Abstract
Epstein-Barr virus (EBV) EBNA-1 is the only EBV-encoded protein that is essential for the once-per-cell-cycle replication and maintenance of EBV plasmids in latently infected cells. EBNA-1 binds to the oriP region of latent EBV plasmids and cellular metaphase chromosomes. In the absence of oriP-containing plasmids, EBNA-1 was highly colocalized with cellular DNA replication foci that were identified by immunostaining S-phase cells for proliferating cell nuclear antigen and replication protein A (RP-A) in combination with DNA short pulse-labeling. For the association of EBNA-1 with the cellular replication focus areas, the EBNA-1 regions of amino acids (aa) 8 to 94 and/or aa 315 to 410, but not the RP-A-interacting carboxy-terminal region, were necessary. These results suggest a new aspect of latent virus-cell interactions.
The replication of Epstein-Barr virus (EBV) plasmid DNAs in latently infected cells proceeds once per cell cycle and in synchrony with cell DNA in latently infected and immortalized cells such as Burkitt's lymphoma, nasopharyngeal carcinoma, and lymphoblastoid cells (44, 47). The EBV nuclear antigen-1 (EBNA-1) protein is the only EBV-encoded protein required for the replication, retention, and partition of the plasmid DNA that carries the EBV oriP region with the sequences for replication and retention (28, 46). EBNA-1 is detected in all types of EBV-infected cells and is the only EBV protein that is detected in cells with type I latency (37). Thus, EBNA-1 represents a nuclear regulatory protein that plays a critical role in the control of the eukaryotic or metazoan DNA replication. The EBNA-1 protein homodimer binds directly to the replication origin region, oriP, of EBV DNA (1, 2, 16, 21, 28). EBNA-1 binds to several cellular proteins (23, 26, 39, 41, 48) and to the metaphase chromosomes (24, 30, 34). The binding of EBNA-1 to metaphase chromosomes is thought to facilitate the partition of latent EBV plasmids (30).
Studies of the essential but obscure role(s) for EBNA-1 in the replication and maintenance of latent EBV plasmids would provide a useful system for investigating the regulation of the DNA replication in metazoan cells where it has been difficult to identify the cellular DNA replication origins (10, 33). We have recently reported that EBNA-1 fused to green fluorescent protein (GFP-EBNA1) colocalizes with newly replicated regions of interphase chromatin in cells that do not contain EBV plasmids (22).
The cell line CEW21N4 stably expressing full-length EBNA-1 was produced by transfecting the Chinese hamster-derived epithelial-like cell line CHO-K1 (American Type Culture Collection) with the EBNA-1-expressing vector pC43 (20, 25). The EBV-negative human B-cell line DG75 was kindly provided by G. Klein (Karolinska Institute). CHO-K1 cells in mitotic phase were released from the dish substratum by shaking and were cultured on glass-bottom dishes for 2 h, and then aphidicolin (10 μg/ml) was added. The cells were incubated for 16 h, supplemented with fresh medium, and then incubated for 10 to 30 min, 3 to 4 h, or 7 to 8 h for synchronous early, middle, or late S-phase cells, respectively. The GFP fusion protein of EBNA-1 that lacks the Gly-Ala copolymer, designated GFP-EBNA1, has been described (25). The enhanced GFP (EGFP) fusion of an EBNA-1 mutant lacking the sequence of amino acids (aa) 1 to 374 was constructed and designated S(Δ1-374) (see Fig. 3A). The two EGFP-EBNA-1 mutant fusions, M4 (containing aa 8 to 94 and aa 315 to 410) and M15 (containing aa 378 to 606), were kindly provided by V. Marechal (Hôpital Rothschild, Paris, France). For indirect immunofluorescence antibody analyses, cells were fixed with cold acetone-methanol (2:1). Antibodies were diluted with phosphate-buffered saline containing 2.0% bovine serum albumin, 0.2% Tween 20, and 10% glycerol. EBNA-1 was stained with the EBNA-1 rat monoclonal antibody 2B4-1 or the negative control antibody KO, which were kindly provided by F. A. Grasser (Universitätskliniken, Homburg, Germany) and Texas red-conjugated donkey anti-rat immunoglobulin G (IgG) (Jackson). The proliferating cell nuclear antigen (PCNA) mouse monoclonal antibody PC10 (Santa Cruz Biotechnology), Texas red-conjugated donkey mouse IgG antibodies (Jackson), or FITC-conjugated rabbit mouse-IgG antibodies (DAKO) were used for PCNA staining. The mouse monoclonal antibody against the p32 (p34) subunit of human replication protein A (RP-A) (Ab-2) (Oncogene) was used for immunostaining of RP-A. β-galactosidase (Zymed) mouse monoclonal antibody was used as negative control in staining for PCNA and RP-A. The preparation of cell extracts and Western blot analyses were described previously (23). The following antibodies were used: EBNA-1 mouse monoclonal antibody OT1x given by Y. Middeldorp (Vrije Universiteit, Amsterdam, The Netherlands), the PCNA mouse monoclonal antibody PC10 (Santa Cruz Biotechnology), the GFP rabbit polyclonal antibody (Santa Cruz, SC8334), the RP-A monoclonal antibodies (Ab-2) (Oncogene), and the human lutenizing hormone mouse monoclonal antibody F476-22-5 (Mochida Pharmaceutical Co., Tokyo, Japan) as a negative control. Cell fractionation was done according to the method of Petit et al. (34) using micrococcal nuclease (Sigma). Confocal laser scanning microscopy (LSM), premature chromosome condensation (PCC) for live-cell observation, and Cy3-dUTP pulse-labeling were described previously (25). For flow cytometry cells were stained with Hoechst dye 33258, and cells were analyzed with a FACS Vantage SE analyzer (Becton Dickinson). GFP-EBNA1-expressing cells were sorted by flow cytometry through dual gating for GFP and for DNA content per cell.
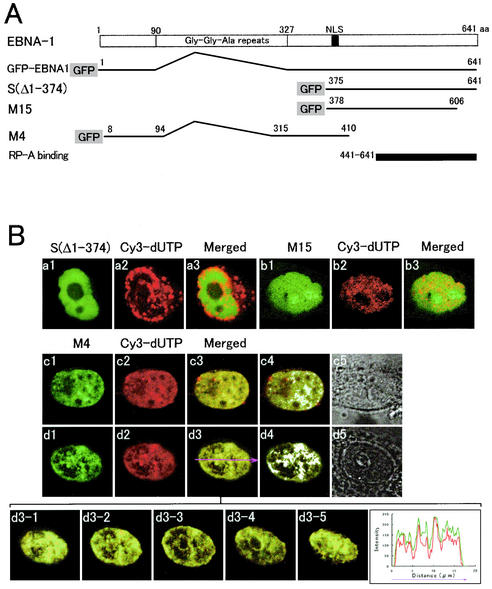
FIG. 3.
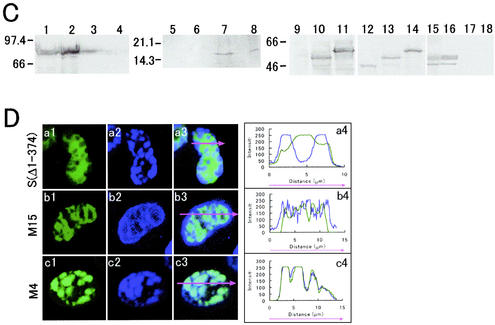
Expression of GFP-EBNA1 and the GFP-EBNA1 mutant proteins and LSM photographs of their localization profile and colocalization with Cy3-dUTP labeled foci. (A) Constructs of GFP-EBNA1, S(Δ1-374), M4, and M15. The domain of EBNA-1 that is essential for interaction with RP-A (48) is depicted on the bottom. (B) Confocal LSM profiles of S(Δ1-374) (a1), Cy3-dUTP pulse-labeling (a2), and their merged image (a3), and those of M15 (b1), Cy3-dUTP pulse-labeling (b2), and the merged image (b3) in living CHO-K1 cells expressing the GFP-EBNA1 mutants in S phase. Also shown are confocal LSM profiles of M4 (c1), Cy3-dUTP pulse-labeling (c2), and their merged image (c3) in a cell synchronously cultured in early S-phase. Also shown are the white spots containing high intensities of both GFP-EBNA1 (green) and Cy3-dUTP (red) selected by the LSM510 colocalization option (c4) and the Nomarski image (c5). Similar profiles of M4 (d1), Cy3-dUTP pulse-labeling (d2), and the merged image (d3) in a synchronously cultured middle-S-phase cell are also shown. Also shown are the images of the cell shown in panel d3 at serial focal planes through the nucleus vertically from the bottom to the top at 1.0 μm (d3-1 to d3-5), the white spots of high intensities of both GFP-EBNA1 and Cy3-dUTP selected by the colocalization option (d4), and the Nomarski image (d5); a graph drawn by the profile option along the horizontal arrow (d3) is also shown on the right. (C) Immunoblots of cell extracts and fractions from GFP-EBNA1-expressing DG75 cells using the EBNA-1 monoclonal antibody 0T1x are shown: cytoplasmic (lane 1), nucleoplasmic (lane 2), chromatin (lane 3), and nuclear matrix (lane 4) fractions. Also shown are immunoblots of the same cell extracts using 15% polyacrylamide gel containing sodium dodecyl sulfate and the histone monoclonal antibody; cytoplasmic (lane 5), nucleoplasmic (lane 6), chromatin (lane 7), and nuclear matrix (lane 8) fractions. Shown separately are immunoblots of M4 (lanes 9 and 12), M15 (lanes 10 and 13), and S(Δ1-374) (lanes 11 and 14) in extracts from cells expressing the GFP-EBNA1 mutant fusion proteins using the EBNA-1 monoclonal antibody OT1x (lanes 9 to 11) and the GFP rabbit polyclonal antibody (lanes 12 to 14), and immunoblots of M15 in cytoplasmic (lane 15), nucleoplasmic (lane 16), chromatin (lane 17), and nuclear matrix (lane 18) fractions using OT1x. (D) LSM profiles of S(Δ1-374), M15, and M4 in living interphase cells in which chromatin had been prematurely condensed by incubating the cells in the presence of calyculin A at 37°C for 60 min are shown in panels a1, b1, and c1, respectively. DNA profiles of these cells that were stained with Hoechst dye 33258 before the confocal LSM are shown in panels a2, b2, and c2, respectively. The merged images are shown in panels a3, b3, and c3, respectively. Graphs drawn by the LSM510 profile option along the magenta arrows that show the direction of profile (a3, b3, and c3) are shown in panels a4, b4, and c4, respectively.
A GFP-EBNA1 protein of 84 kDa was expressed in the absence of EBV plasmid DNAs in the CHO-K1 cell lines, in which chromosomes are clearly visible because of their small number (22). In this study, the interaction of EBNA-1 with components of replication foci was examined.
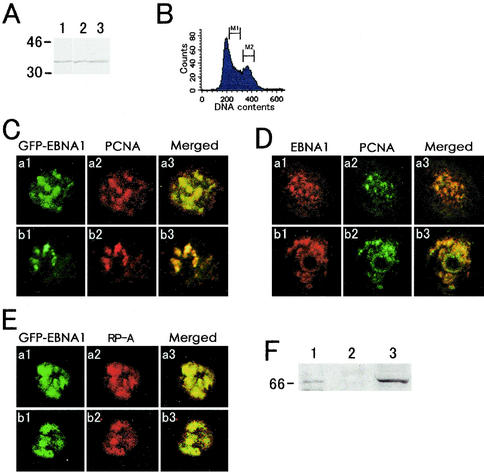
PCNA, a homotrimeric clamp at replication forks that has been well-studied by microscopic observations in CHO cells, is a widely used marker for replication foci (4-6, 27, 29, 36). GFP-EBNA1-expressing CHO-K1 cells were sorted by fluorescence-activated cell sorter, fixed, and immunostained for PCNA, using PCNA monoclonal antibody (Santa Cruz), which showed a single major band of PCNA in Western blots of CEW21N4, DG75, and Raji cells (Fig. 1A, lanes 1 to 3). The confocal microscopy analyses of PCNA and GFP-EBNA1 in the S-phase cells (Fig. 1B, M1 fraction) suggested that GFP-EBNA1 highly colocalized with PCNA foci (Fig. 1C). The PCNA foci were not observed in the G2- and M-phase cells (Fig. 1B, M2 fraction) (data not shown). We also examined the colocalization of the nonfused full-length EBNA-1 with PCNA in CEW21N4 cells by a double indirect immunofluorescence method, using the EBNA-1 and PCNA monoclonal antibodies. The merged images show that the full-length EBNA-1 protein also colocalizes with the PCNA foci (Fig. 1D). EBNA-1 and PCNA proteins, however, were not coimmunoprecipitated from Raji cell extracts using the anti-EBNA-1 antibody OT1x or the anti-PCNA mouse monoclonal antibody (data not shown).
FIG. 1.
Colocalization of spots of EBNA-1 with PCNA foci and with RP-A foci in S-phase cells. (A) Immunoblots of PCNA in lysates from DG75 (lane1), Raji (lane 2), and CEW21N4 (lane 3) cells using the PCNA mouse monoclonal antibody that was used for LSM. (B) Flow cytometry of GFP-EBNA1-expressing CHO-K1 cells. The cells were sorted by flow cytometry through dual gating for GFP and for DNA. M1 and M2 indicate S-phase cells and G2/M-phase cells, respectively. (C) Photomicrographs of GFP-EBNA1 and immunostained PCNA in GFP-EBNA1-expressing CHO-K1 cells in S phase, or in the M1 fraction shown in panel B, that had been sorted by flow cytometry. Profiles of two cells of GFP-EBNA1 (a1 and b1) and PCNA (as and b2), and the merged images (a3 and b3) are shown, respectively. (D) Double immunostaining of native EBNA-1 and PCNA proteins in CEW21N4 cells. Shown are EBNA-1 immunostained using the 2B4-1 anti-EBNA-1 antibody and Texas red-conjugated donkey anti-rat IgG antibodies (a1 and b1), PCNA immunostained using an anti-PCNA mouse monoclonal antibody and FITC-conjugated secondary antibody (a2 and b2), and the merged images (a3 and b3). (E) Photomicrographs of GFP-EBNA1 and immunostained RP-A in GFP-EBNA1-expressing CHO-K1 cells in S phase that had been sorted by flow cytometry. Profiles of GFP-EBNA1 in two fixed cells are shown (a1, and b1). These cells were immunostained using an anti-RP-A monoclonal antibody and Texas red-conjugated secondary antibody and are shown in panels a2 and b2, respectively. Merged images of a1 with a2 and of b1 with b2 are shown in panels a3 and b3, respectively. (F) Coimmunoprecipitation of EBNA-1 and RP-A. Raji cell extracts (310 μl) were incubated with the anti-RP-A mouse monoclonal antibody (lane 1) or with the negative control mouse monoclonal antibody F476-22-5 (lane 2), and the immunoprecipitates were analyzed by Western blot with the anti-EBNA-1 monoclonal antibody OT1x. The Western blot of EBNA-1 in 15 μl of the Raji cell extracts detected with the antibody OT1x is shown as a positive control in lane 3.
RP-A (hSSB) is another constituent of the cellular replication machinery (42) that exists in a soluble and chromatin-bound form in the nuclei (12). A high degree of colocalization between RP-A and PCNA at replication foci throughout S phase has been reported (12, 14). Confocal LSM showed that GFP-EBNA1 also colocalized with foci of RP-A in GFP-EBNA1-expressing S-phase CHO-K1 cells sorted by fluorescence-activated cell sorter (Fig. 1E). We next examined whether EBNA-1 bound RP-A by coimmunoprecipitation of cell lysates. RP-A was minimally coimmunoprecipitated with EBNA-1 from Raji cell extracts using the EBNA-1 monoclonal antibody OT1x (data not shown). EBNA-1 was clearly coimmunoprecipitated with RP-A from the Raji cell extracts using an RP-A mouse monoclonal antibody (Fig. 1F).
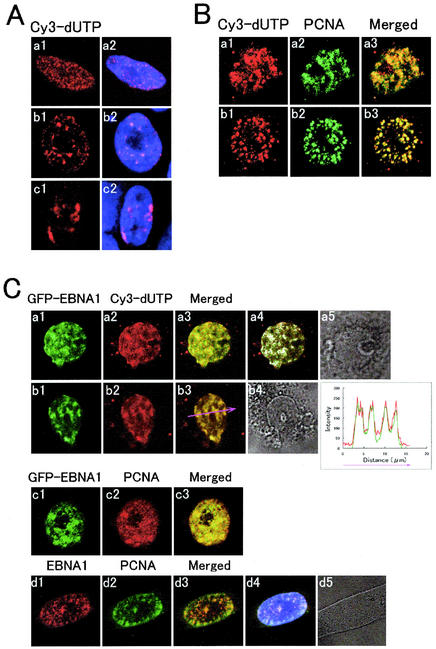
We pulse-labeled cellular DNAs in synchronously growing CHO-K1 cells in early (10 to 30 min), middle (3 to 4 h), and late (7 to 8 h) S phase with Cy3-dUTP (Fig. 2A). Patterns of Cy3-dUTP-labeled foci on the chromatin in the fixed cells varied throughout the nucleus. Foci that were typical of early S phase, i.e., DNA labeling patterns 1 and 2, as described in CHO cells by O'Keefe et al. (32), are shown in Fig. 2A, panels a. Middle S-phase foci that are larger, more discrete, and large patches or elongated areas, i.e., patterns 2 or 3, are shown in Fig. 2A, panels b. The foci that are larger in size and fewer in number, which are typical of late S phase or patterns 3, 4, and 5, are shown in Fig. 2A, panels c. Foci of PCNA colocalized precisely with the Cy3-dUTP-pulse-labeled foci as expected (Fig. 2B), confirming that the spots of PCNA or of pulse-labeled DNAs presented above are replication foci. In the cells that had been synchronized in early and middle S phase, many GFP-EBNA1 foci colocalized with both the Cy3-dUTP-labeled replication foci and the PCNA-immunostained replication foci (Fig. 2C, panels a, b, and c). We further examined whether nontagged full-length EBNA-1 showed the same colocalization profiles. In CEW21N4 cells that had been synchronized in early S phase, full-length EBNA-1 foci also colocalized with PCNA replication foci (Fig. 2C, panels d).
FIG. 2.
GFP-EBNA1 colocalized with replication foci identified by Cy3-dUTP pulse-labeling and immunostaining for PCNA in early-middle S phase. (A) LSM profiles of Cy3-dUTP-pulse-labeled replication foci in synchronously cultured fixed CHO-K1 cells in early (a1), middle (b1), and late (c1) S phase during the replication cycle. The merged images of the profiles of the Cy3-dUTP-labeled foci and Hoechst dye-stained DNAs are shown in panels a2, b2, and c2, respectively. (B) Confocal LSM profiles of Cy3-dUTP (a1 and b1), PCNA (a2 and b2), and the merged images (a3 and b3) of two different CHO-K1 cells that had been synchronously cultured, Cy3-dUTP pulse-labeled, acetone-methanol fixed, and then immunostained for PCNA using the anti-PCNA mouse monoclonal antibody and the FITC-conjugated rabbit anti-mouse IgG antibodies are shown. The cell shown in panels a1, a2, and a3 is in early S phase, and the other one shown in panels b1, b2, and b3 is in middle S phase. (C) Confocal LSM profiles of GFP-EBNA1, Cy3-dUTP pulse-labeling, and the merged image of living GFP-EBNA1-expressing and synchronously cultured early-middle S-phase CHO-K1 cells. Shown are profiles of GFP-EBNA1 (a1), Cy3-dUTP pulse-labeling (a2), and the merged image (a3) and the Nomarski image (a5) of a living GFP-EBNA1-expressing and synchronously cultured early S-phase cell. The white spots selected by the LSM150 colocalization option indicate those in which the intensities of GFP-EBNA1 (green) and Cy3-dUTP (red) were both high (a4). The images in panels b show a middle-S-phase cell: GFP-EBNA1 (b1), Cy3-dUTP pulse-labeling (b2), and the merged image (b3) and the Nomarski image (b4). To the right of panel b4 is a graph drawn by the LSM510 profile option along the magenta arrow (b3), which shows the direction of profile. Also shown are profiles of GFP-EBNA1 (c1), PCNA (c2), and the merged image (c3) in a synchronously cultured early-S-phase GFP-EBNA1-expresssing cell and profiles of nontagged full-length EBNA-1 immunostained with the monoclonal antibody 2B4-1 (d1), PCNA (d2), the merged image (d3), the merged image overlapped with Hoechst dye 33258 profile (d4), and the Nomarski image (d5) in a synchronously cultured early-S-phase CEW21N4 cell.
The mutant S(Δ1-374), which is deficient in binding to mitotic chromosomes (Fig. 3A; Fig. 3C lanes 11 and 14), and the M15 mutant (Fig. 3A; Fig. 3C, lanes 10 and 13) were both distributed in a diffuse fashion in the nuclei of synchronously cultured S-phase cells, in contrast to GFP-EBNA1 (Fig. 3B, panels a and b). By contrast, the mutant M4 (Fig. 3A; Fig. 3C lanes 9 and 12), appeared in the forms of spots and colocalized with the foci identified by Cy3-dUTP-pulse-labeling (Fig. 3B, panels c and d) and PCNA immunostaining (data not shown), in synchronously cultured early and middle S-phase cells as well as GFP-EBNA1. These results indicate that the localization of GFP-EBNA1 on cellular replication foci requires the EBNA-1 regions of aa 8 to 94 and/or aa 315 to 410, but not the region aa 441 to 641, which was shown to interact directly with RP-A (48).
GFP-EBNA1 was recovered in the interphase histone-containing chromatin fractions that were identified on Western blots using a histone monoclonal antibody (Chemicon) (7) (Fig. 3C, lanes 1 to 8). In contrast to GFP-EBNA1, the M15 mutant that did not colocalize with cellular replication foci was not detected in the interphase chromatin fraction (Fig. 3C, lane 17). To confirm these results, taking into consideration the well-known difficulty of biochemically isolating intact chromatin (9), we performed chemical PCC, which is calyculin A-induced condensation of interphase chromatin (22). We recently have shown that the GFP-EBNA1 protein binds to interphase chromatin by the PCC method (22). Neither the M15 nor the S(Δ1-374) mutant protein was associated with prematurely condensed interphase chromatin, whereas the M4 mutant protein was associated with the prematurely condensed chromatin (Fig. 3D). These results indicate that the domain that is essential for EBNA-1 colocalization with replication foci and the one that is required for binding to interphase chromatin are both located in aa 8 to 94 and/or aa 315 to 410.
The key findings in this report are that EBNA-1 colocalizes with the microscopically detected cellular replication foci or replication factories (8, 10, 19, 20, 27, 31), in the absence of EBV plasmids. This colocalization depends on the EBNA-1 region that includes the domains essential for interactions with p32 (41), EBNA1-binding protein 2 (EBP2) (39) and mitotic chromosome (30), and the domain for transactivation (35, 45).
EBNA1 proteins were highly colocalized with the PCNA foci in S-phase cells in the absence of oriP plasmid (Fig. 1). Replication foci containing PCNA are detected only in S phase (11-13, 27) and are heterogeneous during their lifetime (27). Although we have shown that EBNA1 colocalizes with PCNA in cellular replication foci, EBNA-1 was not coimmunoprecipitated with PCNA. This is in agreement with the report that human PCNA does not interact with the C-terminal 200 aa of EBNA-1 (48).
The RP-A heterotrimeric complex stabilizes the single-stranded DNA and is required at the replication fork (40). RP-A interacts with simian virus 40 T-antigen and the E2 protein of bovine papillomavirus (17), both of which share some structural and functional similarity with EBNA-1 (16). A surface plasmon resonance study (48) showed that the carboxyl region (aa 441 to 641) of EBNA-1 interacted directly with the 70-kDa subunit of RP-A. The results in this study have suggested that EBNA-1 colocalizes, and possibly interacts, with RP-A in cellular replication foci. However, the confocal LSM of the EBNA-1 mutants has suggested that the RP-A-interaction domain of EBNA-1 is not necessary for the colocalization with cellular replication foci.
Another EBNA-1-interacting cellular protein, EBP2, is important for stable segregation of EBV plasmids (39, 43), but EBP2 is not necessary for the replication of EBV episomes (39, 43). EBNA-1 has recently been shown to be associated with the origin recognition complex in vivo (38). EBNA-1 may colocalize with cellular replication foci through possible interaction with prereplication complexes (3) that contain the origin recognition complex.
Given the oriP-binding property of EBNA-1 (15, 16, 18, 21, 44) and the colocalization of EBNA-1 with oriP plasmids in the form of dots in the nuclei (25), the EBNA-1 proteins concentrated at cellular replication focus areas might bring EBV plasmids close to the cellular active replication sites and facilitate the replication and maintenance of EBV plasmids in concert with cellular DNAs and under the strict control of the cell cycle. Thus, the association of the EBNA-1 with the cellular DNA replication focus area in the absence of the viral plasmids suggests a new strategy for the long-lasting virus-cell interactions.
Acknowledgments
We thank Friedrich A. Grasser, Universitätskliniken, for the anti-EBNA-1 rat monoclonal antibody 2B4-1; Yaap Middeldorp, Vrije Universiteit, for the anti-EBNA-1 mouse monoclonal antibody OT1x; and Vincent Marechal, Hôpital Rothschild, for the GFP-fusions M4 and M15.
Financial support for this research was provided by a grant-in-aid from the Ministry of Health and Welfare.
REFERENCES
- 1.Avolio-Hunter, T. M., and L. Frappier. 1998. Mechanistic studies on the DNA linking activity of Epstein-Barr nuclear antigen 1. Nucleic Acids Res. 26:4462-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 3.Bogan, J. A., D. A. Natale, and M. L. Depamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 4.Bravo, R. 1986. Synthesis of the nuclear protein cyclin (PCNA) and its relationship with DNA replication. Exp. Cell Res. 163:287-293. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, R., and H. Macdonald-Bravo. 1985. Changes in the nuclear distribution of cyclin (PCNA) but not its synthesis depend on DNA replication. EMBO J 4:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo, R., and H. Macdonald-Bravo. 1987. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J. Cell Biol. 105:1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brimmell, M., J. S. Burns, P. Munson, L. McDonald, M. J. O'Hare, S. R. Lakhani, and G. Packham. 1999. High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. Br. J. Cancer 81:1042-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, P. R. 1999. The organization of replication and transcription. Science 284:1790-1795. [DOI] [PubMed] [Google Scholar]
- 9.Cook, P. R. 2001. Principles of nuclear structure and function. Wiley-Liss, New York, N.Y.
- 10.DePamphilis, M. L. 1999. Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21:5-16. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrova, D. S., and D. M. Gilbert. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4:983-993. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., and D. M. Gilbert. 2000. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp. Cell Res. 254:321-327. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrova, D. S., and D. M. Gilbert. 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat. Cell Biol. 2:686-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frappier, L., and M. O'Donnell. 1991. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J. Biol. Chem. 266:7819-7826. [PubMed] [Google Scholar]
- 16.Fujita, T., M. Ikeda, S. Kusano, M. Yamazaki, S. Ito, M. Obayashi, and K. Yanagi. 2001. Amino acid substitution analyses of the DNA contact region, two amphipathic alpha-helices and a recognition-helix-like helix outside the dimeric beta-barrel of Epstein-Barr virus nuclear antigen 1. Intervirology 44:271-282. [DOI] [PubMed] [Google Scholar]
- 17.Han, Y., Y. M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hearing, J., Y. Mulhaupt, and S. Harper. 1992. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J. Virol. 66:694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hozak, P., A. B. Hassan, D. A. Jackson, and P. R. Cook. 1993. Visualization of replication factories attached to nucleoskeleton. Cell 73:361-373. [DOI] [PubMed] [Google Scholar]
- 20.Hozak, P., D. A. Jackson, and P. R. Cook. 1994. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci. 107:2191-2202. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, N., S. Harada, T. Honma, T. Kitamura, and K. Yanagi. 1991. The domain of Epstein-Barr virus nuclear antigen 1 essential for binding to oriP region has a sequence fitted for the hypothetical basic-helix-loop-helix structure. Virology 182:84-93. [DOI] [PubMed] [Google Scholar]
- 22.Ito, S., E. Gotoh, S. Ozawa, and K. Yanagi. 2002. Epstein-Barr virus nuclear antigen-1 is highly colocalized with interphase chromatin and its newly replicated regions in particular. J. Gen. Virol. 83:2377-2383. [DOI] [PubMed] [Google Scholar]
- 23.Ito, S., M. Ikeda, N. Kato, A. Matsumoto, Y. Ishikawa, S. Kumakubo, and K. Yanagi. 2000. Epstein-barr virus nuclear antigen-1 binds to nuclear transporter karyopherin alpha1/NPI-1 in addition to karyopherin alpha2/Rch1. Virology 266:110-119. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, W. Q., V. Wendel-Hansen, A. Lundkvist, N. Ringertz, G. Klein, and A. Rosen. 1991. Intranuclear distribution of Epstein-Barr virus-encoded nuclear antigens EBNA-1, -2, -3 and -5. J. Cell Sci. 99:497-502. [DOI] [PubMed] [Google Scholar]
- 25.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusano, S., K. Tamada, H. Senpuku, S. Harada, S. Ito, and K. Yanagi. 2001. Epstein-Barr virus nuclear antigen-1-dependent and -independent oriP-binding cellular proteins. Intervirology 44:283-290. [DOI] [PubMed] [Google Scholar]
- 27.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madsen, P., and J. E. Celis. 1985. S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication? FEBS Lett. 193:5-11. [DOI] [PubMed] [Google Scholar]
- 30.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Keefe, R. T., S. C. Henderson, and D. L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116:1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 35.Polvino-Bodnar, M., J. Kiso, and P. A. Schaffer. 1988. Mutational analysis of Epstein-Barr virus nuclear antigen 1 (EBNA 1). Nucleic Acids Res. 16:3415-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raska, I., K. Koberna, M. Jarnik, V. Petrasovicova, J. Bednar, K. Raska, Jr., and R. Bravo. 1989. Ultrastructural immunolocalization of cyclin/PCNA in synchronized 3T3 cells. Exp. Cell Res. 184:81-89. [DOI] [PubMed] [Google Scholar]
- 37.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 38.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 15:18-29. [DOI] [PubMed] [Google Scholar]
- 42.Wold, M. S. 1997. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 43.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates, J. L. 1996. Epstein-Barr virus DNA replication, p. 751-774. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Yates, J. L., and S. M. Camiolo. 1988. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 46.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, D., L. Frappier, E. Gibbs, J. Hurwitz, and M. O'Donnell. 1998. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 26:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]