Abstract
We describe a mutational analysis of the 3′ nontranslated RNA (3′NTR) signals required for replication of subgenomic hepatitis C virus (HCV) RNAs. A series of deletion mutants was constructed within the background of an HCV-N replicon that induces the expression of secreted alkaline phosphatase in order to examine the requirements for each of the three domains comprising the 3′NTR, namely, the highly conserved 3′ terminal 98-nucleotide (nt) segment (3′X), an upstream poly(U)-poly(UC) [poly(U/UC)] tract, and the variable region (VR) located at the 5′ end of the 3′NTR. Each of these domains was found to contribute to efficient replication of the viral RNA in transiently transfected hepatoma cells. Replication was not detected when any of the three putative stem-loop structures within the 3′X region were deleted. Similarly, complete deletion of the poly(U/UC) tract abolished replication. Replacement of a minimum of 50 to 62 nt of poly(U/UC) sequence was required for detectable RNA replication when the native sequence was restored in a stepwise fashion from its 3′ end. Lengthier poly(U/UC) sequences, and possibly pure homopolymeric poly(U) tracts, were associated with more efficient RNA amplification. Finally, while multiple deletion mutations were tolerated within VR, each led to a partial loss of replication capacity. The impaired replication capacity of the deletion mutants could not be explained by reduced translational activity or by decreased stability of the RNA, suggesting that each of these mutations may impair recognition of the RNA by the viral replicase during an early step in negative-strand RNA synthesis. The results indicate that the 3′-most 150 nt of the HCV-N genome [the 3′X region and the 3′ 52 nt of the poly(U/UC) tract] contain RNA signals that are essential for replication, while the remainder of the 3′NTR plays a facilitating role in replication but is not absolutely required.
Hepatitis C virus (HCV) is prevalent throughout the world. Most acute infections with this hepatotropic human virus lead to persistent infection and various forms of associated liver injury, including chronic hepatitis, cirrhosis, and even hepatocellular carcinoma (1, 38). Although recombinant alpha interferon, either alone or in combination with ribavirin, is widely used to treat HCV infection, the efficacy of these drugs in eliminating the infection is less than 50% overall (27, 31). The development of more effective drugs for the treatment of chronic hepatitis C is urgently awaited but has been hampered by the lack of cell culture systems that are fully permissive for viral replication and by the absence of small-animal models of the disease. Recently, however, the development of subgenomic HCV replicons that are capable of replication in human hepatoma (Huh7) cells has facilitated drug discovery efforts while providing new approaches to characterizing the molecular processes involved in replication of the viral RNA (28).
HCV is the sole member of the Hepacivirus genus within the family Flaviviridae. The genome of this virus is a single-stranded RNA molecule with positive-sense polarity and a length of ∼9.6 kb (3, 8, 30). The genome includes a 5′ nontranslated RNA (5′NTR) segment of ∼342 nucleotides (nt) and a 3′NTR that is somewhat shorter, about 225 nt in length. Between these terminal nontranslated RNA sequences, there is a single large open reading frame that encodes at least 10 proteins, including (in a 5′-to-3′ direction) several structural proteins (core, E1, E2, and p7) and at least six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The polypeptides spanning NS3 to NS5B in the polyprotein are required for replication of the viral RNA, and most likely all contribute to a membrane-bound, macromolecular replicase complex (3, 8). The NS5B protein is an RNA-dependent RNA polymerase (RdRp) and forms the catalytic core of this complex. The structure of this RdRp has been well characterized, and its enzymatic activities have been studied in detail by using recombinant proteins (10, 29, 33). NS5B is capable of primer-independent initiation of RNA synthesis on an RNA template. The details of this process are not known, however, and the mechanisms by which the RdRp (replicase complex) recognizes the terminal sequences of the viral RNA for proper and specific initiation of RNA transcription are controversial (15, 22, 34, 41).
Although different genotypes of HCV vary considerably in their nucleotide sequences, the sequences of the 5′ and 3′ NTRs are relatively well conserved (6, 24, 42, 45). This conservation is likely due to the need to maintain RNA structures that are required either for the initiation of translation of the viral polyprotein or for recognition of the positive- and negative-strand forms of the viral RNA by the replicase during the process of RNA replication. In support of the latter possibility, several recent studies suggest that both the 3′- and 5′-terminal RNA segments contain specific RNA sequences that are required for viral RNA replication, either in chimpanzees inoculated with synthetic genome-length RNA (25, 48) or in cell cultures supporting the replication of subgenomic viral RNAs (11, 12, 23).
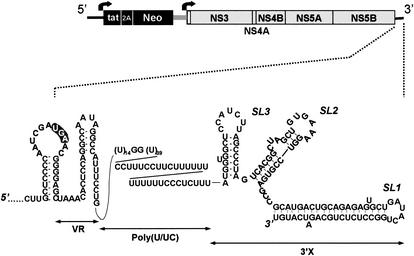
The 5′NTR of HCV folds into a complex structure containing four distinct domains (14). Domains II to IV comprise an internal ribosome entry site (IRES) that is responsible for cap-independent initiation of the translation of the viral polyprotein (36, 47), while domains I, II, and III contain overlapping replication signals that are required for optimal replication of the viral RNA in cultured cells (12, 23). The essential 5′ RNA replication signals in HCV thus appear to extend over a considerably longer segment of the 5′-terminal sequences than has been shown to be the case in pestiviruses and flaviviruses, other members of the family Flaviviridae (13, 21, 52). The 3′NTR also contains significant predicted RNA structure with three structurally distinct domains: a very highly conserved, 98-nt 3′ terminal segment (3′X) that putatively forms three stem-loop structures (designated SL1, SL2, and SL3, in the 3′-to-5′ direction), a lengthy poly(U)-poly(UC) [poly(U/UC)] tract, and an upstream variable region (VR) within which substantial sequence diversity is evident between different genotypes of HCV (Fig. 1) (17, 24, 45). The poly(U/UC) tract and downstream 3′X segment are both required for infectivity of synthetic, genome-length RNA inoculated into chimpanzees (25, 48). In addition, in a recently published paper, Friebe and Bartenschlager (11) have shown that the VR segment also contributes to efficient RNA replication by using subgenomic replicons derived from the Con1 strain of HCV.
FIG. 1.
Schematic of the organization of the dicistronic Ntat2ANeo(SI) replicon RNA (49), within which deletion mutations were created in the 3′NTR. The two open reading frames are represented by boxes, while the bold arrows indicate the translational initiation sites controlled by the HCV (left) and EMCV (right) IRES elements (see text). An expanded view of the 3′NTR sequence below the map of the replicon shows the secondary structure of the 3′NTR and the immediately upstream sequence, as suggested by Blight and Rice (5), with modifications suggested by MFOLD (version 3.1). The 3′NTR is comprised of three distinct domains, namely, VR, the poly(U/UC) tract, and the 3′X tail, which contains three putative stem-loop structures, SL1, SL2, and SL3.
We have carried out an analysis of the role of these various 3′NTR domains in replication of subgenomic RNAs derived from a second genotype 1b strain of HCV, HCV-N (2, 16). We show here the importance of conserved sequences within each of these different segments of the 3′NTR for replication of this RNA in cultured Huh7 cells.
MATERIALS AND METHODS
Cells.
En5-3 cells are a clonal cell line derived from Huh7 cells by stable transformation with the plasmid pLTR-SEAP, which expresses secreted alkaline phosphatase (SEAP) under control of the long terminal repeat (LTR) promoter of the human immunodeficiency virus (HIV) (49). These cells were cultured in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal calf serum, 2 μg (each) of blasticidin per milliliter (Invitrogen), penicillin, and streptomycin. Cell lines were passaged once or twice per week.
Plasmids.
The plasmid pNtat2ANeo(SI) contains a T7 transcriptional unit encoding a replication-competent, dicistronic subgenomic HCV RNA that expresses the HIV Tat protein from its upstream cistron (Fig. 1) (49). It was used to generate a series of mutants with deletions within the 3′NTR as follows. First, we excised the large ClaI fragment from pHCV-N (12) that spans most of the polyprotein coding region in this full-length clone. The resulting subclone, pHCV-NΔCla, retains the entire 3′NTR. The 3′NTR mutant ΔSL123 was constructed by PCR mutagenesis of this subclone by using primer pairs in which the forward primer contained the ClaI site that is located within the NS5B coding sequence; the reverse primer contained the XbaI site at the 3′ end of the HCV sequence. This PCR fragment was digested with ClaI and XbaI and inserted into pHCV-NΔCla. The resulting DNA was sequenced throughout the entire 3′NTR region to verify the deletion and then digested with ClaI and XbaI. The resulting fragment was ligated to pNtat2ANeo(SI) following digestion with ClaI and XbaI, and the sequence of the 3′NTR again was verified. A similar PCR mutagenesis strategy was used for construction of the Δ3′VR-23A, Δ3′VR-23B, Δ3′VR-14, and ΔSL1 mutants. QuikChange (Stratagene) site-directed mutagenesis was used for construction of the other 3′NTR mutants, including ΔSL2, ΔSL3, Δ[U/UC], [U/UC]-14, [U/UC]-30, [U/UC]-50, [U/UC]-62, and 3m5B, by using the subclone pHCV-NΔCla as the template. The resulting QuikChange products were verified by sequencing of the entire 3′NTR region, and the 3′NTR fragments were ligated directly into pNtat2ANeo(SI) at the ClaI and XbaI site.
The plasmid pU-BciVI is a modified version of pNtat2ANeo(SI) that contains a BciVI restriction endonuclease recognition sequence downstream of the HCV sequence, permitting the runoff transcription of synthetic RNAs with 3′ termini that are identical to the authentic HCV 3′ terminus. It was constructed by using QuikChange (Stratagene) mutagenesis by first introducing the BciVI site downstream of the HCV sequence in pHCV-NΔCla. Two BciVI sites, located in the Neo and NS4B coding regions of pNtat2ANeo(SI), were similarly removed by site-directed mutagenesis without changing the amino acid sequence of the encoded proteins. This involved a G-to-C substitution at nt 5657 of the HCV-N sequence. The final pU-BciVI construct was subsequently assembled by a three-way ligation of fragments containing each of these changes. The DNA sequences of the manipulated regions were verified in the final construct before its use in transcription reactions for replication assays.
ΔGDD mutants contain a 10-amino-acid deletion within the NS5B protein that removes the GDD RNA polymerase motif. This mutation, and single nucleotide substitutions described within the text, were constructed by QuikChange mutagenesis, as reported previously (16).
RNA transcription and transfection.
RNA was synthesized with T7 MEGAScript reagents (Ambion) after the plasmids were linearized with XbaI, except in the case of pU-BciVI, where BciVi was used to linearize the plasmid. Following treatment with RNase-free DNase to remove template DNA and precipitation of the RNA with lithium chloride, the RNA was transfected into En5-3 cells by electroporation. Briefly, 5 μg of RNA was mixed with 2 × 106 cells suspended in 500 μl of phosphate-buffered saline in a cuvette with a gap width of 0.2 cm (Bio-Rad). Electroporation was performed with two pulses of current delivered by the Gene Pulser II electroporation device (Bio-Rad), set at 1.5 kV, 25 uF, and maximum resistance. Cells were transferred to two wells of a six-well plate; the medium was changed at 24-h intervals, with the cells being washed twice with phosphate-buffered saline prior to the addition of fresh medium. Collected media samples were stored at 4°C until assayed for SEAP activity. Where noted, transfected cells were cultured in the presence of 250 μg of Geneticin/ml in an effort to force selection of G418-resistant cell colonies containing replicating HCV RNAs (16).
Alkaline phosphatase assay.
SEAP activity was measured in 20-μl aliquots of supernatant fluids from transfected cell cultures by using the Phospha-Light chemiluminescent reporter assay (Tropix) with the manufacturer's suggested protocol reduced 1/3 in scale. The luminescent signal was read by using a TD-20/20 luminometer (Turner Designs, Inc.). Since the culture medium was replaced every 24 h in the transient-transfection assays, the SEAP activity measured in these fluids reflected the daily production of SEAP by the cells.
Nucleotide sequence of replicating HCV RNAs.
HCV RNA was extracted from cells, converted to cDNA, and amplified by PCR as described previously (12). First-strand cDNA synthesis was carried out with Superscript II reverse transcriptase (Gibco-BRL), and PFU-Turbo DNA polymerase (Stratagene) was used for PCR amplification of the DNA. The amplified DNAs were subjected to direct sequencing by using an ABI 9600 automatic DNA sequencer. To obtain the sequence of the 3′-terminal region, first-strand cDNA synthesis was carried out with a primer (TTCTCCATCCTTCTAGCTCT) targeting nt 8901 to 8918 in the negative-strand RNA. The cDNA was purified by using the QIAquick PCR purification kit (Qiagen) to remove the nucleotides, and it was tailed with dCTP by using recombinant terminal deoxynucleotidyl transferase (Gibco-BRL). The poly(C)-tailed cDNA was amplified by PCR by using a primer targeting nt 8991 to 9023 in the HCV sequence and a second primer called Abridged-Xba (GCTCTAGAGCGGGIIGGGIIGGGIIG).
Real-time RT-PCR analysis of HCV RNA.
Quantitative RT-PCR assays were carried out by using TaqMan chemistry on a PRISM 7700 instrument (ABI). For detection and quantitation of HCV RNA, we used primers complementary to the 5′NTR region of HCV (44), with in vitro transcribed HCV RNA being included in the assays as a standard. Results were normalized to the estimated total RNA content of the sample, as determined by the abundance of cellular GAPDH mRNA detected in a similar real-time RT-PCR assay with reagents provided with Taqman GAPDH control reagents (human) (Applied Biosystems).
Northern analysis of HCV RNA.
G418-resistant, replicon-bearing cells were seeded into 10-cm-diameter culture dishes at a density of 5 × 105 cells/dish, with the RNA subsequently being harvested at day 4. Total cellular RNAs were extracted with TRizol reagent (Gibco-BRL) and quantified by spectrophotometry at 260 nm. A total of 30 μg of the total RNA extracted from each well was loaded onto a denaturing agarose-formaldehyde gel, subjected to electrophoresis, and transferred to positively charged Hybond-N+ nylon membranes (Amersham-Pharmacia Biotech) by using reagents provided with the NorthernMax kit (Ambion). RNAs were immobilized on the membranes by UV-cross-linking. The membrane was hybridized with a 32P-labeled antisense riboprobe complementary to the 3′ end of NS5B sequence (HCV nt 8990 to 9275), and the hybridized probe was detected by exposure to X-ray film.
RESULTS
HCV RNA replication in transiently transfected cells.
We characterized replication signals within the 3′NTR of HCV by using a modified subgenomic, dicistronic HCV replicon that expresses the heterologous HIV Tat protein (49). The autonomous replication of this replicon can be monitored in stably transformed or transiently transfected Huh7 cells expressing secreted alkaline phosphatase (SEAP) under control of the HIV LTR promoter (En5-3 cells) by measuring the SEAP activity secreted into the media bathing the cells. The details of this reporter replicon system have been described elsewhere (49). In brief, pNtat2ANeo is a plasmid with a T7 transcriptional unit containing a dicistronic, subgenomic HCV replicon in which the upstream cistron is under control of the HCV IRES and encodes a minipolyprotein consisting of Tat fused to the 2A proteinase of foot-and-mouth disease virus and neomycin phosphotransferase (Neo) sequences (Fig. 1). The 2A proteinase directs the self-cleavage of this minipolyprotein, resulting in the production of tat2A and Neo (49). The downstream cistron encodes the sequences from NS3 to NS5B of the HCV-N strain of HCV, under the translational control of the encephalomyocarditis virus (EMCV) IRES, and is followed by the full-length 3′NTR of HCV. The HCV-N nonstructural proteins support efficient replication of the dicistronic, subgenomic RNA due to the presence of a natural 4-amino-acid insertion within the NS5A protein (16).
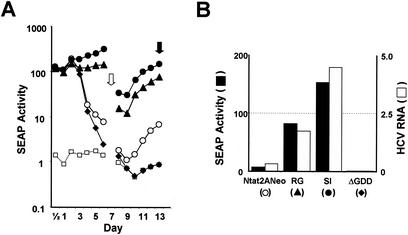
The transfection of synthetic RNA transcripts produced from pNtat2ANeo into En5-3 cells leads to expression of the tat2A fusion protein and results in an immediate and substantial induction of SEAP secretion that is evident as early as 12 h after transfection (Fig. 2A). SEAP production continues for approximately 3 days and then begins to decline, reaching a nadir around 8 to 9 days before once again beginning to increase. Control RNAs that are similar in sequence to Ntat2ANeo but that contain a deletion within the HCV NS5B sequence including the GDD polymerase motif (ΔGDD mutant) result in a similar, early stimulation of SEAP secretion. However, the SEAP expression following the transfection of these replication-incompetent RNAs does not follow a biphasic pattern, and there is no late increase in SEAP activity (Fig. 2A). Thus, the early expression of SEAP (during the first 48 to 72 h after transfection) reflects translation of the input RNAs, while the late increase in SEAP activity (at day 7 and beyond) is due to autonomous amplification of the RNA. This has been confirmed by direct assay of the viral RNA (49). Although these RNAs encode the Neo selectable marker, no G418 was included in the culture media in transient-transfection experiments such as that shown in Fig. 2A.
FIG. 2.
Transient transfection of Ntat2ANeo replicons into En5-3 cells. (A) SEAP activity in culture supernatants collected at 12- to 24-h intervals following electroporation of the cells with Ntat2ANeo (○), Ntat2ANeo(RG) (▴), Ntat2ANeo(SI) (•), and Ntat2ANeo(ΔGDD) (♦). See the text for descriptions of the R-to-G mutation in NS5B and the S-to-I mutation in NS5A. Also shown is the level of SEAP expression from normal En5-3 cells (□). (B) SEAP activity secreted into supernatant fluids (solid columns) and intracellular RNA abundance (stippled columns) 13 days after transfection with Ntat2ANeo and the indicated related mutated RNAs. RNA abundance was determined by quantitative TaqMan RT-PCR analysis (see Materials and Methods for details). Other studies have shown a close correlation between SEAP secretion and intracellular replicon abundance (49).
The replication of Ntat2ANeo RNA can be enhanced by the introduction of either of two mutations, an S-to-I substitution in NS5A or an R-to-G substitution in NS5B, that have been shown previously to enhance the replication of HCV RNAs in transfected Huh7 cells (4, 16, 26). These RNAs induce a similar, early level of SEAP expression following transfection into En5-3 cells, but there is no subsequent decline in SEAP activity due to the robust replication of the RNA provided by these adaptive mutations (Fig. 2A). Because Ntat2ANeo(SI) transcripts have reproducibly superior replication capacity in En5-3 cells than either Ntat2ANeo(RG) or unmodified Ntat2ANeo transcripts, Ntat2ANeo(SI) was the basis for the subsequent studies reported here. SEAP expression associated with the replication of this RNA can be clearly discriminated from that due to the translation of nonreplicating RNA containing the ΔGDD mutation within 4 to 5 days of transfection (Fig. 2A).
With each of the RNAs evaluated in the transient-transfection assay shown in Fig. 2A, intracellular RNA abundance correlated closely with the level of ongoing SEAP secretion 13 days following transfection (Fig. 2B). A close statistical correlation between SEAP expression and replicon RNA abundance has been demonstrated formally in other experiments reported elsewhere (49).
Reconstitution of the authentic HCV 3′-terminal nucleotide sequence in replicating RNAs.
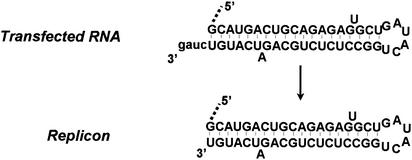
The RNAs transfected into En5-3 cells in the experiments shown in Fig. 2 were derived by runoff transcription from plasmid DNAs that had been digested with XbaI. These RNAs all contain an additional four nt downstream of the 3′-terminal residues of the HCV sequence, similar to replication-competent subgenomic HCV-N RNAs that have been described previously (16) (Fig. 3). To ascertain the fate of these additional 3′ nucleotides following transfection and replication of these RNAs, we determined the nucleotide sequence of the extreme 3′ end of the replicon RNAs present in stable G418-resistant cell clones selected following RNA transfection. This was accomplished as described in Materials and Methods. In multiple cell clones, these sequencing results indicated that the RNAs had regained the authentic 3′ terminus of the HCV genomic RNA by removal of these exogenous bases (Fig. 3). This indicates that the HCV replicase possesses a precise mechanism for selecting the terminal RNA sequence that is capable of distinguishing and excluding from amplification exogenous bases that are not part of the native genome.
FIG. 3.
3′ terminal sequence of the transfected Ntat2ANeo(SI) RNA obtained by runoff transcription of XbaI-digested plasmid DNAs (top) and the replicon RNA (bottom) present in stable, G418-resistant En5-3 cells obtained following transfection.
The conserved 3′X sequences are required for RNA replication in cultured cells.
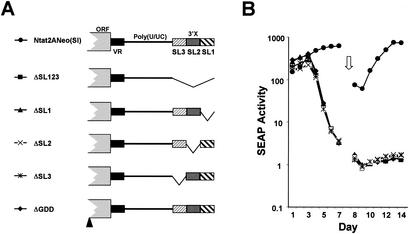
Yanagi et al. (48) demonstrated that the ability of synthetic HCV RNA to initiate infection in a naïve chimpanzee was destroyed by the deletion of sequences comprising either all three putative stem-loops within the 3′X segment of the 3′NTR (SL1, SL2, and SL3), SL1 alone, or a combination of SL2 and SL3. Freibe and Bartenschlager (11) also reported that these sequences were essential for amplification of replicons derived from the Con1 strain of HCV. To determine whether such deletions would cause a similar defect in the replication of HCV-N-derived subgenomic replicons in En5-3 cells, we assessed the replication capacity of Ntat2ANeo(SI) mutants in which each of the stem-loops was individually deleted (Fig. 4A, ΔSL1, ΔSL2, and ΔSL3) or in which all three stem-loops were deleted (Fig. 4A, ΔSL123). Since we have previously shown that RNA replication capacity correlated strongly with SEAP expression (Fig. 2B) (49), we monitored SEAP activity as a marker of the efficiency of replication of these RNAs in En5-3 cells. As shown in Fig. 4B, each of the mutant RNAs induced immediate and high levels of SEAP activity following transfection, indicating that the 3′NTR deletions had no profound effect on translation directed by the HCV IRES. However, none of the RNAs with deletions within the 3′X region elicited a subsequent pattern of SEAP expression that was consistent with amplification of the viral RNA. We conclude that the sequences contributing to each of these three putative stem-loops play a vital role in RNA replication.
FIG. 4.
Transient transfection of En5-3 cells with Ntat2ANeo(SI) and related mutants containing deletions within the 3′X segment located at the 3′ end of the 3′NTR. (A) Schematic showing locations of deletion mutations within the 3′X segment. (B) SEAP activity present in supernatant culture fluids collected at 24-h intervals following transfection of En5-3 cells with Ntat2ANeo(SI) (•) or ΔSL123 (▪), in which the entire 3′X segment was deleted, or with ΔSL1 (▴), ΔSL2 (×), or ΔSL3 (*), in which individual stem-loops were removed. Results following transfection of the replication-incompetent ΔGDD mutant (⧫) are also shown.
Permissive and nonpermissive point mutations within SL2 and SL3.
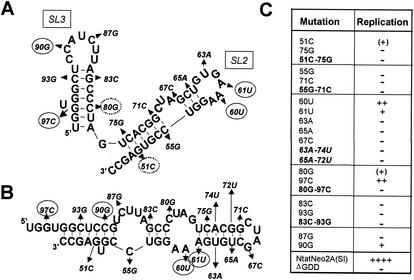
Computerized predictions of the structures present within the 3′X domain were obtained with use of the MFOLD program. Multiple possible folds of this RNA segment consistently include the 3′-most stem-loop structure, SL1, but indicate that there are several alternative folds possible for the sequences contributing to SL2 and SL3. As noted previously (5), these alternative folds are predicted to have free energies approximately equivalent to that shown in Fig. 1. Although the results of nuclease and chemical mapping of RNA secondary structure were consistent with the existence of SL2 and SL3 (5), a high degree of sequence conservation within this segment of the viral RNA has precluded any determination of the favored fold based on the presence of covariant nucleotide substitutions. In an effort to determine more precisely the structure in this region and whether any degree of sequence variation would be tolerated, we constructed a series of mutants with single point substitutions at different positions within the SL2 and SL3 sequences (Fig. 5A). Most of these point mutations had a lethal effect on replication. While four mutants were capable of replication in the transient-transfection assay, each produced a substantially reduced level of SEAP activity compared with Ntat2ANeo(SI) RNA (Fig. 5C). Notably, three of these four mutations (60A, 61A, and 90G) were within the predicted single-stranded loop segments of SL2 and SL3, while the remaining permissive mutation (97C) prevented formation of a Watson-Crick base pair adjacent to an unpaired U residue within the stem of SL3 (Fig. 5A).
FIG. 5.
Point mutations introduced into the sequences of SL2 and SL3 and their effects on replication of the subgenomic RNA. (A) MFOLD (version 3.1) prediction of the structures of SL2 and SL3, representing a modification of the structure proposed by Blight and Rice (5). The locations and natures of the point mutations introduced into Ntat2ANeo(SI) are indicated; those with evidence of replication in the transient-transfection assay are encircled with a solid line, while pseudolethal mutations that produced small numbers of G418-resistant mutants but failed to induce SEAP secretion in the transient-transfection assay are encircled with a dotted line. The mutations are labeled according to the base position from the 3′ end of the genome. The predicted free energy of the 3′X structure containing the stem-loops shown was −44.6 kcal. (B) An alternative fold of the SL1 and SL2 sequences, showing the locations of point mutations indicated in panel A. The predicted free energy of 3′X containing this structure was −46.4 kcal. (C) Tabular summary of transient replication assays carried out with the indicated mutants. Mutant RNAs with dual, potentially compensatory mutations are indicated in boldface type; those that are compensatory in the structure shown in panel A are in boldface type, while those that are compensatory in the structure shown in panel B are in italicized boldface type. ++, +, and ++++, SEAP activities of <25%, <10%, and 100% of that achieved following transfection of the parent Ntat2ANeo(SI) RNA, respectively; −, SEAP activity indistinguishable from that of the ΔGDD mutant RNA; (+), pseudolethal mutation that failed to replicate in the transient assay but remained capable of transducing the selection of G418-resistant cell colonies with reversion of the sequence to the wild-type 51G and 80C sequences.
To ascertain whether lethal point mutations within the predicted duplex stems blocked replication by virtue of a destabilizing effect on duplex formation, we constructed several additional mutants with paired, compensatory mutations that were predicted to restore to the stems of SL2 (mutants 51C-75G and 55G-71C) and SL3 (mutants 80G-97C and 83C-93G) base pairing that had been eliminated by the single mutations. Interestingly, none of these dual mutants were viable in the replication assay. We also examined paired, compensatory mutations (63A-74U and 65A-72U) that restored the predicted base pairing within an alternative RNA fold with approximately equal free energy (Fig. 5B). These compensatory mutations also failed to rescue replication capacity, however, indicating that the lethal effect of the single base substitutions, 63A and 65A, was not due to disruption of the predicted base pairing.
The results of these experiments thus provide no proof for the existence of SL2 or SL3, but they do indicate that there are stringent requirements for the sequence in this region and that even single point mutations are tolerated poorly within this segment of the viral RNA. These results are consistent with a BLASTN search of the GenBank database for this sequence that identified 80 HCV sequences. Only four differed from the query NtatNeo2A(SI) sequence (data not shown). In each case, the difference was due to a single base substitution, none occurring at sites we examined by mutagenesis.
As might be expected from the results of the transient-transfection assays (Fig. 5C), it was possible to select G418-resistant colonies from cells transfected with the 60U, 61U, 90G, and 97C mutants. Although we did not rigorously quantify the efficiency of colony formation, the number of surviving cell colonies was greatest for 60U (∼103 colonies/106 cells), followed by 97C, 61U, and 90G (∼10 colonies/106 cells). When the 3′NTR sequences of the RNAs replicating in these G418-resistant colonies were examined, the point mutations were found to be present and not accompanied by other mutations. Despite a lack of evidence for replication in the transient- transfection assay, it was also possible to select G418-resistant cell colonies from cells transfected with 51C (one colony/106 cells) and 80G (three colonies/106 cells) (Fig. 5C). Analysis of the sequence of the replicon RNA in these G418-resistant cells showed that the introduced mutations had reverted to the parental sequence. This indicates that the 51C and 80G mutations are pseudolethal and allow only sufficient replication for reversion of the mutated base. The other mutants shown in Fig. 5 did not give rise to G418-resistant colonies, suggesting that these mutations were completely lethal to replication.
Poly(U/UC) tract requirements for RNA replication.
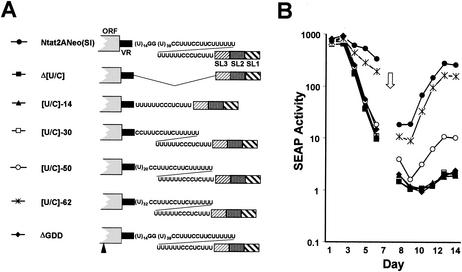
We next examined the polypyrimidine tract that exists between the nonconserved VR region and the highly conserved 3′X tail of the 3′NTR (Fig. 1). Yanagi et al. (48) found that the deletion of this sequence destroyed the infectivity of synthetic RNA derived from the genotype 1a, Hutchinson strain of HCV in chimpanzees. Freibe and Bartenschlager (11) also found this sequence to be essential for replication of genotype 1b replicons. When the poly(U/UC) tract present in the Con1 sequence was replaced with variable lengths of pure, homopolymeric poly(U) sequence, they found that a minimal poly(U) tract of 26 nt was required for amplification of replicons. We found that the complete deletion of the poly(U/UC) tract from Ntat2ANeo(SI) (Fig. 6A, Δ[U/C] mutant) similarly abrogated the ability of Ntat2ANeo(SI) to replicate in En5-3 cells, as indicated by a lack of sustained SEAP expression following transfection of the RNA (Fig. 6B). Again, the deletion did not result in defective IRES-directed translation, as the amount of SEAP expressed immediately after transfection was similar to that observed with the replication-competent, parental Ntat2ANeo(SI) RNA. These results confirm that the 3′ untranslated region poly(U/UC) tract is indeed critically important for HCV RNA replication.
FIG. 6.
Transient transfection of En5-3 cells with Ntat2ANeo(SI) and related mutants containing deletions within the poly(U/UC) tract. (A) Schematic showing locations of deletion mutations within the poly(U/UC) tract. (B) SEAP activity present in supernatant culture fluids collected at 24-h intervals following transfection of En5-3 cells with Ntat2ANeo(SI) (•) or Δ[U/UC] (▪), in which the entire poly(U/UC) segment was deleted, or with [U/UC]-14 (▴), [U/UC]-30 (□), [U/UC]-50 (○), or [U/UC]-62 (*), in which various lengths of the poly(U/UC) segment were restored from the 3′ end of the deletion. Results following transfection of the replication-incompetent ΔGDD mutant (♦) are also shown.
To gain an understanding of the minimal length of polypyrimidine sequence required for replication of the viral RNA, we constructed a series of additional mutants in which the poly(U/UC) tract deleted in Δ[U/C] was replaced in a stepwise fashion from its 3′ end (Fig. 6A). The parental RNA, Ntat2ANeo(SI) has an 85-nt-long poly(U/UC) tract with the sequence (U)14GG(U)39(C)2(U)3(C)2(U)2C(U)12(C)3UC(U)3. Replacing this sequence with the segment representing its 3′-most 14 nt, (U)6(C)3UC(U)3, or the 3′-most 30 nucleotides, (C)2(U)3(C)2(U)2C(U)12(C)3UC(U)3 (Fig. 6A, mutants [U/C]-14 and [U/C]-30, respectively) did not restore replication competence (Fig. 6B). Thus, the 3′ 30 nt of the 85-nt polypyrimidine tract in Ntat2ANeo(SI) are insufficient to support RNA replication. In contrast, the restitution of a 20-nt pure poly(U) tract immediately upstream of the mixed polypyrimidine tract in [U/C]-30 (Fig. 6B, mutant [U/C]-50) did restore replication competence to the RNA, although the level of SEAP expression (and thus the imputed abundance of viral RNA) was less than 10% of that observed with the parental RNA. Increasing the pure poly(U) tract to 32 nt, and the total length of the poly(U/UC) tract to 62 nt (Fig. 6A, [U/C]-62 mutant), resulted in a level of SEAP expression that was close to that of the parental RNA (Fig. 6B). These results suggest that the minimal length of the polypyrimidine tract required for efficient replication of this RNA is on the order of 50 to 62 nt, although slightly greater replication competence is seen with the 85-nt poly(U/UC) tract in the parental RNA. While the results shown in Fig. 6 can also be interpreted as indicating that a pure poly(U) segment is important for RNA replication, additional mutants will need to be studied in order to distinguish this possibility from a simple requirement for overall length of the poly(U/UC) tract (see Discussion).
Role of the VR segment in RNA replication.
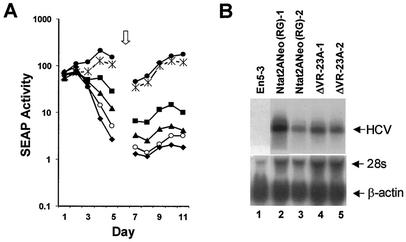
The VR segment within the 3′NTR of the genotype 1b Ntat2ANeo(SI) RNA is 40 nt in length. Its sequence differs at 11 positions from the VR segment of the infectious molecular clone of the genotype 1a HCV-H virus in which deletion of the 5′ 24 nt of the VR segment did not abrogate the infectivity of synthetic HCV RNA in the chimpanzee model (48). To assess the effect of a comparable deletion on the replication of Ntat2ANeo(SI) RNA in Huh7 cells, we removed the 5′ 23 nt of the VR segment (Fig. 7A, mutant ΔVR-23A). Although this deletion destroyed two potential stem-loop structures within the VR (Fig. 7B), this deletion mutant was capable of replication in transfected En5-3 cells (Fig. 8A). However, its replication was very impaired compared with that of the parental replicon, as it induced SEAP expression at levels that were less than 5% of that induced by the parental RNA. Nonetheless, we were able to select G418-resistant cell colonies following transfection of En5-3 cells with this RNA. Northern analysis demonstrated that the abundance of replicon RNA in these cells was similar to that in colonies selected following transfection with RNA containing the intact 3′NTR, Ntat2ANeo(RG), although we have noted substantial variation in the RNA abundance in such cells, as shown in Fig. 8B.
FIG. 7.
Deletion mutations within the VR segment of the 3′NTR. (A) Alignment of the sequences of the VR segment in Ntat2ANeo(SI) and related VR deletion mutants. (B) Impact of deletions in the VR segment on predicted secondary structure in this segment of the 3′NTR. Nucleotide substitutions in the NS5B-coding sequence upstream of the VR segment in the 3m5B mutant are indicated by underlining. The symbols shown under each mutant are as described in the legend to Fig. 8A. The structures are those predicted by MFOLD for the VR and upstream NS5B coding-region segment shown.
FIG. 8.
(A) Transient transfection of En5-3 cells with Ntat2ANeo(SI) and related mutants containing deletions within the VR segment or multiple point mutations in the upstream NS5B-coding region (see Fig. 7). SEAP activity was measured in supernatant culture fluids collected at 24-h intervals following transfection of En5-3 cells with Ntat2ANeo(SI) (•) or with ΔVR-23A (○), ΔVR-23B (▴), or ΔVR-14 (▪), which contain deletions in the VR segment, or with 3m5B (*), in which multiple point mutations destabilize a predicted stem-loop involving the polyprotein termination codon. Results following transfection of the replication-incompetent ΔGDD mutant (♦) are also shown. (B) Northern analysis of HVC RNA present in G418-resistant cells lines selected following transfection with replicon RNAs. Lane 1, normal En5-3 cells; lanes 2 and 3, cell lines selected following transfection with the parental Ntat2ANeo(RG) RNA; lanes 4 and 5, two independent cell lines selected following transfection with the ΔVR-23A mutant (see Fig. 7). The lower panel shows hybridization with a probe specific for β-actin as a loading control.
Sequencing of the replicon RNAs present in G418-resistant cell lines selected following transfection with the ΔVR-23A mutant demonstrated retention of the 23-nt deletion (data not shown). Yanagi et al. (48) found that the replication of the genotype 1a VR deletion mutant was accompanied by a change in the termination codon from UGA to UAA as early as 2 weeks following the inoculation of the chimpanzee with RNA. However, we found no changes in the sequence of the mutated 3′NTR or within the immediately adjacent upstream sequence in ΔVR-23A RNAs recovered from stable G418-resistant cell clones. These data suggest that the VR sequences are not essential for replication but that they do play an accessory role in promoting the efficiency of replication. Thus, the VR segment is distinct from the 3′X segment, in which deletion cannot be made without complete loss of replication capacity (Fig. 4).
We created several additional mutations within the VR in an effort to better characterize the requirements for sequence in this region and possibly to define the reasons for the significant loss of replication capacity in the ΔVR-23A mutant. In the mutant ΔVR-14, we restored the 9 nt at the 5′ end of the deletion in the ΔVR-23A mutant (Fig. 7A), thereby reconstituting the small stem-loop structure that is predicted to contain the polyprotein termination codon within its loop (Fig. 7B). Although the base-paired nucleotides are different in the genotype 1a HCV-H virus, the related sequence in this virus also has the capacity to form a very similar stem-loop structure in this region, albeit one with a smaller loop segment (48). As shown in Fig. 8A, ΔVR-14 was clearly more active in inducing SEAP secretion (RNA replication) than the ΔVR-23A mutant, although it remained less active than the parental sequence. This could reflect either the restoration of the stem-loop or the presence of a lengthier (31- versus 17-nt) VR segment in ΔVR-14. To address this question, we constructed an additional mutant in which the deletion in ΔVR-14 was extended downstream to include a total of 23 nt (Fig. 7A, mutant ΔVR-23B). This mutant retained the stem-loop structure that contains the termination codon (Fig. 7B), but it was intermediate in its replication capacity, which was somewhat greater than that of ΔVR-23A but significantly less than that of ΔVR-14, as indicated by the level of SEAP expression (Fig. 8A). These results are consistent with, but not proof of, a potential role for this stem-loop in RNA replication. We further tested this possibility by introducing three silent nucleotide substitutions within the 3′-terminal NS5B coding region that effectively disrupt the stem-loop, shortening the stem and altering the predicted loop sequence (Fig. 7B, mutant 3m5B). Since all three substitutions in this mutant are upstream of the 3′NTR, the VR sequence was unchanged from that of the parental Ntat2ANeo(SI) RNA. As shown in Fig. 8, these nucleotide substitutions caused only a slight decrease in the efficiency of RNA replication compared to that of the parent RNA. Thus, while a minor role for this stem-loop in promoting the efficiency of replication cannot be excluded, the structure does not appear to be an important determinant of RNA replication.
Role of 3′NTR sequences in promoting IRES-directed translation and RNA stability.
Since Ito et al. (19) suggested that sequences within the HCV 3′NTR may positively influence the translation-initiating activity of an upstream IRES, it was important to consider whether any of the deletions in the 3′NTR could have impaired the translational activity of the HCV IRES. As pointed out above, none of the 3′NTR deletions that significantly reduced replication resulted in consistent decreases in the levels of SEAP expressed within the first 24 to 48 h after transfection of En5-3 cells (Fig. 4B, 6B, and 8A). The SEAP that is produced during this period of time reflects translation of the input RNA and not replication, since there is no difference between the amount of SEAP produced by Ntat2ANeo and a related NS5B ΔGDD mutant at these early time points (Fig. 2A). Because SEAP is produced as a result of the HIV Tat protein's action on the LTR promoter in the transfected En5-3 cells, and since the translation of the Tat polypeptide is under control of the HCV IRES in these RNAs, the absence of significant differences in SEAP activity 24 to 48 h after transfection indicates that these 3′NTR deletions do not result in a reduction of the activity of the upstream HCV IRES.
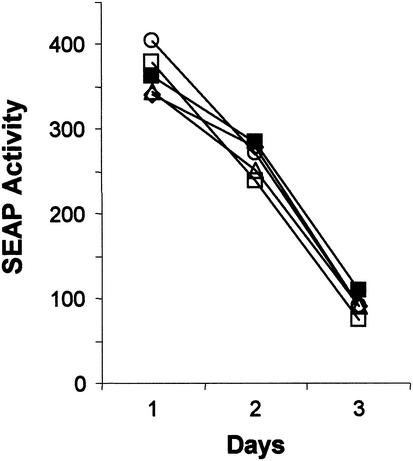
An additional possibility is that the decreased replication capacity observed with some 3′NTR deletion mutants is due to a reduction in the stability of the RNA within the cell. This might be expected if the highly structured 3′ end of the genome served to protect the RNA from nuclease attack. Were this the case, deletions in the 3′NTR should cause an accelerated rate of decline in the quantity of SEAP produced over successive 24-h periods following transfection of a replication-incompetent RNA (see the ΔGDD mutant, Fig. 2A). To examine this possibility more closely, we created ΔGGD mutations within the NS5B sequences of several 3′NTR mutants with impaired replication phenotypes. These dual mutants were thus completely replication incompetent. We then carefully assessed the rate of decay in the 24-h SEAP expression rate, following transfection of these RNAs into En5-3 cells (Fig. 9). These results were compared with the kinetics of SEAP expression following transfection of cells with a similar NS5B ΔGDD mutant RNA derived from pU-BciVi, a modified version of pNtat2ANeo(SI) in which a unique 3′ BciVi recognition site placed downstream of the HCV sequence allows the production of synthetic RNAs with authentic 3′ termini. As shown in Fig. 9, ΔGDD mutants constructed from Δ[U/C]-50 or Δ[U/C]-62 demonstrated a pattern of SEAP expression similar to that of U-BciVi-ΔGDD, even though their cognate replication-competent RNAs were very different in their abilities to undergo RNA replication (Δ[U/C]-50 was markedly impaired in replication, as shown in Fig. 6). Similarly, ΔGDD mutants constructed from ΔVR-14 and 3m5B (Fig. 6) also showed parallel SEAP expression kinetics following transfection into En5-3 cells, despite substantial differences in the replication competence of the parent mutants and a significant defect in the ability of ΔVR-14 to replicate. These results provide strong evidence that the defect in the ability of the original mutants to replicate is due to the removal of a structure or sequence within the 3′NTR that is essential for RNA replication and not to decreased stability of the RNA following transfection.
FIG. 9.
SEAP activity present in supernatant culture fluids collected at 24-h intervals following transfection of En5-3 cells with ΔGDD mutants created within the background of U-BciVI (▪) [identical to Ntat2ANeo(SI) but with the authentic 3′ terminus of HCV], [U/UC]-62 (□), [U/UC]-50 (⋄) (see Fig. 6), ΔVR-14 (○), or 3m5B (▵) (see Fig. 7).
DISCUSSION
The experiments reported here confirm the importance of each of the three major domains of the HCV 3′NTR to viral RNA replication. Our results are consistent with those published recently by Friebe and Bartenschlager (11), who used subgenomic replicons derived from the Con1 strain of HCV to characterize the 3′NTR sequences required for replication of subgenomic viral RNAs in Huh7 cells. We analyzed the effects of deletions within the 3′NTR by using an independent HCV-N replicon with a sequence that is distinct from that of the Con1 strain (16, 49). Our results, like those of Friebe and Bartenschlager (11), are also generally consistent with earlier observations concerning the requirements for these sequences for successful rescue of infectious virus following inoculation of synthetic, genome-length HCV RNAs into the livers of HCV-naïve chimpanzees (25, 48). Thus, although 3′NTR sequences have been shown to be less than completely essential for the replication of some other positive-strand RNA viruses (46), these reports provide a compelling body of evidence for an essential role for the 3′NTR sequences in HCV RNA replication, both in vivo in chimpanzee liver as well as in vitro in cultured cells. They indicate that earlier reports describing the replication of nearly full-length HCV RNAs lacking the 3′NTR in cultured hepatoma cells should be discounted (7, 50).
Ito et al. (18, 19) suggested that the 3′NTR sequences of HCV enhance translation directed by the HCV IRES. However, neither the failure of the 3′X deletion mutants (Fig. 4B) to replicate nor the impaired replication phenotypes of the poly(U/UC) or VR deletion mutants (Fig. 6B and 8A) can be related to inefficient translation of the upstream cistron in these replicons. Since the transient-transfection experiments were carried out in the absence of G418 selection, the protein products of the first cistron played no role in the replication of these RNAs (Fig. 1). Furthermore, the initial levels of SEAP that were produced by cells within the first 48 to 72 h of transfection with the mutated RNAs were similar to those induced by either the parental Ntat2ANeo(SI) replicon or the ΔGDD control RNA (Fig. 4B, 6B, and 8A). Thus, the 3′NTR deletions in these mutants appear to have had no biologically significant effect on the function of the upstream HCV IRES in these cells. Perhaps this is not too surprising, since other studies have failed to confirm that the 3′NTR positively regulates translation. For example, Murakami et al. (32) reported that the 3′NTR sequences down regulate the activity of the IRES, while Fang and Moyer (9) found that the 3′NTR sequences had no effect on the translation of a genome-length RNA under optimized conditions in vitro. These latter results are consistent with our findings reported here, as well as those described recently by Friebe and Bartenschlager (11). Together, they confirm the importance of carrying out such analyses with complete, genome-length RNAs, rather than truncated or internally deleted reporter transcripts.
Like Fang and Moyer (9) and Friebe and Bartenschlager (11), we also found no evidence that deletions within the 3′NTR accelerated the degradation of synthetic HCV RNA transcripts (Fig. 9). Thus, the negative impact of deletions within this segment of the viral RNA is likely to result from the loss of an essential RNA signal that participates in the replication process. It seems likely that such a signal might be involved in the recognition of the 3′ end of the viral genome by the RNA replicase complex. How this occurs remains shrouded in mystery, however, as relatively little is known about the specifics of the initiation of transcription of the negative-strand RNA during HCV RNA replication. Oh et al. (33, 34) have shown that the 3′NTR is required for specific recognition of genome-length RNA by a purified NS5B RNA-dependent RNA polymerase in vitro. Furthermore, the de novo synthesis of genome-length, negative-strand RNA in this in vitro system was associated with the binding of NS5B to sequences within the 3′X region, the poly(U/UC) tract, and the NS5B-coding sequence (33, 34).
Oh et al. (34) also reported that the transcription of the negative-strand RNA was initiated within the loop sequence of the 3′X stem-loop I, approximately 21 nt from the 3′ end of the RNA (34). More recently, Kim et al. (22) reported that transcription initiated further downstream but still internally, within the 3′ stem sequence of SL1, in a similar in vitro system. However, Hong et al. (15) have proposed that a unique β-hairpin within the thumb domain of the NS5B polymerase positions the terminal sequences of the genome so as to initiate de novo transcription from the 3′ terminal nucleotides. They proposed that the β-hairpin ensures the initiation of de novo RNA synthesis at the 3′ terminus by preventing movement of the 3′ end of the single-stranded RNA template into the active site of the enzyme. Such a mechanism is consistent with the requirement for conservation of the 3′-terminal sequences during replication. However, it leaves unresolved the mechanism by which RNAs with an authentic 3′ end are generated following transfection of synthetic RNAs with exogenous nucleotides at the 3′ terminus, as summarized in Fig. 3. Clearly the process of negative-strand initiation must be more complex than that which is modeled in these in vitro systems. This is not surprising, since several nonstructural proteins other than NS5B are likely to contribute to the functional replicase complex (3, 8).
Like Friebe and Bartenschlager (11), we found that a minimal poly(U/UC) tract was necessary for replication of subgenomic HCV RNAs in Huh7 cells. These results are also consistent with the earlier findings of Yanagi et al. (48), who noted the failure of synthetic RNAs lacking this element of the 3′NTR to replicate in chimpanzees. However, there are significant differences in the results we obtained in these experiments and those reported recently by Friebe and Bartenschlager (11). While these investigators replaced the deleted mixed polypyrimidine tract normally present in the Con1 replicon with a pure homopolymeric poly(U) sequence, we replaced the deleted poly(U/UC) tract in Ntat2ANeo(SI) in a stepwise fashion, rebuilding the native sequence from the 3′ end of the deleted segment (Fig. 6A). This led us to determine that a minimal poly(U/UC) sequence of 50 nt was required for detectable replication, while a 62-nt poly(U/UC) tract provided for significantly better replication (Fig. 6B). In contrast, only 26 nt of pure poly(U) sequence were required for replication of Con1 replicons (11).
It seems unlikely that this difference reflects a difference in the sensitivity of the assays employed to monitor replication of the RNAs in these studies. Rather, it is more likely that the variance is due to the difference in the sequences used to replace the deleted poly(U/UC) segments. It is interesting to note that replication was regained in our experiments only after the inclusion of the pure, 20-nt-long poly(U) tract that is present within the native poly(U/UC) tract of Ntat2ANeo(SI) (Fig. 6). In contrast, only a 12-nt-long poly(U) segment is present in the nonreplicating [U/C]-30 mutant. Furthermore, replication was substantially enhanced when this pure poly(U) segment was increased to 32 nt in [U/C]-62. Thus, the variance that is evident between our results and those of Friebe and Bartenschlager (11) could be explained by the possibility that a pure poly(U) sequence is more effective in this position than a mixed poly(U/UC) sequence, rather than by different requirements for the length of the polypyrimidine tract in HCV-N and Con1 replicons. However, further experiments will be required to prove this.
Although these results show the importance of the poly(U/UC) sequence, how this polypyrimidine tract functions in RNA replication remains a mystery. One possibility is that the sequence assists in circularizing the viral genome through the binding of a cellular protein, such as polypyrimidine tract-binding protein (PTB), to both the 5′ and 3′ ends of the genome (17). Circularization has been shown to be important for efficient replication of the RNA of other flaviviruses (20), but there are no RNA sequences within the termini of the HCV genome that have the potential to circularize the viral RNA through base pairing (5). Thus, a protein bridge that would bring together the two ends of the genome, mediated in part by protein binding to the 3′ poly(U/UC) tract, is an attractive possibility. Alternatively, it is possible that the poly(U/UC) tract functions directly in recognition of the 3′ end of the viral RNA by the replicase complex during an early step in the initiation of negative-strand RNA synthesis (34).
It is interesting to note that the poly(U/UC) tract of HCV is relatively unique within the family Flaviviridae. The 3′NTR of GBV-B virus, an unclassified hepatotropic flavivirus that is very closely related to HCV and that replicates to high titer in the liver of tamarins, contains a pure poly(U) tract that is 27 nt in length (43). However, such lengthy polypyrimidine tracts are lacking within the 3′NTRs of the classical flaviviruses, such as yellow fever virus (35), or in pestiviruses, such as bovine viral diarrhea virus (51), representatives of the two genera that, together with the genus Hepacivirus (HCV), comprise this virus family. Thus, there is no general requirement for such a 3′NTR sequence that is shared by all flaviviruses. The positive-strand RNA genome of hepatitis A virus, a picornavirus that, like HCV, replicates within the hepatocytes of humans and chimpanzees, possesses a poly(U/UC) tract that is ∼40 nt in length. However, in contrast to the poly(U/UC) tract of HCV, the pyrimidine-rich pY1 tract of hepatitis A virus is located within the viral 5′NTR, just upstream of the IRES, and can be deleted completely without loss of replication capacity in cultured cells or in primate liver in vivo (39, 40). Thus, although its function remains unknown, it appears to be distinct from that of the poly(U/UC) tract in HCV.
Consistent with the results of others (11, 48), we found that sequences within the variable region of the 3′NTR, upstream of the poly(U/UC) tract, are not essential for replication of the RNA. However, they do contribute to this process, since deletions in this region impair the efficiency of amplification of subgenomic replicons. Our results also suggest that a putative stem-loop that involves the 5′ part of the variable sequence and that includes the termination codon for the HCV polyprotein is not required for RNA replication (Fig. 7 and 8A).
Taken together, the results of the experiments presented here suggest that the 3′ terminal RNA signals that contribute to efficient HCV RNA replication extend upstream approximately 225 nt from the 3′ terminus of the genome. The 3′-most 150 nt of the genome [3′X region and 3′ 52 nt of the poly(U/UC) tract] are essential for replication of subgenomic RNAs, while the remaining upstream region of the 3′NTR plays a facilitating role in replication but is not absolutely required (Fig. 10). These results suggest an interesting symmetry in the 5′ and 3′ terminal RNA replication signals, as studies by others indicate that the 5′-most domains I and II of the 5′NTR are essential for replication, while sequences lying further downstream within the 5′NTR and comprising domain III help to facilitate replication but are not absolutely required (12, 23) (Fig. 10). Such a notion is strengthened by the suggestion that the 3′ ends of the negative- and positive-strand RNAs may share common structural features (37). However, further studies will be needed to determine the extent to which this apparent symmetry reflects common mechanisms of template recognition by the HCV replicase during negative- and positive-strand RNA synthesis.
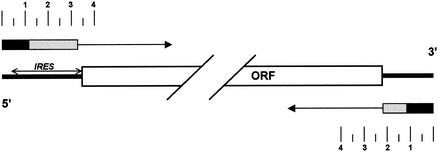
FIG. 10.
Schematic comparing the location of RNA replication signals located within the 5′NTR (12, 23) and 3′NTR (11; this study) segments of the HCV genome. The large open box represents the single large open reading frame that is present in the genomic RNA of HCV, with expanded views of the nontranslated regions at both ends of the RNA. The location of the IRES is depicted by the double-headed arrow. Adjacent to each end of the genome schematic, unidirectional arrows indicate the direction of transcription from the termini of the RNA, marked with a scale in hundreds of nucleotides. The terminal RNA segments that have been shown to be absolutely required for initiation of transcription from each end of the replicon RNA are indicated by the black boxes, while the contiguous downstream (5′ end) and upstream (3′ end) accessory sequences that promote replication efficiency but that are not absolutely required for replication of the RNA are indicated by the shaded boxes. Within the 3′ end, the essential sequences include the entire 3′X segment and 3′ half of the poly(U/UC) track (see Fig. 1), while the accessory sequences extend upstream to the polyprotein termination codon. The figure emphasizes both the overlapping nature of the replication and translation signals within the 5′NTR and the apparent symmetry of the RNA replication signals located within each end of the genome.
Acknowledgments
This work was supported in part by a grant from the National Institute of Allergy and Infectious Diseases (U19-AI40035).
We thank Masanori Ikeda and Ren Rijnbrand for helpful discussions and reviews of the manuscript.
REFERENCES
- 1.Alter, M. J., E. E. Mast, L. A. Moyer, and H. S. Margolis. 1998. Hepatitis C. Infect. Dis. Clin. North Am. 12:13-26. [DOI] [PubMed] [Google Scholar]
- 2.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, K. E. Reed, E. V. Agapov, and C. M. Rice. 1998. Molecular virology of hepatitis C virus: an update with respect to potential antiviral targets. Antivir. Ther. 3:71-81. [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 7.Dash, S., A. B. F. Halim, H. Tsuji, N. F. Hiramatsu, and M. A. Gerber. 1997. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am. J. Pathol. 151:363-373. [PMC free article] [PubMed] [Google Scholar]
- 8.De Francesco, R., P. Neddermann, L. Tomei, C. Steinkuhler, P. Gallinari, and A. Folgori. 2000. Biochemical and immunologic properties of the nonstructural proteins of the hepatitis C virus: implications for development of antiviral agents and vaccines. Semin. Liver Dis. 20:69-83. [DOI] [PubMed] [Google Scholar]
- 9.Fang, J. W., and R. W. Moyer. 2000. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 33:632-639. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., and M. M. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., S. M. F. Tahara, and M. M. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 8789-8796. [DOI] [PMC free article] [PubMed]
- 20.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, M., H. Kim, S. P. Cho, and M. K. Min. 2002. Template requirements for de novo RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase on the viral X RNA. J. Virol. 76:6944-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y. K. F., C. S. F. Kim, S. H. F. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 24.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsay, K. L., C. Trepo, T. Heintges, M. L. Shiffman, S. C. Gordon, J. C. Hoefs, E. R. Schiff, Z. D. Goodman, M. Laughlin, R. Yao, and J. K. Albrecht. 2001. A randomized, double-blind trial comparing pegylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology 34:395-403. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 30.Major, M. E., and S. M. Feinstone. 1997. The molecular virology of hepatitis C. Hepatology 25:1527-1538. [DOI] [PubMed] [Google Scholar]
- 31.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 32.Murakami, K., M. Abe, T. Kageyama, N. Kamoshita, and A. Nomoto. 2001. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729-741. [DOI] [PubMed] [Google Scholar]
- 33.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 35.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 36.Rijnbrand, R. C., and S. M. Lemon. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:85-116. [DOI] [PubMed] [Google Scholar]
- 37.Schuster, C., C. Isel, I. Imbert, C. Ehresmann, R. Marquet, and M. P. Kieny. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J. Virol. 76:8058-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeff, L. B. 1997. Natural history of hepatitis C. Hepatology 26:21S-28S. [DOI] [PubMed] [Google Scholar]
- 39.Shaffer, D. R., E. A. Brown, and S. M. Lemon. 1994. Large deletion mutations involving the first pyrimidine-rich tract of the 5′ nontranslated RNA of hepatitis A virus define two adjacent domains associated with distinct replication phenotypes. J. Virol. 68:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaffer, D. R., S. U. Emerson, P. C. Murphy, S. Govindarajan, and S. M. Lemon. 1995. A hepatitis A virus deletion mutant which lacks the first pyrimidine-rich tract of the 5′ nontranslated RNA remains virulent in primates after direct intrahepatic nucleic acid transfection. J. Virol. 69:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmonds, P., F. McOmish, P. L. Yap, S.-W. Chan, C. K. Lin, G. Dusheiko, A. A. Saeed, and E. C. Holmes. 1993. Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J. Gen. Virol. 74:661-668. [DOI] [PubMed] [Google Scholar]
- 43.Simons, J. N., T. J. Pilot-Matias, T. P. Leary, G. J. Dawson, S. M. Desai, G. G. Schlauder, A. S. Muerhoff, J. C. Erker, S. L. Buijk, M. L. Chalmers, C. L. Van Sant, and I. K. Mushahwar. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. USA 92:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, T., N. Kato, M.-J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of the hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 46.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi, M., F. Bodola, and S. M. Lemon. 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304:197-210. [DOI] [PubMed] [Google Scholar]
- 50.Yoo, B. J., M. J. Selby, J. Choe, B. S. Suh, S. H. Choi, J. S. Joh, G. J. Nuovo, H.-S. Lee, M. Houghton, and J. H. Han. 1995. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J. Virol. 69:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, H., C. W. Grassmann, and S. E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, H., O. Isken, C. W. Grassmann, and S. E. Behrens. 2000. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J. Virol. 74:5825-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]