Abstract
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) has a bisegmented negative-strand RNA genome. Each segment carries two viral genes in opposite orientation and separated by an intergenic region (IGR). The RNA-dependent RNA polymerase (RdRp) L of LCMV produces subgenomic mRNA and full-length genomic and antigenomic RNA species in two different processes termed transcription and replication, respectively. It is widely accepted that intracellular nucleoprotein (NP) levels regulate these two processes. Intracellular NP levels increase during the course of the infection, resulting in the unfolding of secondary RNA structures within the IGR. Structure-dependent transcription termination at the IGR is thereby attenuated, promoting replication of genome and antigenome RNA species. To test this hypothesis, we established a helper-virus-free minigenome (MG) system where intracellular synthesis of an S segment analogue from a plasmid is driven by RNA polymerase I. Cotransfection with two additional plasmids expressing the minimal viral trans-acting factors L and NP under control of RNA polymerase II allowed for RNA synthesis mediated by the intracellularly reconstituted LCMV polymerase. Both processes, transcription and replication, were strictly dependent on NP. However, both were equally enhanced by incrementally increasing amounts of NP up to levels in the range of those in LCMV-infected cells. Our data are consistent with a central role for NP in transcription and replication of the LCMV genome, but they do not support the participation of NP levels in balancing the two processes.
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is an enveloped virus with a bisegmented, negative-strand RNA genome. The two genomic RNA segments are called large (L) and short (S) and have sizes of 7.2 and 3.4 kb, respectively (27-29). Each segment directs the synthesis of two gene products, encoded in ambisense orientation. The two open reading frames of each segment are separated by an intergenic region (IGR), with a sequence predicted to form a hairpin structure of high stability (29). The S segment encodes the virus nucleoprotein (NP; ca. 63 kDa) and glycoprotein (GP) precursor, whereas the L segment encodes a large polypeptide (L protein; ca. 200 kDa) and a small RING finger protein (Z; ca. 11 kDa), whose function in the virus life cycle is poorly understood (29). The NP and L genes are carried in antigenome polarity 3′ of the IGR in the S and L segments, respectively. NP encapsidates the genome segments and represents the most abundant protein in virions and infected cells (2). The L protein (ca. 200 kDa) exhibits the characteristic hallmarks common to the RNA-dependent RNA polymerases (RdRp) of negative-strand RNA viruses (27). GP and Z are encoded in genome polarity 5′ of the IGR in the S and L segments, respectively. GP is synthesized as precursor polypeptide GP-C (75 kDa), which is posttranslationally processed into GP-1 (40 to 46 kDa) and GP-2 (35 kDa) (3). These two proteins remain noncovalently associated, form club-shaped projections on virions, and mediate cell entry.
RNA synthesis by the arenavirus RdRp is thought to comprise two fundamentally different processes. The transcriptase activity associated with the RdRp synthesizes capped (20, 25), unencapsidated (25) mRNA, allowing for translation of viral proteins. The 3′ ends of these subgenomic mRNA molecules are heterogeneous and not polyadenylated, and they have been mapped within the predicted hairpin in the IGR (21, 30). The replicase function of the RdRp directs synthesis of mostly encapsidated (25), uncapped (17) full-length antigenomic and genomic RNA species, which are used as templates for further mRNA transcription. Moreover, newly synthesized full-length genomic viral RNA is used for formation of new particles. A widely accepted model for regulation of transcription and replication of negative-strand RNA viruses (19) is based on early studies of the rhabdovirus vesicular stomatitis virus and the paramyxovirus Sendai virus (SeV). It assumes that both processes require the same polymerase complex according to the following two postulates (16). First, the soluble NP protein is required for RNA replication, i.e., soluble NP must be available in sufficient quantities to encapsidate nascent viral RNA so as to allow for the process of replication to occur. Second, the abundance of available NP for encapsidation acts as a switch to shift the polymerase from transcription to replication, and thereby NP acts as a transcription antiterminator. This, in turn, allows NP to regulate its own synthesis, analogous to the attenuation signals in the tryptophan and histidine operons of procaryotes (1).
Several lines of evidence support the first postulate for arenaviruses also: (i) full-length genomic and antigenomic RNA species are found almost exclusively in encapsidated form (25), (ii) arenavirus RNA replication is dependent on ongoing protein synthesis and can be inhibited by translation inhibitors (10, 30), and (iii) introduction of the NP protein can restore viral replication in cells treated with a translation inhibitor (30). The second postulate originates from early studies using cycloheximide to block translation in vesicular stomatitis virus- or SeV-infected cells. This treatment resulted in inhibition of replication. In addition, some investigators found a concomitant modest increase in production of subgenomic-size viral RNA (24, 32) though others did not (4, 26, 31). The activity of negative regulatory proteins (e.g., C protein for SeV [7]) and the modulation of polymerase processivity by NP (31) may also contribute to modulate the balance between the transcriptase and replicase activities of RdRps. The latter is of particular importance since the above-mentioned studies could not distinguish between the formation of bona fide viral mRNA and subgenomic-size viral RNA resulting from premature termination of replication (24, 32). Moreover, a recent study using a minireplicon system for respiratory syncytial virus could not confirm this second postulate (8).
We have described a reverse genetic system for direct manipulation of virus cis-acting signals and trans-acting factors involved in the control of LCMV RNA synthesis (14). This system is based on the use of a synthetic LCMV model genome or minigenome (MG) that allows for the expression of the chloramphenicol acetyltransferase (CAT) reporter gene mediated by the expression of viral proteins provided from cloned cDNAs. Intracellular coexpression of MG RNA and viral trans-acting polypeptides was done with the vaccinia virus T7 RNA polymerase expression system (11). Using this system we showed that NP and the L are the minimal trans-acting factors required for viral transcription and replication. Moreover, the viral 5′ and 3′ untranslated regions (UTRs) together with the IGR represent sufficient cis-acting signals for RNA synthesis mediated by the LCMV polymerase. Initial attempts to address the role of intracellular NP levels in regulation of transcription and replication of LCMV with our previously described model genome system were hampered by the following two factors: (i) L protein-dependent CAT activity in the absence of NP was, although extremely low, consistently over background, rendering it difficult to accurately assess effects of very low intracellular NP levels on viral transcription and (ii) amplification of plasmid-supplied MG RNA by the viral RdRp was very limited (less than twofold), indicating that the system did not promote multiple rounds of replication. The possibility that vaccinia virus infection was at the root of these findings led us to consider an alternative approach for intracellular expression of the LCMV MG. However, other arenaviruses may differ in their specific requirements for efficient MG amplification (18).
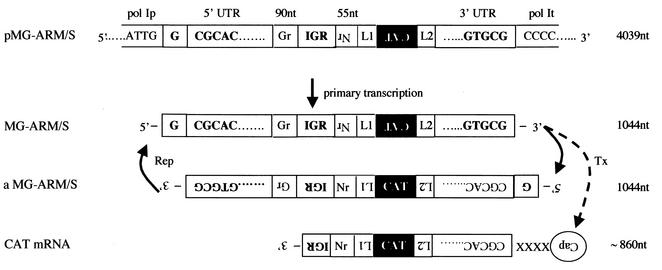
A recent report described a reverse genetic system for the bunyavirus Uukuniemi virus that was based on RNA polymerase I (pol-I)-driven intracellular synthesis of MG RNA (9). Bunyaviruses also are segmented negative-strand RNA viruses with a cytoplasmic life cycle. Therefore, we established a pol-I/pol-II-based system for intracellular reconstitution of LCMV replication and transcription. For this purpose the LCMV Armstrong (ARM) S segment MG (14) was cloned in the pol-I expression vector pRF42 (9). We sequenced the entire MG and verified that all cis-acting elements were precisely as described for the LCMV ARM S segment (28). In the resulting construct, pMG-ARM/S (Fig. 1), pol-I initiates transcription after the murine pol-I promoter with a nontemplated G residue (12) and transcribes the 5′ UTR, 90 nucleotides (nt) of the sequence encoding the GP C terminus, the S segment IGR (64 nt), 55 nt of the sequence encoding the NP C terminus, the short polylinker L1, the CAT reporter gene in antisense polarity, short polylinker L2, and the 3′ UTR. Thereafter it terminates when reaching the murine pol-I terminator located downstream of the 3′ UTR sequences (34). This obviates the need for a cis-acting ribozyme to generate a correct 3′ end. Upon encapsidation of the primary pol-I transcript, the LCMV polymerase is expected to use it as a template for synthesis of full-length antiminigenome (aMG) RNA (replicate) and subgenomic CAT mRNA (transcript) (Fig. 1).
FIG. 1.
Schematic of the pol-I-driven LCMV MG, showing also transcription and replication intermediates. pMG-ARM/S contains the elements described for the MG-ARM/S construct (14) flanked by the murine pol-I promoter (pol Ip) and terminator sequences (pol It) of pRF42 (9). Transcription of pMG-ARM/S by the cellular pol-I (primary transcription) generates MG-ARM/S RNA (MG), detectable by a CAT sense riboprobe. Replication of the MG (solid right arrow) yields an aMG RNA, detectable by a CAT antisense riboprobe. This aMG serves as a template for synthesis of more MG RNA by the virus polymerase (replication [Rep]; solid left arrow). Both MG and aMG RNA species are assumed to have a nontemplated G at their 5′ ends (12). Transcription (Tx; dashed arrow) of the MG RNA by the virus polymerase yields a subgenomic-length CAT mRNA that terminates within the IGR and that is detectable by a CAT antisense riboprobe. Subgenomic mRNA species are assumed to have 5′ end cap structures, containing 4 or 5 nt not derived from viral template sequences (XXXX) (20). cis-Acting sequences are in bold. IGR, intergenic region; Gr and Nr, sequences within the GP and NP ORFs, respectively.
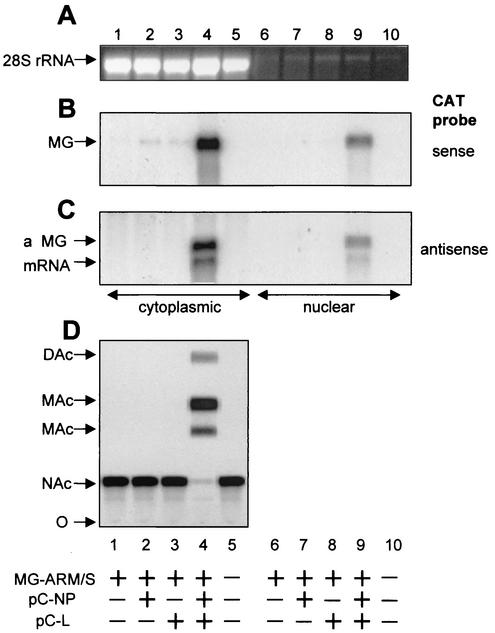
Transfection of pMG-ARM/S alone resulted in intracellular expression of MG RNA, detected as a faint band of the expected size by hybridization with a CAT sense probe in a Northern blot (Fig. 2B, lane 1), while no aMG RNA or mRNA was detected with an antisense CAT probe (Fig. 2C, lane 1). For intracellular expression of LCMV NP and L proteins in the absence of vaccinia virus T7 RNA polymerase, the corresponding open reading frames were subcloned from pCITE-NP and pGEM-L (14) into the plasmid pCAGGS (pC) (22), a pol-II-based expression vector. Cotransfection of either pC-NP or pC-L together with pMG-ARM/S did not significantly change the pattern of RNA detected compared to transfection with pMG-ARM/S only (Fig. 2B and C, lanes 2 and 3 versus 1). In contrast, cotransfection of pMG-ARM/S with plasmids encoding both minimal viral trans-acting factors, NP and L, resulted in amplification of MG RNA (Fig. 2B, compare lane 4 with 1 to 3) along with formation of an aMG replicative intermediate and mRNA species of the expected sizes (Fig. 2C, lane 4). This confirmed our previous finding that NP and L represent the minimal viral trans-acting factors needed for transcription and replication (14) and validated the pol-I/pol-II-driven MG system. Since pol-I is a nuclear enzyme and since the LCMV life cycle is cytoplasmic, we assessed the subcellular localization of LCMV MG RNA amplification and transcription. Phosphorimager analysis showed five times more signal in the cytoplasmic fraction than in the nuclear fraction for amplified MG (Fig. 2B, lane 4 versus 9) as well as for aMG and mRNA species (Fig. 2C, lane 4 versus 9). This finding indicated that the LCMV MG RNA initially synthesized in the nucleus was exported to the cytoplasm, where it was transcribed and replicated by the intracellularly reconstituted LCMV polymerase. A 70-fold amplification of plasmid-supplied MG upon cotransfection of NP and L (Fig. 2B, compare lane 4 with 1 to 3) indicated that the pol-I-derived LCMV MG RNA went through multiple rounds of replication. Consistent with the RNA data, CAT activity was detected only upon cotransfection of pMG-ARM/S, pC-NP, and pC-L plasmids (Fig. 2D).
FIG. 2.
Subcellular distribution of plasmid-supplied LCMV MG RNA and derived RNA species and MG-derived CAT activity. BHK-21 cells in six-well plates (80% confluent) were transfected with pMG-ARM/S (0.5 μg), pC-NP (0.8 μg), and pC-L (1 μg) in the combinations indicated at the bottom by using Lipofectamine as described previously (23). At 72 h posttransfection cells were harvested. For each sample, one-half of the cells were used to prepare nuclear and cytoplasmic RNA as described previously (6). Ten percent of the total cytoplasmic (lanes 1 to 5) or nuclear (lanes 6 to 10) RNA obtained from each sample was analyzed on duplicate blots by Northern hybridization, thereby normalizing for the amount of RNA on a per-cell basis. (A) Ethidium bromide staining of 28S rRNA showed comparable total RNA amounts loaded for samples 1 to 5 and 6 to 10. (B) Hybridization to a CAT sense riboprobe. (C) Hybridization to a CAT antisense riboprobe. (D) The other half of each cell sample was processed for CAT assay as described previously (5). O, origin of sample application; NAc, nonacetylated chloramphenicol; MAc, monoacetylated chloramphenicol; DAc, diacetylated chloramphenicol.
We next tested the hypothesis that intracellular NP levels determine the balance between the replicase and transcriptase mode of the LCMV polymerase. For this, fixed amounts of pMG-ARM/S and pC-L were cotransfected with amounts of pC-NP ranging from 6 to 800 ng. Additional “empty” plasmid pC was added to normalize for the total amount of DNA transfected in each sample. This allowed analysis of transcription and replication intermediates formed by the viral RdRp in the presence of various amounts of NP under otherwise-standardized conditions. We have shown that MG-derived mRNA species are, similar to bona fide LCMV mRNAs, nonpolyadenylated (14). Therefore, MG-derived transcript and replicate RNA species can be easily distinguished by a Northern blot assay based on their predicted lengths (Fig. 1). In addition, we attempted to distinguish encapsidated antigenome molecules from nonencapsidated mRNA based on their resistance to micrococcal nuclease (MNase). Several independent experiments using low concentrations of MNase yielded inconclusive results. Unlike the situation described for bunyaviruses (13), the arenavirus nucleocapsid may not provide encapsidated RNAs with protection against low levels of nucleases, which would explain our results. As an alternative approach we tried to immunoprecipitate the RNPs containing the MG and aMG RNA species by using a guinea pig (gp) polyclonal serum against LCMV. However, in control experiments we observed that the gp serum also precipitated mRNA species under the experimental conditions required to preserve the integrity of RNPs.
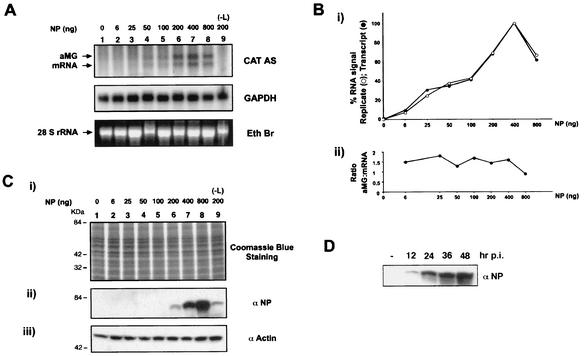
Neither aMG (1,044 nt) nor CAT mRNA (860 nt) was detected in the absence of NP or L, respectively (Fig. 3A, lanes 1 and 9). Phosphorimager analysis detected specific hybridization signals for both CAT mRNA and aMG RNA species upon cotransfection of as little as 6 ng of pC-NP (Fig. 3B). Consistent with this result, longer exposure time for the blot shown in Fig. 3A revealed the presence of two bands in lanes 2 and 3 that corresponded to the CAT mRNA and aMG RNA species (not shown). A steady increase of aMG and CAT mRNA was observed up to about 0.4 μg of pC-NP transfected (Fig. 3A and B). Further increases in the amount of pC-NP to 0.8 μg resulted in similar reductions of both transcription and replication (Fig. 3A, lane 8). This observation was highly reproducible and even more prominent with pC-NP doses up to 1.6 μg (data not shown). Ethidium bromide staining of the 28S rRNA and results of hybridization with the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe excluded any significant differences among samples with respect to the amount of RNA loaded (Fig. 3A). CAT activity paralleled the levels of MG-derived CAT mRNA and aMG (Fig. 4). The ratio of aMG (replication) to CAT mRNA (transcription) remained constant over a large range (>50-fold) of increased RNA synthesis by the virus polymerase, with no trend toward transcription or replication observed (Fig. 3B). Six independent experiments indicated that intracellular NP levels had no effect on the aMG/CAT mRNA ratio obtained within one experiment, but we observed some variability in the specific ratios obtained in independent experiments (data not shown). We found that 0.8 μg of pC-NP produced levels of NP in transfected cells that were comparable to the highest levels of NP detected in LCMV-infected cells (Fig. 3C and D). Maximum levels of MG transcription and replication were reached with plasmid-supplied NP levels that seemed to be slightly below those observed during the peak of virus replication in infected cells. This may be explained by differences between the numbers of transfected and infected cells. In transfection assays only 20 to 40% of the total number of cells expressed NP as determined by immunofluorescence (IF) (data not shown) while the majority (over 95%) of the cells in an LCMV-infected cell population were NP positive by IF. Accordingly, our Western blot analysis most likely underestimated the amount of NP expressed on a per-transfected-cell basis. An alternative and not mutually exclusive explanation may be that the amounts of intracellularly reconstituted functional viral polymerase and RNP complexes in transfected cells are significantly lower than those generated by infection, with an accordingly lower saturation threshold for NP.
FIG. 3.
Influence of intracellular NP levels on transcription and replication of the LCMV MG. BHK-21 cells in six-well plates (80% confluent) were transfected with pMG-ARM/S (0.5 μg), pC-L (0.2 μg), the amount of pC-NP indicated, and empty pC to normalize the total amount of plasmid DNA transfected in each case to 2.3 μg. Cells were harvested 40 h posttransfection. For each sample 50% of the cells were used for isolation of total cellular RNA and the other 50% were used to determine NP expression levels by Western blotting. (A) Analysis of RNA synthesis mediated by the LCMV polymerase. Equal amounts of RNA of each sample, as determined by ethidium bromide (Eth Br) staining of the 28S rRNA, were analyzed by Northern blot hybridization with a CAT antisense probe to detect CAT mRNA and aMG RNA species. After hybridization with the CAT antisense (AS) probe and phosphorimager analysis of the blot, the membrane was stripped and hybridized with a probe to the housekeeping gene encoding GAPDH to confirm that the membrane contained similar amounts of RNA in each lane. One representative experiment of six is shown. (B) The intensities of aMG and CAT mRNA bands shown in panel A were assessed by phosphorimager. The hybridization signal obtained in the absence of NP (A, lane 1) was considered nonspecific and subtracted from all the samples. We observed similar background hybridization signals in the absence of L (A, lane 9). Phosphorimager values for each sample were normalized with respect to the corresponding GAPDH hybridization signals. Normalized signals were depicted as percentages of replication (aMG RNA) or transcription (CAT mRNA) with respect to the values (100%) obtained with 0.4 μg of pC-NP. The replicate (aMG)-to-transcript (CAT mRNA) ratio for each sample (bottom) was calculated based on the normalized phosphorimager values obtained for the corresponding bands shown in panel A. One representative experiment of six is shown. (C) Expression levels of NP protein in transfected cells. Equal protein amounts for each sample, as determined by Coomassie blue staining (i), were analyzed by Western blotting using a gp polyclonal serum against LCMV that recognizes the virus NP (ii). The membrane was stripped and probed with rabbit serum against actin to verify that the membrane contained similar protein amounts in each lane (iii). (D) Expression levels of NP protein in LCMV-infected cells. BHK-21 cells were infected with LCMV (multiplicity of infection, 1), and at the indicated times after infections cell extracts were prepared for analysis of protein expression by Western blotting. Equal amounts of protein were loaded for each sample and analyzed for NP expression levels as described for panel C. One representative experiment of two is shown. p.i., postinfection.
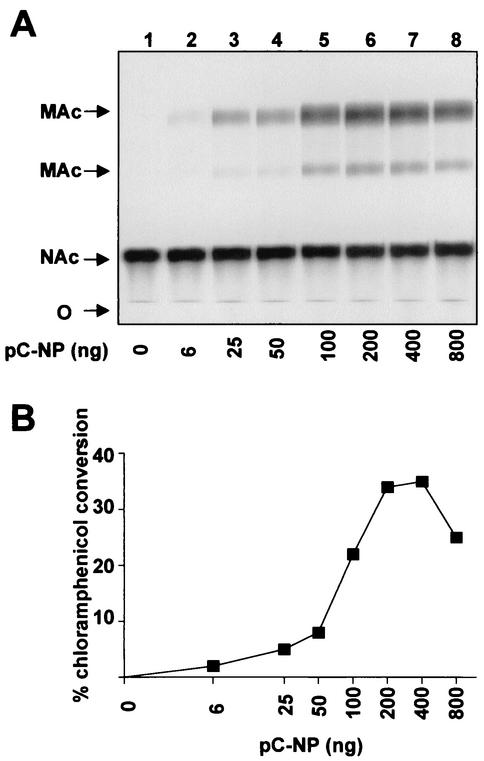
FIG. 4.
Influence of intracellular NP levels on CAT activity. BHK-21 cells were seeded and transfected as described for Fig. 3A and B. Total cell extracts were processed for CAT assay as described previously (5). (A) Autoradiography of the thin-layer chromatography plate. Abbreviations: O, origin of sample application; NAc, nonacetylated chloramphenicol; MAc, monoacetylated chloramphenicol. No double-acetylated form of chloramphenicol was generated. (B) Chloramphenicol conversion was quantitated by phosphorimager analysis.
As for other negative-strand RNA viruses (16), our data also support a requirement for NP encapsidation of LCMV RNA templates for transcription and replication. Unlike what was done in previous studies using infection of cycloheximide-treated cells (10, 30), we were unable to demonstrate transcription in the absence of NP. This is not necessarily contradictory. Infection of cycloheximide-treated cells delivers a fully functional RNP into a cellular environment which is unable to support any NP production. But at this stage the viral template RNA is already encapsidated. Under these selected conditions transcription may proceed in complete absence of replication. In our MG system the pol-I-derived MG RNA is initially not encapsidated. Only a pool of intracellularly produced NP can encapsidate the MG RNA to make it a template suitable for RNA synthesis by the viral polymerase. Therefore we cannot recreate a situation with an MG RNP in the absence of soluble NP to parallel the aforementioned situation involving LCMV infection of cycloheximide-treated cells.
Our results showed that replication and transcription of LCMV MG RNA are equally enhanced by an increment in intracellular NP levels, but this was not associated with a “switch” from transcription to replication. Based on these observations we favor the model in which two distinct polymerase complexes, which are committed to either transcription or replication, may exist (8). Accordingly, the two processes would not compete. This view is supported by the finding that LCMV mRNAs are capped while genomes and replicative intermediates are not (20), suggesting that the replicase is already committed to either process when initiating at the 3′ end of the template by using either a cap oligonucleotide primer or a prime-and-realign mechanism (12). Therefore, it seems unlikely that the fate of nascent RNA molecules as either mRNA or genome and antigenome RNA species will be determined only when the polymerase complex reaches the IGR where mRNAs terminate. The mapping of the 3′ ends of LCMV mRNAs within the strong secondary structures found in the IGR (20, 30) is reminiscent of intrinsic or rho-independent termination in procaryotes (30, 33) and represents strong support for the IGR as the termination signal for transcription complexes. However, no clear implications for the role of NP in antitermination can be derived from this. Assuming that unencapsidated viral RNA cannot serve as a template for either transcription or replication (14, 18; present study), the postulated antitermination mechanism of NP through interaction with the IGR of the template RNA (30) would require that template RNA (genome and antigenome species) exhibit different degrees of encapsidation, which seems unlikely and which is unprecedented.
Our observation of a reduction in both transcription and replication when a certain level of NP is surpassed was unexpected though not unprecedented (8). It most likely indicates that correct stoichiometry rather than large amounts of each viral protein is required for optimal function of the LCMV polymerase complex. The observed similar reductions in transcription and replication, with a concomitant decrease in reporter gene activity (Fig. 4), further argue against a switch of the polymerase to a replicase mode of action.
Intrinsic termination in bacteria can be attenuated if formation of secondary RNA structures in the nascent transcript is prevented (33). Therefore, a more likely role for NP in antitermination may be through interaction with the IGR sequences of the nascent RNA. The presence of a 5′ cap structure on nascent LCMV mRNAs may prevent their encapsidation by NP. When the transcription complex reaches the IGR of the template RNA, the resulting hairpin structure at the 3′ end of the nascent transcript may cause termination. Nascent RNA species lacking a 5′ cap structure would be encapsidated if soluble NP is available, and encapsidation may attenuate the efficiency of termination signals contained within the IGR sequences of the nascent RNA. If replicase complexes are not intrinsically resistant to the termination signal, the lack of soluble NP in cycloheximide-treated cells may cause inhibition of replication through premature termination. The RNA generated in such a process would be of subgenomic size though not a bona fide transcript due to the lack of a 5′ cap structure. As an alternative model one could envisage that replicase complexes are uniform and that initiation with either a cap oligonucleotide primer or through a prime-and-realign mechanism determines the sensitivity or resistance, respectively, to termination signals contained in the IGR. These signals could be provided by a secondary structure formed in the nascent transcript. The finding that LCMV transcription and replication are equally enhanced by increasing intracellular NP levels is consistent with the reported NP-mediated enhancement of polymerase processivity in SeV (31). During LCMV persistent infection steady-state levels of NP remain high and an accumulation of genome RNA molecules over mRNAs can frequently be observed. In the light of our results, this may, however, be related to the longer half-lives of encapsidated genomes and replicative intermediates rather than to biased de novo synthesis in the presence of high intracellular NP levels. Our conclusions are in part based on the assumption that the cis-acting elements (5′ UTR, 3′UTR, and IGR) function as a cassette element in the MG comparable to their function in the LCMV S genomic RNA segment. Currently, we cannot formally exclude the existence of long-range RNA interactions involved in transcription termination and template recognition other than those involving the above-mentioned cis-acting elements. The absence of a subgenomic-size RNA species in the reverse genetic system of the arenavirus Tacaribe virus (18), which uses an MG devoid of IGR sequences, and our preliminary results with an MG lacking the IGR sequences, however, support our assumption that the IGR itself acts as transcription termination signal. We are currently investigating the role of the IGR as a transcription terminator in our MG system. The correct functioning of the cis-acting elements was further supported by the ability of the MG described herein to form virus-like particles, which could be generated either by providing the viral proteins GP and Z as previously described (15) or by infecting RNP MG-expressing cells with LCMV, resulting in CAT activity upon passage of supernatant onto fresh cells.
Acknowledgments
This work was supported by a fellowship of the Gebert Rüf Stifung, Switzerland, to Daniel Pinschewer and NIH grant AI47140 to J. C. de la Torre.
We thank Ana B. Sanchez for helpful discussions and Rosalia Garcia for excellent technical assistance.
REFERENCES
- 1.Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837-845. [DOI] [PubMed] [Google Scholar]
- 2.Buchmeier, M. J., J. H. Elder, and M. B. Oldstone. 1978. Protein structure of lymphocytic choriomeningitis virus: identification of the virus structural and cell associated polypeptides. Virology 89:133-145. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, M. J., P. J. Southern, B. S. Parekh, M. K. Wooddell, and M. B. Oldstone. 1987. Site-specific antibodies define a cleavage site conserved among arenavirus GP-C glycoproteins. J. Virol. 61:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsen, S. R., R. W. Peluso, and S. A. Moyer. 1985. In vitro replication of Sendai virus wild-type and defective interfering particle genome RNAs. J. Virol. 54:493-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubitt, B., and J. C. de la Torre. 1994. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J. Virol. 68:1371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran, J., J. B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 8.Fearns, R., M. E. Peeples, and P. L. Collins. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188-201. [DOI] [PubMed] [Google Scholar]
- 9.Flick, R., and R. F. Pettersson. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J. Virol. 75:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franze-Fernandez, M. T., C. Zetina, S. Iapalucci, M. A. Lucero, C. Bouissou, R. Lopez, O. Rey, M. Daheli, G. N. Cohen, and M. M. Zakin. 1987. Molecular structure and early events in the replication of Tacaribe arenavirus S RNA. Virus Res. 7:309-324. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, D., and D. Kolakofsky. 1990. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J. Virol. 64:6196-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacker, D., R. Raju, and D. Kolakofsky. 1989. La Crosse virus nucleocapsid protein controls its own synthesis in mosquito cells by encapsidating its mRNA. J. Virol. 63:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de la Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leppert, M., L. Rittenhouse, J. Perrault, D. F. Summers, and D. Kolakofsky. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18:735-747. [DOI] [PubMed] [Google Scholar]
- 17.Leung, W. C., H. P. Ghosh, and W. E. Rawls. 1977. Strandedness of Pichinde virus RNA. J. Virol. 22:235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López, N., R. Jácamo, and M. T. Franze-Fernández. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, B. J., J. C. de la Torre, and P. J. Southern. 2002. Arenaviruses: genomic RNAs, transcription, and replication. Curr. Top. Microbiol. Immunol. 262:139-157. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, B. J., and P. J. Southern. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J. Virol. 67:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, B. J., and P. J. Southern. 1994. Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J. Virol. 68:7659-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 23.Perez, M., M. Watanabe, M. A. Whitt, and J. C. de la Torre. 2001. N-terminal domain of Borna disease virus G (p56) protein is sufficient for virus receptor recognition and cell entry. J. Virol. 75:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portner, A., and D. W. Kingsbury. 1972. Identification of transcriptive and replicative intermediates in Sendai virus-infected cells. Virology 47:711-725. [DOI] [PubMed] [Google Scholar]
- 25.Raju, R., L. Raju, D. Hacker, D. Garcin, R. Compans, and D. Kolakofsky. 1990. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174:53-59. [DOI] [PubMed] [Google Scholar]
- 26.Robinson, W. S. 1971. Sendai virus RNA synthesis and nucleocapsid formation in the presence of cycloheximide. Virology 44:494-502. [DOI] [PubMed] [Google Scholar]
- 27.Salvato, M., E. Shimomaye, and M. B. Oldstone. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169:377-384. [DOI] [PubMed] [Google Scholar]
- 28.Salvato, M., E. Shimomaye, P. Southern, and M. B. Oldstone. 1988. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, clone 13 (CTL−). Virology 164:517-522. [DOI] [PubMed] [Google Scholar]
- 29.Salvato, M. S., and E. M. Shimomaye. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Tortorici, M. A., C. G. Albarino, D. M. Posik, P. D. Ghiringhelli, M. E. Lozano, R. Rivera Pomar, and V. Romanowski. 2001. Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res. 73:41-55. [DOI] [PubMed] [Google Scholar]
- 31.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertz, G. W., and M. Levine. 1973. RNA synthesis by vesicular stomatitis virus and a small plaque mutant: effects of cycloheximide. J. Virol. 12:253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]
- 34.Zobel, A., G. Neumann, and G. Hobom. 1993. RNA polymerase I catalysed transcription of insert viral cDNA. Nucleic Acids Res. 21:3607-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]