. Moriya, Y. Yamamoto, K. Fukutake, J. Mimaya, A. Ajisawa, M. Taki, K. Yamada, and Y. Nagai. 1999. Identification of insertion mutations in HIV-1 reverse transcriptase causing multiple drug resistance to nucleoside analogue reverse transcriptase inhibitors. J. Hum. Virol. 2:146-153. [PubMed] [Google Scholar]
- 31.Tamalet, C., N. Yahi, C. Tourrès, P. Colson, A.-M. Quinson, I. Poizot-Martin, C. Dhiver, and J. Fantini. 2000. Multidrug resistance genotypes (Insertions in the β3-β4 finger subdomain and MDR mutations) of HIV-1 reverse transcriptase from extensively treated patients: incidence and association with other resistance mutations. Virology 270:310-316. [DOI] [PubMed] [Google Scholar]
- 32.Tan, C.-K., J. Zhang, Z.-Y. Li, W. G. Tarpley, K. M. Downey, and A. G. So. 1991. Functional characterization of RNA-dependent DNA polymerase and RNase H activities of a recombinant HIV reverse transcriptase. Biochemistry 30:2651-2655. [DOI] [PubMed] [Google Scholar]
- 33.Terai, C., and D. A. Carson. 1991. Pyrimidine nucleotide and nucleic acid synthesis in human monocytes and macrophages. Exp. Cell Res. 193:375-381. [DOI] [PubMed] [Google Scholar]
- 34.Tong, W., C.-D. Lu, S. K. Sharma, S. Matsuura, A. G. So, and W. A. Scott. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36:5749-5757. [DOI] [PubMed] [Google Scholar]
- 35.Ueno, T., and H. Mitsuya. 1997. Comparative enzymatic study of HIV-1 reverse transcriptase resistant to 2′,3′-dideoxynucleotide analogs using the single-nucleotide incorporation assay. Biochemistry 36:1092-1099. [DOI] [PubMed] [Google Scholar]
- 36.van Vaerenbergh, K., K. van Laethem, J. Albert, C. A. B. Boucher, B. Clotet, M. Floridia, J. Gerstoft, B. Hejdeman, C. Nielsen, C. Pannecouque, L. Perrin, M. F. Pirillo, L. Ruiz, J.-C. Schmit, F. Schneider, A. Schoolmeester, R. Schuurman, H. J. Stellbrink, L. Stuyver, J. van Lunzen, B. van Remoortel, E. van Wijngaerden, S. Vella, M. Witvrouw, S. Yerly, E. de Clercq, J. Desmyter, and A.-M. Vandamme. 2000. Prevalence and characteristics of multinucleoside-resistant human immunodeficiency virus type 1 among European patients receiving combinations of nucleoside analogues. Antimicrob. Agents Chemother. 44:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winters, M. A., K. L. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, R. W. Shafer, D. A. Katzenstein, and T. C. Merigan. 1998. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Investig. 102:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]