Abstract
Integrase can insert retroviral DNA into almost any site in cellular DNA; however, target site preferences are noted in vitro and in vivo. We recently demonstrated that amino acid 119, in the α2 helix of the central domain of the human immunodeficiency virus type 1 integrase, affected the choice of nonviral target DNA sites. We have now extended these findings to the integrases of a nonprimate lentivirus and a more distantly related alpharetrovirus. We found that substitutions at the analogous positions in visna virus integrase and Rous sarcoma virus integrase changed the target site preferences in five assays that monitor insertion into nonviral DNA. Thus, the importance of this protein residue in the selection of nonviral target DNA sites is likely to be a general property of retroviral integrases. Moreover, this amino acid might be part of the cellular DNA binding site on integrase proteins.
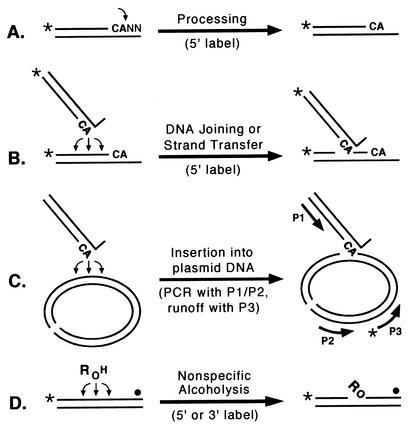
Integration of a DNA copy of the retroviral genome into cellular DNA is a critical step in the retrovirus life cycle and in the pathogenesis of retrovirus infections. Formation of an integrated provirus requires that the viral integrase act on its two DNA substrates with different levels of specificity. When preparing the viral DNA for integration, two nucleotides that follow conserved CA bases at the 3′ ends of each DNA strand are removed by integrase; this site-specific endonuclease reaction is referred to as processing. In contrast, integrase can insert the processed viral DNA ends into almost any site in cellular DNA; this second endonuclease reaction is referred to as DNA joining or strand transfer. Both of these actions can be modeled in vitro by using purified integrase and oligonucleotides that represent the viral DNA ends (Fig. 1A and B) (8, 17, 20). Although any accessible site in nonviral DNA can be used as the target for viral DNA insertion, preferences are noted in vitro and in vivo (7, 19, 32, 33, 39, 43). Characteristics of cellular DNA that affect the susceptibility of target sites were reviewed recently (6, 16, 19). For example, integration preferentially occurs into phosphodiester bonds at areas of DNA distortion on the outside of DNA bends; DNA sequence might play an additional minor role in susceptibility. Understanding how integrase recognizes these features of cellular DNA and identifying the part or parts of integrase responsible for any selectivity in choosing target sites are important for modeling integration, for developing methods of targeted gene delivery that are based on retroviral integration, and for designing a new class of antiretroviral agents that interfere with these enzyme-substrate interactions.
FIG. 1.
Integrase assays. The names of the assays are shown above the horizontal arrows, and the key aspects of the readouts (e.g., the position of the radiolabel or the pairing of PCR primers) are shown below the arrows. The CA bases near the 3′ ends of viral DNA are shown in boldface, the terminal nucleotides are indicated by NN, asterisks or solid circles indicate 32P groups, and P1, P2, and P3 refer to primers. (A) Processing, during which integrase makes a site-specific nick to form a labeled product two nucleotides shorter than the substrate. (B) DNA joining or strand transfer, during which integrase inserts processed viral DNA ends into any of various sites on either DNA strand to yield labeled products longer than the substrate. (C) Insertion, during which integrase inserts processed viral DNA ends into any of various sites in plasmid DNA (which is shown in its linear form); the joined products are then identified by PCR with primers P1 and P2, followed by a nested runoff reaction with 5′-radiolabeled primer P3. (D) Nonspecific alcoholysis, during which integrase uses certain nucleophilic alcohols (shown as ROH) to nick nonviral DNA at any of various sites; the labeled products will be simple oligonucleotides if the substrate is labeled at the 5′ end but will have attached R groups if the substrate is labeled near the 3′ end.
Studies using chimeric integrases involving human immunodeficiency virus type 1 (HIV-1), feline immunodeficiency virus, and visna virus indicate that the central domain of integrase plays a major role in selecting the target sites for insertion of viral DNA ends. This conclusion was based on results obtained with the standard oligonucleotide joining assay (Fig. 1B) (24) as well as with a PCR-based assay that monitors insertion of viral DNA ends into a longer plasmid DNA target (Fig. 1C) (2, 12, 25, 38). Moreover, the central domain of integrase was solely responsible for the selection of nonviral target sites when chimeric integrases used exogenous alcohols, rather than processed viral DNA ends, as the nucleophilic donor for nicking nonviral DNA (Fig. 1D) (22, 24, 25). In fact, the isolated central fragment of HIV-1 integrase (from residues 50 to 186) exhibited the same target site preferences in this nonspecific alcoholysis assay, which has many similarities to the joining reaction (25), as did the full-length 288-amino-acid HIV-1 protein. Thus, this region of approximately 140 amino acids is capable of binding and positioning nonviral DNA for nucleophilic attack.
Within the central domain of integrase, many amino acids can be replaced without affecting target site preferences (2, 13). However, we recently identified residue 119, the second amino acid in the α2 helix in the central domain of HIV-1 integrase, as strongly affecting the choice of nonviral target DNA sites (13). This residue was identified by a novel approach that involved screening a large set of patient-derived HIV-1 integrase variants for alterations in nonviral target site selection, comparing the sequences of proteins that exhibited similar target site preferences, and using this information to guide mutagenesis of a laboratory HIV-1 integrase (13). In fact, HIV-1 integrases with any of five different amino acids (Ser, Thr, Gly, Ala, or Lys) at position 119 exhibited five different patterns of target site selection in nonviral DNA. To test the hypothesis that these results are generalizable to other retroviral integrases, we have now assessed the role of the analogous protein residue in the integrases from a nonprimate lentivirus (visna virus) and a more distantly related alpharetrovirus (Rous sarcoma virus [RSV], formerly classified in the Oncovirinae retrovirus subfamily). Because the preferred sites of viral DNA insertion can differ depending on whether Mn2+ or Mg2+ is present during reactions (13), it was important that each of these divalent metal cations be used for these analyses.
To introduce amino acid substitutions into integrase, we used the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, Calif.) with pQE-30 plasmids (Qiagen, Chatsworth, Calif.) that encoded the wild-type integrases, as described previously (13, 41). The entire integrase-coding region for all proteins was confirmed by DNA sequencing, and proteins were purified from M15[pREP4] bacteria (Qiagen) (13). The purified proteins were tested under conditions known to optimize activity for visna virus and RSV integrase, including the use of oligonucleotides derived from the U3 end of viral DNA (21, 22, 42). Conditions for the standard oligonucleotide-based assays (Fig. 1A, B, and D) were as described previously (13) but included either 10 mM Mn2+ or 5 mM Mg2+. In the case of visna virus integrase and its derivatives, reactions that included Mg2+ were supplemented with 30% dimethyl sulfoxide (DMSO) because of our previous demonstration that this maneuver enhances the Mg2+-dependent activity of visna virus integrase (31). For the plasmid insertion assays (Fig. 1C) (26, 34), double-stranded 30/32-mers representing the preprocessed U3 end of viral DNA were incubated under the same reaction conditions but in the presence of 0.5 μg of φX174 DNA (Invitrogen, Carlsbad, Calif.) that had been linearized with SstII. The insertion reactions were stopped by fivefold dilution into 40 μl of 10 mM Tris-HCl (pH 8.0)-1 mM EDTA, and 7 μl was transferred to a tube for PCR. PCR was conducted in 25-μl reaction mixtures that contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.2 or 0.4 mM each deoxynucleoside triphosphate, 2 mM MgCl2, 1 U of Taq polymerase (Fisher, Pittsburgh, Pa.), and 15 pmol each of primer P1, which matched the viral donor DNA strand (for visna virus, 5′CAGGGTAGGCATTTGTTCTCTGTCCTGACA3′; for RSV, 5′AAGACTACAAGAGTATTGCATAAGACTACA3′), and primer P2, which was derived from φX174 (5′GGCGACCATTCAAAGGATAAACAT3′). The reaction mixtures underwent 35 cycles at 95°C for 45 s and 65°C for 3 min, with a final extension at 72°C for 10 min. Subsequently, 3 μl of each PCR was transferred to a 10-ul nested runoff reaction that contained 0.24 pmol of 5′ 32P-labeled φX174 primer P3 (5′GGCAGTCGGGAGGGTAGTCGG3′) and 1 U of Taq polymerase under the same buffer conditions as for PCR but for one cycle of 95°C for 2.5 min, 55°C for 4 min, and 72°C for 20 min. All reactions were analyzed by autoradiography after electrophoresis on denaturing polyacrylamide gels (20% gels for the processing, joining, and alcoholysis assays and 6% gels for the PCR-based insertion assay).
Effects of amino acid substitutions in visna virus integrase.
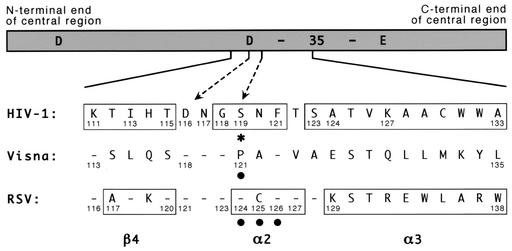
Visna virus integrase shares 89 (32%) of its 281 amino acids with HIV-1 integrase (22). Although there is no structural information available for the ovine protein, there is particularly strong sequence homology near the region that aligns with the α2 helix of HIV-1 integrase, with four of the six amino acids at or near this helix being identical (Fig. 2). We therefore made single-amino-acid substitutions at visna virus integrase position 121, which corresponds to HIV-1 integrase position 119, to examine the effects of these changes on target site selection. In particular, we changed the proline at position 121 to alanine (P121A) or glycine (P121G), both of which were included in the original set of five amino acids at HIV-1 position 119 that we had studied earlier.
FIG. 2.
Amino acid numbering for the central region of different integrases. A schematic of the central domain of HIV-1 integrase is shown with the locations of the 3 acidic residues of the active-site D,D-35-E motif indicated (the number 35 refers to the spacing between the last two conserved residues of this motif). Amino acid sequences of the indicated region from HIV-1, visna virus, and RSV are shown, with dashes indicating identity to the HIV-1 sequence. Numbers that mark landmarks, or are referred to in the text or in this legend, are provided. Boxes indicate the β4 strand, α2 helix, and α3 helix as defined in the original papers that described the HIV-1 and RSV crystal structures (4, 9). The dashed arrows highlight HIV-1 residues D116 and S119; the latter also is marked with an asterisk. Solid circles indicate amino acids in the visna virus and RSV proteins for which substitutions were made. The HIV-1 sequence shown and used for these studies is that of BH10, which is related to HXB2 but has an S instead of G at position 123 and a K instead of R at position 127 (35, 36). It differs from the NL4-3 strain used for the HIV-1 crystal structure described by Dyda et al. (9) by having an I rather than V at position 113 and an A rather than T at position 124. The source of the visna virus and RSV integrases have been described previously (41), and the sequences presented are invariant between known strains.
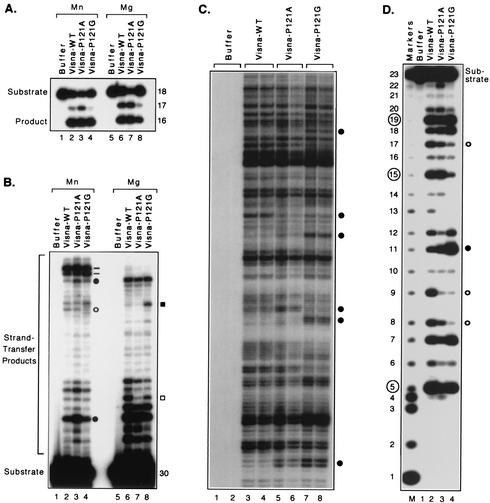
We first established that the P121A and P121G proteins, in similarity to the wild-type visna virus integrase, efficiently catalyzed processing by specifically removing two nucleotides from the 18-mer viral DNA substrate to create a 16-mer product, whether Mn2+ or Mg2+ was present during the reaction (Fig. 3A). To compare target site preferences during DNA joining, we used a longer, preprocessed substrate that represented the terminal 32 bp of one end of visna virus DNA but with the two nucleotides 3′ to the conserved CA already removed (Fig. 3B). With this substrate, the wild-type visna virus integrase created a distinct pattern of slower-migrating products in which certain bands are more prominent, reflecting preferential insertion into particular target sites. Such patterns are known to be independent of the extent or duration of the reaction (13), but as mentioned above, the relative usage of various target sites differed depending on which metal was present during the reaction (Fig. 3B, lanes 2 and 6). The mutant proteins also were active in this assay with either divalent cation. However, although several target sites were used with similar frequencies by the different proteins, the patterns created by the mutant proteins were distinguishable from that of the wild type and from each other at several positions. The strand-transfer patterns shown in Fig. 3B and the differences between them were highly reproducible as they were observed in multiple reactions and with different preparations of each enzyme. For example, in reactions with Mn2+, the P121A protein produced several bands darker than those seen in the lane with the wild-type enzyme, even though the intensities of other bands from the mutant protein were comparable to that of the wild-type pattern (Fig. 3B; compare lanes 2 and 3). In particular, quantitation by densitometric scanning showed that the two bands in lane 3 that are indicated by the filled circles in Fig. 3B had approximately two to three times greater intensity than those in the similar positions in lane 2, whereas the bands in lanes 2 and 3 at the position indicated by the open circle had equal intensities. Similarly, the strand-transfer pattern produced by the visna virus P121G protein differed from those of the wild-type and P121A proteins by the relative intensities of the two prominent bands that form a doublet near the top of the gel (Fig. 3B [dashes]); densitometry showed that the upper band of this doublet was two to three times more intense than the lower band in lanes 2 and 3 but that these bands had equal intensities in lane 4. Another distinguishing characteristic of the P121G pattern is the weaker band at the position indicated by the open circle (Fig. 3B, lane 4). Distinct patterns of strand-transfer products also were evident under Mg2+-dependent conditions. In particular, the strand-transfer pattern made by the P121A protein reproducibly had a lighter band (Fig. 3B, lane 7 [open square]) than did the pattern of the wild-type enzyme (lane 6), reflecting a sixfold difference in utilization of the target site that yields this band. Similarly, the P121G protein (Fig. 3B, lane 8) differed from the wild-type and P121A proteins by reproducibly creating a darker band at the position indicated by the filled square, reflecting a four- to sevenfold preference for this site compared with the other two proteins.
FIG. 3.
Target site selection by visna virus integrases. Autoradiograms of denaturing polyacrylamide gels are shown. (A) Processing assay. Double-stranded 18-mers derived from the U3 end of visna virus DNA were 5′ labeled on the strand that contains the conserved CA and incubated with protein buffer (lanes 1 and 5), wild-type (WT) visna virus integrase (lanes 2 and 6), or visna virus integrases that contained the indicated amino acid substitutions (lanes 3, 4, 7, and 8). Reactions 1 to 4 were conducted with 10 mM Mn2+, and reactions 5 to 8 contained 5 mM Mg2+ and 30% DMSO. Nucleotide sizes are shown at the right, and the 18-mer substrate and 16-mer product are indicated. (B) Joining assay. Double-stranded 30/32-mers representing the preprocessed U3 end of visna virus DNA were 5′ labeled on the strand that contains the conserved CA and incubated with the same proteins and conditions as described for panel A. The 30-mer substrate and strand-transfer products are indicated. Lanes 3 and 4 were loaded with 50% as many counts per minute as the other lanes. (C) Insertion assay. Double-stranded 30/32-mers representing the preprocessed U3 end of visna virus DNA were incubated with plasmid DNA and the joined products were revealed by PCR and a nested runoff reaction. Two PCR results are shown for each of the indicated proteins but only from reactions conducted with Mg2+; similar results, but different patterns, were obtained in reactions with Mn2+. (D) Nonspecific alcoholysis assay. Double-stranded 23-mers of nonviral sequence were 5′ labeled on one strand and incubated with the same proteins as described for panel A in reaction mixtures that included 10 mM Mn2+ and 40% glycerol. Lane M has sequence-specific markers (20); lengths in nucleotides are shown at the left, and the sizes of prominent products created by the wild-type visna virus integrase are circled. Bands indicated by dashes or open or filled circles or squares in panels B, C, and D are discussed in the text.
Because the oligonucleotides used in the standard integrase assay serve both as a viral DNA end and as a surrogate for cellular DNA (Fig. 1B), we also assessed target site preferences when the enzymes catalyzed insertion of viral DNA ends into a much larger plasmid DNA target (Fig. 1C). The joined products in these reactions were amplified by PCR using one primer that was identical to the viral donor strand (primer P1) (Fig. 1C) and another primer that was complementary to an arbitrary but fixed sequence in the target DNA (P2); the joined products were then revealed by a runoff reaction using a radiolabeled nested primer (P3). This assay produces distinct bands that correspond to each site of insertion, and the relative intensities of the bands reflect the utilization of the different sites (26, 34). The P121A and P121G proteins yielded distinct insertion patterns in this assay compared with the wild-type visna virus integrase and with each other, whether Mn2+ was used as the divalent cation (data not shown) or Mg2+ was used (Fig. 3C; some differences are marked by the circles). To highlight the reproducibility and distinctiveness of the target site patterns, the results of two independent PCR amplifications are shown for each enzyme in Fig. 3C; similar results were also obtained when the initial integration reactions were performed in duplicate. Importantly, no bands were evident when integration reactions were conducted with the protein storage buffer in place of the proteins (Fig. 3C, lanes 1 and 2). Thus, visna virus integrases with Pro, Ala, or Gly at position 121 yielded three different patterns of viral DNA insertion, whether joining occurred to other oligonucleotides or to longer DNA substrates and whether Mn2+ or Mg2+ was used as the metal cofactor.
To complement the above results, in which the enzymes used different target DNA molecules but similar nucleophilic donor molecules, we assessed the target site preferences of the proteins under conditions in which they use different nucleophilic donor molecules (Fig. 1D). As previously established (23), wild-type visna virus integrase uses various exogenous alcohols to nick nonviral DNA at every site except those near DNA ends. However, this integrase creates a signature pattern on the 23-mer substrate used in the assay such that the most prominent bands are detected at the 5, 15, and 19 positions (Fig. 3D, lane 2) (these bands are indicated by the circled numbers in the figure). When the P121A and P121G proteins were tested on this substrate, the preferred cleavage patterns were reproducibly different from each other and from that of the wild-type enzyme. For example, these proteins produced bands with greater intensity at the 11 position (Fig. 3D, lanes 3 and 4 [filled circle]) and lower intensities at the 8, 9, and 17 positions (open circles). The 5′-labeled products shown in Fig. 3D arose from the unique alcoholysis activity of integrase and not from contaminating nucleases, because attachment of the attacking nucleophilic alcohol to the appropriate sites was documented in assays that used a 3′-labeled substrate (Fig. 1D and data not shown) (23). The distinct target site preferences of the three visna virus integrases were further confirmed in similar assays that used a different nonviral DNA substrate that was 42 bp in length (data not shown). Although these assays were conducted only with Mn2+ because of the inefficiency of nonspecific alcoholysis with Mg2+, the results strengthen the conclusion that visna virus integrases with any of three amino acids at position 121 exhibit three different patterns of target site preference in nonviral DNA.
Effects of amino acid substitutions in RSV integrase.
To extend these results to a nonlentivirus, we assessed the target site preferences of avian retroviral integrases containing single-amino-acid substitutions. RSV integrase shares 72 (25%) of its 286 amino acids with HIV-1 integrase (1), and there is particularly strong sequence homology near the region that aligns with the α2 helix of HIV-1 integrase, with 8 of the 9 amino acids at or near this helix being identical (Fig. 2). By this alignment, HIV-1 integrase residue 119 corresponds to RSV integrase residue 124. However, unlike the situation with visna virus, crystal structures of the RSV integrase central domain are available (4). Surprisingly, some secondary structures are slightly misaligned relative to the sequence homology (Fig. 2) (1, 4, 9). In particular, HIV-1 residues 118 to 121 align by sequence with RSV amino acids 123 to 126 but RSV amino acids 124 to 127 comprise its α2 helix. Thus, HIV-1 integrase residue 119 also corresponds to RSV residue 125, because each is the second amino acid within this helix. We therefore made single-amino-acid substitutions at each of the RSV positions that correspond to HIV-1 position 119 by changing the serine at position 124 to alanine (S124A, similar to one of the substitutions made for HIV-1 and visna virus) and the cysteine at position 125 to asparagine (C125N [which makes this stretch of nine amino acids identical to those of HIV-1]). We also changed the phenylalanine at RSV position 126 (which is identical in the alignments of all three sequences) to a tyrosine (F126Y).
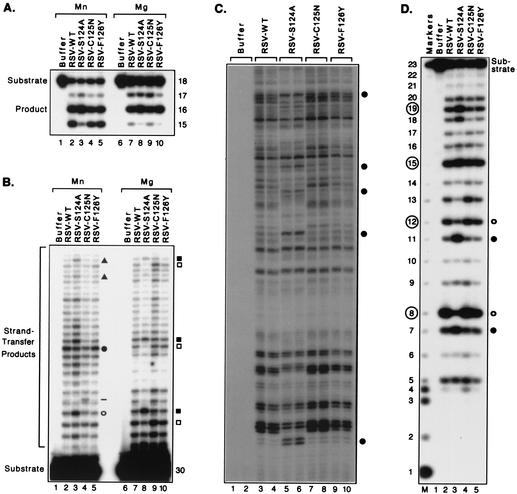
As previously established, RSV integrase removed two or three nucleotides from the 3′ ends of RSV viral DNA ends in the presence of Mn2+ (converting an 18-mer to a 16-mer or 15-mer, as shown in Fig. 4A, lane 2) but specifically removed the terminal two nucleotides in the presence of Mg2+ (Fig. 4A, lane 7) (41). We found that each of the new proteins with single-amino-acid substitutions (S124A, C125N, and F126Y) exhibited a level of processing activity comparable to that of the wild-type RSV integrase, whether reactions were conducted in the presence of Mn2+ or Mg2+ (Fig. 4A). To compare the patterns of joined DNA products, we used a longer, preprocessed substrate derived from the 32 bp at an RSV DNA end but lacking the two nucleotides after the CA (Fig. 4B). As expected, RSV integrase created distinct strand-transfer patterns in reactions with either of the two divalent cations (Fig. 4B, lanes 2 and 7). However, of the three singly substituted proteins, only the S124A protein created novel patterns that clearly differed from those of the wild-type protein. Under Mn2+-dependent conditions, the most prominent band created by the S124A protein (Fig. 4B, lane 3 [open circle]) differed from the most prominent band created by the wild-type, C125N, and F126Y proteins (Fig. 4B, lanes 2, 4, and 5 [filled circle]). This difference was underscored by comparing the intensities of the band at the filled circle to that at the open circle for each protein, which yielded a ratio of only 0.8 for the S124A protein but greater than 1.5 for the other three proteins. Differences in intensities of minor bands near the top of the lanes (Fig. 4B [filled triangles]) also reproducibly distinguished the S124A protein from the wild-type enzyme. In contrast, the C125N and F126Y mutants created patterns of joined products that were very similar to that of the wild-type RSV except for slight enhancement of one minor band with C125N (Fig. 4B, lane 4 [dash]). For reactions performed with Mg2+, differences for the S124A protein again were clearly evident, with certain bands having greater or lesser intensity than those seen with the wild-type pattern (Fig. 4B, lane 8 [some examples are indicated by the filled and open squares, respectively, which mark three sets of adjacent bands]). The distinctiveness of the S124A pattern is underscored by comparing the intensities of the slower-migrating band and the faster-migrating band in each of these sets of bands, yielding ratios of 2.4, 5.6, and 1.8 for the pairs near the top, middle, and bottom, respectively, of lane 8; in contrast, the relative intensities for these three pairs of bands were 0.4, 1.2, and 0.6, respectively, for the wild-type protein. The C125N and F126Y proteins had ratios for these pairs of bands very similar to those for the wild-type enzyme. In fact, the C125N and F126Y proteins (Fig. 4B, lanes 9 and 10) exhibited patterns of target site preferences that were superimposable with that of the wild-type enzyme. The strand-transfer patterns of the four RSV enzymes, both with Mn2+ and with Mg2+, were highly reproducible between multiple experiments.
FIG. 4.
Target site selection by RSV integrases. Details and reaction conditions were as described in the legend to Fig. 3, except that the viral DNA substrates were derived from RSV and reactions conducted with Mg2+ did not contain DMSO. (A) Processing assay. Reactions were incubated with protein buffer (lanes 1 and 6), wild-type RSV integrase (lanes 2 and 7), or RSV integrases with the indicated amino acid substitutions (lanes 3 to 5 and 8 to 10). (B) Joining assay. Lanes 4 and 10 were loaded with approximately twofold more counts per minute than the other lanes. (C) Insertion assay. Two PCR results are shown for each of the indicated proteins but only from reactions conducted with Mg2+; similar results, but different patterns, were obtained in reactions with Mn2+. (D) Nonspecific alcoholysis assay. The prominent products created by the wild-type RSV integrase are circled at the left. Bands indicated by the dash or geometric symbols in panels B, C, and D are discussed in the text.
We also assessed the patterns of target site selection when the RSV enzymes catalyzed insertion of viral DNA ends into a plasmid DNA target (Fig. 1C). As in the oligonucleotide assay, novel patterns of target site preferences for the S124A protein were clearly evident, either with Mn2+ (data not shown) or with Mg2+ (Fig. 4C, lanes 5 and 6 [some differences are highlighted by the circles]), whereas the C125N and F126Y mutants exhibited insertion patterns that closely matched that of the wild-type enzyme with either metal (e.g., Fig. 4C, lanes 7 to 10). Again, the results of two independent PCR amplifications are shown for each enzyme in Fig. 4C to indicate the reproducibility of this system and to highlight any differences between the patterns (similar results also were obtained when the integration reactions were performed in duplicate).
The S124A protein also exhibited altered target site preferences in the nonspecific alcoholysis assay. The signature pattern for wild-type RSV integrase includes frequent nicks at positions that are 8, 12, 15, and 19 nucleotides from the 5′ end of the 23-mer substrate used in this assay (Fig. 4D, lane number 2 [prominent bands are indicated by the circled numbers]). However, the S124A mutant reproducibly exhibited much less preference for the 8 and 12 positions (Fig. 4D, lane 3 [open circles]) but enhanced cleavage at the 7 and 11 positions (lane 3 [filled circles]). In contrast, the nicking patterns created by the C125N and F126Y proteins were indistinguishable from that of the wild-type enzyme (Fig. 4D, lanes 4 and 5). The 5′-labeled products in Fig. 4D were confirmed to have arisen from the unique alcoholysis activity of integrase by the use of a 3′-labeled substrate, as described earlier (data not shown). Similar results were found when the enzymes were tested on a 42-bp nonviral substrate; i.e., the S124A protein made a novel pattern and the C125N and F126Y proteins were indistinguishable from the wild-type enzyme (data not shown). Thus, the S124A substitution altered the patterns of target site selection in several assays (Fig. 4B [with either metal], Fig. 4C [with either metal], and Fig. 4D), whereas the C125N substitution only had a minor effect in one assay (Fig. 4B with Mn2+) and the F126Y protein was indistinguishable from the wild-type protein in all of the assays.
Implications.
In the absence of structural data from crystals of integrase with bound DNA (either viral or nonviral), other approaches are necessary to identify parts of the enzyme that interact with substrate DNA. Accumulating evidence supports a role for the central domain of integrase in interactions with cellular DNA (2, 10, 12, 14, 15, 24, 25, 38). In particular, we recently found that HIV-1 integrases with Ser, Thr, Gly, Ala, or Lys at position 119 exhibited five different patterns of target site selection in nonviral DNA. These results were obtained in oligonucleotide joining assays conducted with Mn2+ or Mg2+ and in the Mn2+-dependent nonspecific alcoholysis assay. It has subsequently been found that these proteins also exhibit five different patterns of viral DNA insertion into plasmid DNA in the PCR-based assay in the presence of both Mn2+ and Mg2+ (A. L. Harper and M. Katzman, unpublished data). HIV-1 integrase with a Pro substitution at position 119 also has been shown in oligonucleotide and plasmid insertion assays to have altered target site preferences (2). The new data demonstrated in the present report extend these findings to other integrases. In particular, visna virus integrases with Pro, Ala, or Gly at the analogous position in its central region exhibited three different patterns of target site preferences in nonviral DNA when tested in all five assays that monitor target site selection (i.e., joining of oligonucleotides in the presence of Mn2+ or Mg2+, insertion of DNA ends into plasmid DNA in the presence of Mn2+ or Mg2+, and nonspecific alcoholysis in the presence of Mn2+). Moreover, changing RSV integrase amino acid 124, which aligns by sequence with HIV-1 position 119, also altered the target site preferences in the five assays. Thus, this position in the central domain of various integrases either interacts with, or influences interactions with, nonviral DNA during catalysis. That these data were obtained with two lentiviruses and an alpharetrovirus suggests that this finding is a general property of retroviral integrases and similar enzymes.
The idea of the importance of the residue analogous to HIV-1 position 119 is strengthened by the finding that substitutions at RSV integrase amino acids 125 and 126 had minimal or no effect on the choice of nonviral target sites. RSV integrase with an F126A substitution was also shown to have wild-type target site preferences by another group (28). In fact, substitutions at more than one-quarter of the HIV-1 integrase central domain residues, including simultaneous replacement of most or all of the amino acids in the α3 or α4 helices, have been shown not to affect target site selection (2, 13). The significance of finding similar roles for HIV-1 and RSV amino acids that align by sequence (i.e., HIV-1 position 119 and RSV position 124), but not for amino acids that align by structure (i.e., HIV-1 position 119 and RSV position 125, each of which is the second amino acid in the α2 helix), is unclear. Perhaps other substitutions at RSV position 125 would reveal a greater influence on target site selection. Alternatively, it should be noted that the available structural information comes from crystals of only parts of these proteins. It is possible that structural elements in the full-length proteins, especially when bound to DNA and in the context of the milieu within a cell nucleus, more closely match the sequence alignment. Additional crystallographic information should help resolve this issue.
Our data show that integrases tolerate several amino acids at the position analogous to that of HIV-1 residue 119 without impairing the efficiency of their catalytic activities, even though their nonviral target site preferences are altered. Future studies should correlate these in vitro findings with the target site preferences of preintegration complexes that mediate integration in vivo. However, it is worth noting that natural HIV-1 isolates contain several amino acids at integrase position 119 (18, 27, 37, 40), and our preliminary data indicate that RSV virions with the S124A substitution replicate at levels similar to that of wild-type virus (Harper and Katzman, unpublished). Indeed, viral fitness is unlikely to be adversely affected by nonviral target site preferences that do not cause frequent integration into sites critical for cell survival.
It is significant that the results demonstrated in this report were obtained with both Mn2+ and Mg2+. Although all integrases exhibit greater activity in standard in vitro assays conducted with Mn2+ and most are inactive with Mg2+ (the RSV and related avian myeloblastosis virus integrases being exceptions), it is generally presumed that Mg2+ is the more relevant divalent metal cation within cells. Moreover, target site preferences in nonviral DNA depend upon which metal is available to integrase. In contrast, specificity for nicking viral DNA during processing generally does not change as a function of the divalent cation (31), although the avian integrases frequently nick between the conserved C and A as well as 3′ to the CA in reactions that use Mn2+. In this regard, it is interesting that the RSV S124A protein was reproducibly more specific at removing the terminal two nucleotides under Mn2+-dependent conditions than was the wild-type enzyme (e.g., Fig. 4A, lane 3 [note the greater selectivity for making the 16-mer rather than 15-mer product]). Thus, under certain conditions, side chains at this protein position can also influence the choice of target sites in viral DNA. This observation underscores the fact that processing and joining use related mechanisms, despite being distinct catalytic events that are separated in time and space (i.e., processing can occur in the cell cytoplasm and joining occurs in the nucleus). Perhaps the amino acid at this position influences the flexibility of the enzyme as a function of the divalent metal in the active site. Such a possibility is likely, given the relative flexibility of the RSV α2 helix (29) and the proximity of RSV residue 124 to residue 121, an active-site residue implicated in binding the metal cofactor (3, 5).
Although other parts of integrase might affect interactions with cellular DNA (12, 22, 24, 25, 38), our data show that one particular residue in the central region strongly affects nonviral target site selection. Whether this effect involves direct binding to nonviral DNA or is mediated indirectly is unknown. However, other data are consistent with the idea that this amino acid is part of the cellular DNA binding site on the integrase protein. In particular, cross-linking studies suggest that the nearby residue 117 in HIV-1 integrase contacts the phosphodiester backbone of cellular DNA (15). Moreover, analyses of crystal structures indicate that residue 119 is very close to the three conserved residues of the D,D-35-E active-site motif and that all four of these residues are on the surface of the protein (9, 11, 30). Thus, the interaction between HIV-1 integrase residue 119 and cellular DNA might provide a new target for antiretroviral agents.
Acknowledgments
This work was supported by Public Health Service grant R21 AI47216 from the National Institute of Allergy and Infectious Diseases and by the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Andrake, M. D., and A. M. Skalka. 1996. Retroviral integrase, putting the pieces together. J. Biol. Chem. 271:19633-19636. [DOI] [PubMed] [Google Scholar]
- 2.Appa, R. S., C.-G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and α helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 276:45848-45855. [DOI] [PubMed] [Google Scholar]
- 3.Bujacz, G., J. Alexandratos, A. Wlodawer, G. Merkel, M. Andrake, R. A. Katz, and A. M. Skalka. 1997. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 272:18161-18168. [DOI] [PubMed] [Google Scholar]
- 4.Bujacz, G., M. Jaskolski, J. Alexandratos, A. Wlodawer, G. Merkel, R. A. Katz, and A. M. Skalka. 1995. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 253:333-346. [DOI] [PubMed] [Google Scholar]
- 5.Bujacz, G., M. Jaskolski, J. Alexandratos, A. Wlodawer, G. Merkel, R. A. Katz, and A. M. Skalka. 1996. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure 4:89-96. [DOI] [PubMed] [Google Scholar]
- 6.Bushman, F. D. 2002. Integration site selection by lentiviruses: biology and possible control. Curr. Top. Microbiol. Immunol. 261:165-177. [DOI] [PubMed] [Google Scholar]
- 7.Carteau, S., C. Hoffmann, and F. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 9.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 10.Gerton, J. L., and P. O. Brown. 1997. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J. Biol. Chem. 272:25809-25815. [DOI] [PubMed] [Google Scholar]
- 11.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulaouic, H., and S. A. Chow. 1996. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 70:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper, A. L., L. M. Skinner, M. Sudol, and M. Katzman. 2001. Use of patient-derived human immunodeficiency virus type 1 integrases to identify a protein residue that affects target site selection. J. Virol. 75:7756-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuer, T. S., and P. O. Brown. 1997. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry 36:10655-10665. [DOI] [PubMed] [Google Scholar]
- 15.Heuer, T. S., and P. O. Brown. 1998. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry 37:6667-6678. [DOI] [PubMed] [Google Scholar]
- 16.Holmes-Son, M. L., R. S. Appa, and S. A. Chow. 2001. Molecular genetics and target site specificity of retroviral integration. Adv. Genet. 43:33-69. [DOI] [PubMed] [Google Scholar]
- 17.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 18.Katzman, M., A. L. Harper, M. Sudol, L. M. Skinner, and M. E. Eyster. 2001. Activity of natural HIV-1 integrases recovered from subjects with varied rates of disease progression. J. Acquir. Immune Defic. Syndr. 28:203-210. [DOI] [PubMed] [Google Scholar]
- 19.Katzman, M., and R. A. Katz. 1999. Substrate recognition by retroviral integrases. Adv. Virus Res. 52:371-395. [DOI] [PubMed] [Google Scholar]
- 20.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzman, M., and M. Sudol. 1994. In vitro activities of purified visna virus integrase. J. Virol. 68:3558-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzman, M., and M. Sudol. 1995. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 69:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzman, M., and M. Sudol. 1996. Nonspecific alcoholysis, a novel endonuclease activity of human immunodeficiency virus type 1 and other retroviral integrases. J. Virol. 70:2598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzman, M., and M. Sudol. 1998. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J. Virol. 72:1744-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katzman, M., M. Sudol, J. S. Pufnock, S. Zeto, and L. M. Skinner. 2000. Mapping target site selection for the non-specific nuclease activities of retroviral integrase. Virus Res. 66:87-100. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura, Y., Y. M. H. Lee, and J. M. Coffin. 1992. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc. Natl. Acad. Sci. USA 89:5532-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiken, C., F. McCutchan, B. Foley, J. W. Mellors, B. Hahn, J. Mullins, P. Marx, S. Wolinsky, and B. Korber. 1999. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 28.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. G. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubkowski, J., Z. Dauter, F. Yang, J. Alexandratos, G. Merkel, A. M. Skalka, and A. Wlodawer. 1999. Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant. Biochemistry 38:13512-13522. [DOI] [PubMed] [Google Scholar]
- 30.Maignan, S., J.-P. Guilloteau, Q. Zhou-Liu, C. Clément-Mella, and V. Mikol. 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 282:359-368. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, A. L., and M. Katzman. 2000. Subterminal viral DNA nucleotides as specific recognition signals for human immunodeficiency virus type 1 and visna virus integrases under magnesium-dependent conditions. J. Gen. Virol. 81:839-849. [DOI] [PubMed] [Google Scholar]
- 32.Pryciak, P. M., H.-P. Muller, and H. E. Varmus. 1995. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. USA 89:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pryciak, P. M., A. Sil, and H. E. Varmus. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11:291-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769-780. [DOI] [PubMed] [Google Scholar]
- 35.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R.-S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequence of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57-69. [DOI] [PubMed] [Google Scholar]
- 36.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. Petteway, Jr., M. L. Pearson, J. A. Lautenberger, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-283. [DOI] [PubMed] [Google Scholar]
- 37.Reinke, R., N. R. Steffen, and W. E. Robinson, Jr. 2001. Natural selection results in conservation of HIV-1 integrase activity despite sequence variability. AIDS 15:823-830. [DOI] [PubMed] [Google Scholar]
- 38.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 272:8361-8369. [DOI] [PubMed] [Google Scholar]
- 39.Shih, C.-C., J. S. Stoye, and J. M. Coffin. 1988. Highly preferred targets for retrovirus integration. Cell 53:531-537. [DOI] [PubMed] [Google Scholar]
- 40.Skinner, L. M., S. L. Lamers, J. C. Sanders, M. E. Eyster, M. M. Goodenow, and M. Katzman. 1998. Analysis of a large collection of natural HIV-1 integrase sequences, including those from long-term nonprogressors. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:99-110. [DOI] [PubMed] [Google Scholar]
- 41.Skinner, L. M., M. Sudol, A. L. Harper, and M. Katzman. 2001. Nucleophile selection for the endonuclease activities of human, ovine, and avian retroviral integrases. J. Biol. Chem. 276:114-124. [DOI] [PubMed] [Google Scholar]
- 42.Vora, A. C., M. L. Fitzgerald, and D. P. Grandgenett. 1990. Removal of 3′-OH-terminal nucleotides from blunt-ended long terminal repeat termini by the avian retrovirus integration protein. J. Virol. 64:5656-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Withers-Ward, E. S., Y. Kitamura, J. P. Barnes, and J. M. Coffin. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 8:1473-1487. [DOI] [PubMed] [Google Scholar]