Abstract
Sendai virus (SeV) C protein functions as an interferon (IFN) antagonist and renders cells unresponsive to both alpha/beta IFN (IFN-α/β) and IFN-γ. We have recently found the physical association of the C protein with signal transducer and activator of transcription 1 (STAT1) in infected cells. However, involvement of the C-STAT1 interaction in the blockade of IFN signaling has remained unclear. We generated here a series of C mutant proteins that retained or lost the STAT1-binding capacity and examined their effects on IFN-α signaling. All of the C mutant proteins with no STAT1-binding capacity lost the ability to inhibit the IFN-α response. In contrast, the C mutant proteins retaining the STAT1-binding capacity suppressed IFN-α-stimulated tyrosine phosphorylation of both STAT2 and STAT1 to various degrees. Remarkably, their anti-IFN-α capacities correlated well with the inhibitory effect on phosphorylation of STAT2 rather than STAT1. In infected cells, the levels of tyrosine-phosphorylated (pY) STAT2 were below the detection level irrespective of duration of IFN-α stimulation, whereas the levels of pY-STAT1 strikingly increased after long-term IFN-α stimulation. These results suggest that the STAT2 activation process is a crucial target for the blockade of IFN-α signaling. An in vitro binding assay with extracts from (STAT1-deficient) U3A and (STAT1-expressing) U3A-ST1 cells suggested the requirement of STAT1 for the C-STAT2 interaction. Furthermore, expression of STAT1 enhanced the inhibitory effect of the C protein on STAT2 activation in U3A cells. The C protein thus appears to participate in the inhibitory process for STAT2 activation through the STAT1 interaction.
All members of the Paramyxovirinae have retained the open reading frame (ORF) for either or both of the accessory proteins, V and C, within the P gene during evolution (26). This finding had suggested crucial roles of the V and C proteins in a virus life cycle, although their roles had remained an enigma for a long time. Several lines of evidence have accumulated, however, demonstrating that the accessory proteins form a group of antagonists against the host immune system (9, 15-17). The Paramyxovirinae family contains three genera: Rubulavirus, Respirovirus, and Morbillivirus. The V protein of rubulavirus simian virus 5 targets a key factor, signal transducer and activator of transcription 1 (STAT1), on interferon (IFN) signaling for proteasome-mediated degradation, thereby inhibiting IFN signal transduction (1, 3, 6, 7, 33, 35, 48, 49). The V proteins of other rubulaviruses, including human parainfluenza virus type 2, mumps virus, and simian virus 41 inhibit IFN signaling likewise by inducing a decrease in the STAT1 or STAT2 level (8, 25, 30, 31, 34, 35, 47, 49). In contrast, the respirovirus Sendai virus (SeV), which possesses both V and C ORFs, has evolved functions of the C protein instead of the V protein so as to block IFN signaling (12, 18). The SeV C ORF produces a nested set of four C proteins, C′, C, Y1, and Y2, which are referred to collectively as the C proteins (5, 14). Translation of C′, C, Y1, and Y2 initiates at different positions (81ACG, 114AUG, 183AUG, and 201AUG, respectively) and terminates at the same position (UAA728). In addition to the C protein, the shorter forms, Y1 and Y2, have the ability to inhibit IFN signaling (11, 22). The C protein, however, does not lead to degradation of any component on the signaling pathway in most cell types (12, 18, 24, 42, 49) except for NIH 3T3 mouse embryo fibroblast (MEF) cells (10, 11). The purpose of the present study was to better understand how the SeV C protein inhibits IFN-α signaling without degrading cellular proteins on the signaling pathway.
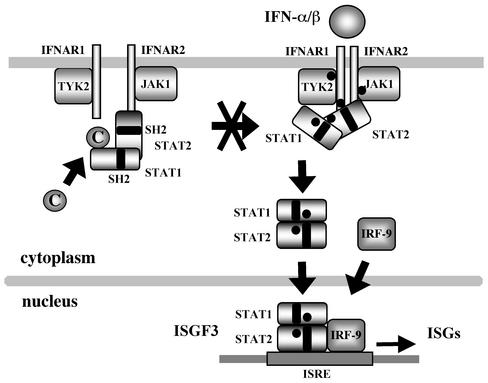
The main pathway of IFN-α/β signaling consists of several components, IFN-α/β receptor subunits (IFNAR1 and IFNAR2), receptor associated kinases (JAK1 and TYK2), two STATs (STAT1 and STAT2), and IFN regulatory factor 9 (p48) (41). Both STAT1 and STAT2 preassociate with the cytoplasmic tail of IFNAR2 in IFN-untreated cells (28). Binding of IFN-α/β to IFN receptor leads to aggregation of IFNAR1 and IFNAR2, causing the cross-activation of TYK2 and JAK1 (32). TYK2 then phosphorylates IFNAR1 on Tyr466 (4), which serves as the docking site for the SH2 domain of STAT2 (45). STAT2 binds to the docking site, followed by the phosphorylation of both STAT2 and STAT1. A current model proposed sequential activation of STAT2 and STAT1 in this order (28). The tyrosine-phosphorylated (pY) STAT2-STAT1 heterodimer then translocates into the nucleus and combines IFN regulatory factor 9 (43) to form IFN-stimulated gene factor 3 (ISGF3) and activates transcription of IFN-stimulated genes (ISGs) by binding to IFN-stimulated response elements (ISREs). Two forms of STAT1—STAT1α and STAT1β—are synthesized in cells. STAT1β, which lacks the COOH-terminal 38 amino acids of STAT1α, is a product of differential splicing (37, 39).
The first important finding regarding a molecular basis for the SeV blockade of IFN signaling was the suppression of tyrosine phosphorylation of STATs, including STAT1, STAT2, and STAT3, in response to short-term (30-min) IFN-α stimulation at the early phase of infection (at 2 h postinfection [p.i.]) (24). This suppression for STAT1 was also observed at the middle phase of infection (42). Long-term IFN-α stimulation, however, caused notable elevation of the pY-STAT1 level, demonstrating incompleteness of the suppression of STAT1 activation (23). Thus, the suppressive effect on STAT1 activation may contribute to the blockade of IFN-α/β signaling but cannot explain entirely the blocking mechanism. It was also reported that serine phosphorylation of STAT1 was suppressed in infected cells (23, 49). However, this cannot account for the mechanism as well, since serine phosphorylation of STAT1 is not required for transactivation function of ISGF3 (44). Another important finding is the association of the C protein with STAT1 in infected cells (42). Remarkably, under low-salt conditions, the C protein and STAT1 form a high-molecular-mass complex. It is likely that this C-STAT1 interaction is involved in the inhibition of IFN signaling, but no evidence has been provided so far for this hypothesis.
The present study was thus designed to explore roles of the STAT1-C interaction in the blockade of IFN-α signaling. To do this, we generated several C mutant proteins that retained or lost the STAT1-binding capacity. We show here a definite correlation among STAT1-binding capacity, the inhibitory activity for IFN-α signaling, and the inhibitory effect on STAT2 activation. Although all of the C mutant proteins that could bind to STAT1 suppressed activation of both STAT1 and STAT2 to greater or lesser degrees, the ability to inhibit the IFN-α response correlated well with the suppressive effect on STAT2 activation rather than STAT1 activation. In infected cells, tyrosine phosphorylation of STAT2 was indeed inhibited under any condition examined. From these results, we conclude that STAT2 activation process is a crucial target for SeV blocking mechanism for IFN-α signaling. The present study further provides evidence for participation of the C protein in this inhibitory process via the interaction with STAT1.
MATERIALS AND METHODS
Cells and virus. HeLa and U3A (STAT1-deficient) cells were maintained in Dulbecco minimum essential medium supplemented with 10% fetal calf serum. SeV (SeVpB) was propagated as described previously (46).
Plasmid constructs.
DNA fragments encoding D1 (amino acids [aa] 85 to 204), D2 (aa 127 to 204), and D3 (aa 30 to 126), in addition to C (aa 1 to 204), Y1 (aa 24 to 204), and Y2 (aa 30 to 204) (Fig. 1A), were amplified by PCR by using the cDNA clone H-33 (38) (a gift from T. Shioda) as a template DNA. The forward primers CF (5′-AAGCTTBamHIggatccAGGCC114ATGCCTTCATTCTTAAAG-3′), Y1F (5′-AAGCTTBamHIggatccAGGCC183ATGTTATCGGATTCCTCG-3′), Y2F (5′-AAGCTTBamHIggatccAGGCC201ATGCTGTCCTGTCGAGTG-3′), D1F (5′-AAGCTTBamHIggatccAGGCCATG370GAGTCTCTGGGAGAACAAG-3′), and D2F (5′-AAGCTTBamHIggatccAGGCC493ATGGAGGAGACTCCGGAATC-3′) and the reverse primer CR (5′-AAGCTTMluIacgcgtCTA728TTACTCTTGCACTATGTG-3′) were used for the PCRs for C, Y1, Y2, D1, and D2. Italics indicate nucleotide sequences that are not involved in ORFs. Restriction enzyme recognition sites are in lowercase. The D3 fragment was amplified with the Y2F primer and another reverse primer, D3R (5′-AAGCTTMluIacgcgtCTATTA491CGCCCAGATCCTGAGATACAG-3′). After digestion with BamHI and MluI, the PCR-amplified DNAs were once cloned into a corresponding site of the multiple cloning site in the Tet-off expression plasmid pTRE2 (Clontech). To generate a DNA fragment (C/FS) encoding the C protein with an amino acid substitution (Phe170 to Ser), a single point mutation (622T to C) was introduced by two-step PCR-based overlap primer extension as described previously (19). Briefly, two DNA fragments with overlapping ends were generated from the pTRE2-C plasmid DNA as a template by using two sets of primer pairs (pTRE2-forward primer [5′-TTAGTGAACCGTCAGATCGC-3′] and CMR [5′-632CCTCTTCTGGGAGTCCTTCAG612-3′]; CMF [5′-612CTGAAGGACTCCCAGAAGAGG632-3′] and pTRE2-reverse primer [5′-CTCACCCTGAAGTTCTCAGC-3′]). The mutations at position 622 in the complementary primers, CMR and CMF, are underlined. The two fragments were combined in the subsequent fusion reaction, digested with BamHI and MluI, and inserted into the corresponding site in pTRE2. pTRE2-C, -Y1, -Y2, -D1, -D2, -D3 and -C/FS were further digested with BamHI and NotI. The C mutant DNA fragments generated by this digestion were then inserted into the corresponding site in pGEX-5X-1 (Amersham Pharmacia Biotech) for production of glutathione S-transferase (GST) fusion proteins in Escherichia coli and in pEFneo vector (2) (a gift from H. Asao) for expression in mammalian cells. To append the RGS-His epitope (RGSH6) tag to the N terminus of the C mutant proteins, two oligonucleotides (5′-TATGAGAGGATCGCATCACCATCACCATCACGG-3′ and 5′-GATCCCGTGATGGTGATGGTGATGCGATCCTCTCATAGTAC-3′) were annealed and inserted into the KpnI and BamHI sites in the pEFneo-C mutant protein plasmids created above.
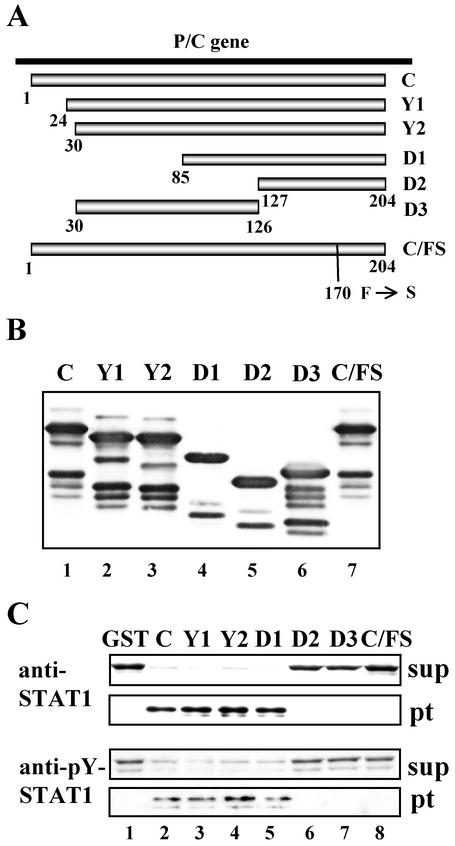
FIG. 1.
C mutant proteins expressed as GST fusion proteins and their STAT1-binding capacities. (A) Schematic diagram of constructs of C, Y1, Y2, D1, D2, D3, and C/FS; (B) Western blot analysis of purified GST-C, -Y1, -Y2, -D1, -D2, -D3, and -C/FS. The purified GST-C mutant fusion proteins were subjected to SDS-10% PAGE and detected by Western blot analyses with anti-C antibody. (C) Binding assay with GST-C mutant proteins. HeLa cells were treated with IFN-α for 1 h and then lysed. The total extracts (∼150 μg) were incubated with GST-C mutant fusion proteins (∼2 μg) conjugated to glutathione-Sepharose beads for 1 h. Each of the mixtures was divided by centrifugation into supernatant (sup) and pellet (pt) fractions. The beads were washed three times with the extraction buffer. Proteins in the supernatant and pellet fractions were separated by SDS-7.5% PAGE, transferred to a nitrocellulose membrane, and detected by Western blot analysis with anti-STAT1 (sc-464) or anti-pY-STAT1 (no. 9171) antibody.
Western blot analysis and EMSA.
Western blot analysis was performed as described previously (18) with anti-pY-STAT1 (no. 9171) (New England Biolabs); anti-pY-STAT2 (no. 07-224; Upstate Biotechnology); anti-STAT1 (sc-346 and sc-464), anti-STAT2 (sc-476), anti-JAK1 (sc-295), anti-TYK2 (sc-169), anti-IFNAR1 (sc-845), and anti-IFNAR2 (sc-704; Santa Cruz Biotechnology); anti-RGS-His (no.34610) antibody (Qiagen); or anti-C rabbit serum (20) (a gift from A. Kato and Y. Nagai). Electrophoretic mobility shift assay (EMSA) was performed with a 32P-labeled ISRE probe under conditions described previously (42). To generate a 32P-labeled ISRE probe, the 5′ terminus of the annealed synthetic DNAs (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′ and 5′-GATCGGCTTCAGTTTCGGTTTCCCTTTCCC-3′) derived from the ISRE of the human ISG15 gene was labeled with [γ-32P]ATP (222 TBq/mmol; Amersham Pharmacia Biotech) by using T4 bacteriophage polynucleotide kinase.
Purification of GST-C mutant fusion proteins expressed in E. coli.
GST-C mutant fusion proteins were expressed in E. coli (JM109 or BL21) transformed with the pGEX-C mutant plasmids by adding IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM. The cells in a sonication buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]) containing protease inhibitor cocktail (Sigma) were lysed by ultrasonic treatment. After the addition of Triton X-100 (final concentration, 1%), insoluble materials were removed by centrifugation. The lysates were then mixed with glutathione-Sepharose beads (Amersham Pharmacia Biotech) and incubated at 4°C for 1 h. After four washes with phosphate-buffered saline containing 0.5% Triton X-100, GST fusion proteins were eluted by adding the sonication buffer containing 14 mM glutathione. The eluate was dialyzed overnight in 50 mM Tris-HCl (pH 7.5)-5 mM MgCl2-100 mM NaCl-10% glycerol-5 mM 2-mercaptoethanol and stored at −80°C until use.
Preparation of a mammalian cell extract.
Cells were lysed in an extraction buffer (10 mM HEPES [pH 7.9], 300 mM NaCl, 0.25% NP-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM DTT) containing protease inhibitor cocktail (Sigma) as described previously (42). After centrifugation, the clarified supernatant was stored as the total extract at −80°C.
Binding assay with GST-C mutant fusion proteins or using His-C mutant proteins.
The GST-C mutant fusion protein was mixed with glutathione-Sepharose beads and incubated for 1 h with gentle rotation. After centrifugation, the beads were further incubated with cell extracts at 4°C for 1 h. The beads were then washed four times with the extraction buffer. Bound proteins were eluted by adding sodium dodecyl sulfate (SDS)-gel loading buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and analyzed by Western blotting. For binding assay with Ni-nitrilotriacetic acid (NTA) beads and His-C mutant proteins, the established cell lines were lysed in a lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 0.25% NP-40, 5 mM 2-mercaptoethanol) containing protease inhibitor cocktail. After centrifugation, the clarified lysates (250 μg) were incubated with Ni-NTA beads at 4°C for 1 h. The beads were then washed four times with the lysis buffer containing 20 mM imidazole. Bound proteins were eluted in the lysis buffer containing 250 mM imidazole and separated by SDS-7.5% PAGE for Western blot analyses.
Establishment of HeLa cell clones that constitutively express each of mutant C proteins and a U3A-ST1 clone that expresses hemagglutinin (HA)-tagged STAT1α.
To obtain cell lines expressing each of His-C mutant proteins, HeLa cells were transfected with either pEFneo-His-C, pEFneo-His-Y1, pEFneo-His-Y2, pEFneo-His-D1, pEFneo-His-D2, pEFneo-His-D3, or pEFneo-His-C/FS by using PolyFect transfection reagent (Qiagen) according to the manufacturer's protocol. At 3 days after transfection, media were replaced with those containing 0.8 mg of G418/ml. When colonies resistant to G418 became visible, 30 to 50 colonies were transferred into 24-well plates. About 30 clones per each transfection were tested for expression of the C mutant proteins by Western blot analysis with anti-RGS-His antibody. Three independent clones that exhibited high expression levels were stored in liquid nitrogen. To generate U3A-ST1 that expresses HA-tagged STAT1α, U3A cells were doubly transfected with pEF-BOS-HA-STAT1 (29) (a gift from T. Hirano) and pcDNA3 (Invitrogen). A clone (U3A-ST1) that expresses HA-STAT1α was isolated from G418-resistant colonies as described above.
Reporter gene assay.
Cells in a 24-well plate were transfected with pISRE-TA-Luc (0.5 μg; Clontech) together with pRL-TK-luc (0.03 μg; Clontech) by using Polyfect transfection reagent. At 20 h posttransfection, medium was replaced with fresh medium or medium supplemented with human recombinant IFN-α-2a (1,000 IU/ml; Takeda Chemical Industries, Osaka, Japan) and incubated further for 6 h. Luciferase activities in the cell lysates were measured by dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol. The relative activity was expressed by a ratio of firefly luciferase activity to renilla luciferase activity.
RESULTS
The C-terminal half of the C protein is responsible for the interaction with STAT1.
We have recently found that the C protein physically associates with STAT1 in infected cells (42). However, several questions have remained unanswered. What is the region responsible for the interaction with STAT1? Does the C protein interact with the other components on the signaling pathway? Is the C-STAT1 interaction really involved in the blockade of IFN-α signaling? To answer these questions, four C mutant proteins in addition to C, Y1, and Y2 were generated as GST fusion proteins (Fig. 1A and B). The D1 and D2 were N-terminally truncated Y2 fragments encompassing aa 85 to 204 and aa 127 to 204, respectively. The D3 was a C-terminally truncated Y2 fragment encompassing aa 30 to 126. C/FS was the C protein with a single amino acid substitution (Phe170 to Ser). It was previously reported that this mutation resulted in loss of the anti-IFN ability (12). The GST-C mutant fusion proteins were coupled to glutathione-Sepharose beads and incubated with extracts from IFN-α-treated HeLa cells. Each of the mixtures was divided into two fractions, supernatant and pellet fractions, by centrifugation. Proteins in the two fractions were then separated by the SDS-PAGE, followed by Western blot analyses with anti-STAT1 or anti-pY-STAT1 antibody. As shown in Fig. 1C, not only C but also Y1, Y2, and D1 could efficiently bind to both STAT1 and pY-STAT1 without the aid of the other viral proteins (Fig. 1C, lanes 2 to 5), whereas D2, D3, and C/FS could not (Fig. 1C, lanes 6 to 8). The binding did not appear to depend on the phosphorylation status of STAT1 because STAT1 was pulled down with similar efficiency from even IFN-untreated cell extracts (data not shown). These results demonstrated that the C-terminal half (aa 85 to 204) was responsible for the STAT1 interaction and further suggested that Phe170 at the C-terminal half was an important residue for maintaining the integrity of the STAT1 binding structure.
Establishment of HeLa cell lines that constitutively express each of the C mutant proteins.
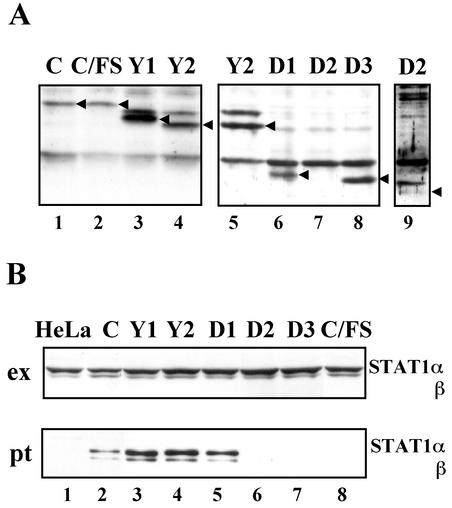
To examine effect of these C mutant proteins on IFN-α signaling, HeLa cell clones constitutively expressing each of the mutant C proteins were isolated as described in Materials and Methods. To compare their expression levels to one another, the RGS-His epitope tag was appended to the N terminus of these C mutant proteins. The expression level of D2 was extremely low (Fig. 2A, lanes 7 and 9), possibly due to its lability. Y1 was expressed the largest in amount (Fig. 2A, lane 3), whereas the levels of C and C/FS were the lowest of all except for that of D2 (Fig. 2A, lanes 1 and 2). The expression levels of the other mutant proteins were comparable to one another (Fig. 2A, lanes 4 to 6 and lane 8). Expression of these C mutant proteins did not affect the STAT1 levels (Fig. 2B, ex), confirming that the C protein did not target STAT1 for degradation. RGS-His-tagged proteins expressed in these cell lines were precipitated by using Ni-NTA agarose beads. STAT1 was coprecipitated with the C, Y1, Y2, and D1 (Fig. 2B, pt, lanes 2 to 5), indicating that the C, Y1, Y2, and D1 interacted with STAT1 in the established cell lines.
FIG. 2.
Establishment of HeLa cell lines, which constitutively express either of C mutant proteins. (A) Extracts (40 μg) from the established cell lines were subjected to SDS-13% PAGE, followed by Western blot analyses with anti-RGS-His antibody. Arrowheads indicate positions of C, Y1, Y2, D1, D2, and D3 proteins. D2 was detected only after long exposure (lane 9). (B) Binding assay with Ni-NTA beads and extracts from the established cell lines. The cells were lysed in the extraction buffer. Proteins in the extracts (ex) and those bound to Ni-NTA beads (pt) were separated by SDS-7.5% PAGE, followed by Western blot analyses with anti-STAT1 (sc-464) antibody.
C-STAT1-binding is required for the inhibitory effect on the IFN-α response.
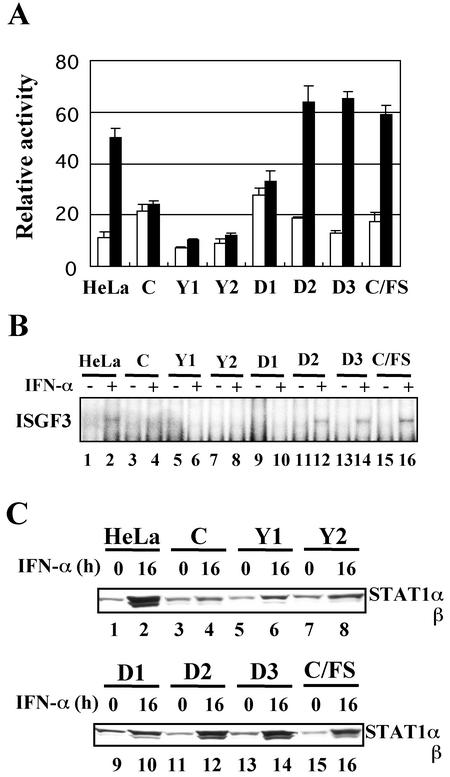
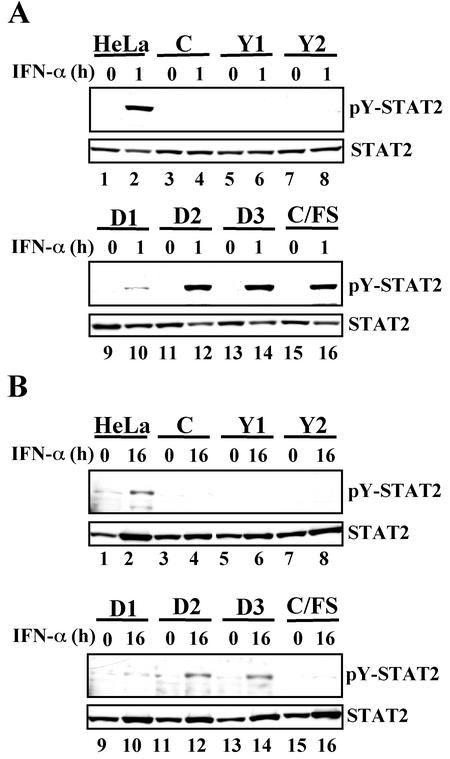
We next examined effects of these mutant C proteins on the IFN-α response. To do this, we used a reporter plasmid, pISRE-TA-luc, in which the firefly luciferase gene is driven under the control of an IFN-α-responsive promoter. The cells were transfected with pISRE-TA-luc and an internal control plasmid pRL-TK-luc, which contains the herpes simplex virus thymidine kinase promoter upstream of the renilla luciferase gene. At 20 h posttransfection, the cells were either treated or mock treated with IFN-α for 6 h. Relative levels of promoter activation were expressed as a ratio of firefly luciferase activity to renilla luciferase activity. In the cell line expressing either C, Y1, Y2, or D1 the activation of an IFN-α-responsive promoter was suppressed (Fig. 3A). In contrast, the cell line expressing either D2, D3, or C/FS responded normally to IFN-α stimulation (Fig. 3A). Similar results were obtained from transient-expression experiments, in which the C mutant proteins without RGS-His tag were transiently expressed by transfection of HeLa cells with each of pEF-RGS-His tag minus C mutant plasmids, together with pISRE-TA-luc and pRL-TK-luc (data not shown). Thus, RGS-His tag did not affect the capability of inhibiting the IFN-α response. Second, ISGF3 formation was examined in these cell lines by EMSA (Fig. 3B). The cells were treated or mock treated with IFN-α for 1 h and then lysed. The extracts were then subjected to EMSA with a 32P-labeled ISG15 ISRE probe. As was expected, the ISGF3 formation was not observed in the cells expressing either C, Y1, Y2, or D1 (Fig. 3B, lanes 3 to 10) but seen in the cells expressing either D2, D3, or C/FS (Fig. 3B, lanes 11 to 16). These results suggested that the C-STAT1 binding was required for the inhibitory effect on the IFN-α response. STAT1 is not only a key factor on IFN signaling but also one of ISG products (27). As shown in Fig. 3C, IFN-α-mediated elevation of the STAT1 level was suppressed in the C-, Y1-, Y2-, and D1-expressing cells (Fig. 3C, lanes 3 to 10), supporting the conclusion presented above. It should be noted that the STAT1 level at 16 h after IFN-α stimulation in the cells expressing D1 was slightly but obviously higher than those in the cells expressing either C, Y1, or Y2. This reproducible result suggested that there existed signaling leak in the cells expressing D1 greater than that in the cells expressing either C, Y1, or Y2. The D1 thus appeared to be inferior to the C, Y1, or Y2 in the ability to inhibit the IFN-α response. Since the expression level of D2 was extremely low (Fig. 2A, lanes 7 and 9), the possibility could not be excluded that the lack of the anti-IFN activity in His-D2-HeLa cells might be attributable to its low expression level.
FIG. 3.
Effects of the C mutant proteins on IFN-α-stimulated gene activation and on ISGF3 formation. (A) The established cell lines were doubly transfected with pISRE-TA-luc and pRL-TK-luc. At 20 h posttransfection, the cells were treated or mock treated with IFN-α (1,000 IU/ml) for 6 h and then lysed. The firefly luciferase activity in the extracts, expressed in relative light units, was normalized to renilla luciferase activity. Data represent the mean values of the normalized luciferase activities from triplicate samples. (B) Confluent monolayers of the established cell lines were treated or mock treated with IFN-α (1,000 IU/ml) for 1 h. The extracts from the cells were subjected to EMSA with a 32P-labeled ISG15 ISRE probe. (C) Confluent monolayers of the established cell lines were treated with IFN-α (1,000 U/ml) for 0 or 16 h. Extracts from the cells were subjected to SDS-6.5% PAGE, followed by Western blot analyses with anti-STAT1 (sc-464) antibody.
The ability to suppress STAT1 activation does not correlate with the inhibitory effect on the IFN-α response.
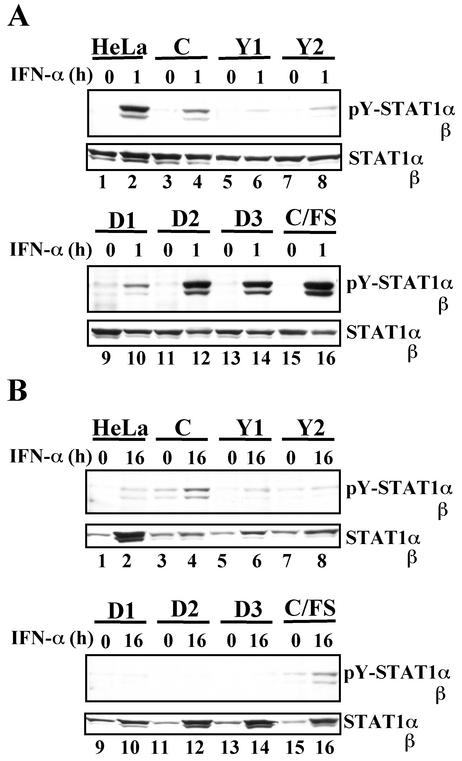
The C protein suppresses tyrosine phosphorylation of STAT1 in immediate response to IFN-α (23, 42). To determine whether this ability would correlate with the inhibitory effect on the IFN-α response, effects of the C mutant proteins on STAT1 activation were examined under the two conditions: short-term (1 h) (Fig. 4A) and long-term (16 h) (Fig. 4B) IFN-α stimulations. Tyrosine phosphorylation of STAT1 in response to short-term IFN-α stimulation was strikingly suppressed in the cells expressing either C, Y1, Y2, or D1 (Fig. 4A, lanes 4, 6, 8, and 10). A little but noteworthy elevation of the pY-STAT1 level, however, was reproducibly observed in the C- and D1-expressing cell lines (Fig. 4A, lanes 4 and 10). Long-term IFN-α stimulation caused moderate accumulation of pY-STAT1 in C- and C/FS-expressing cells (Fig. 4B, lanes 4 and 16). The C and D1 were thus inferior to the Y1 and Y2 in the ability to suppress IFN-α-stimulated tyrosine phosphorylation of STAT1. These results demonstrated that the suppressive effect on STAT1 activation did not strictly correlate with the inhibitory effect on the IFN-α response.
FIG. 4.
Effect of the C mutant proteins on tyrosine phosphorylation of STAT1 in response to short (A)- or long (B)-term stimulation with IFN-α. Confluent monolayers of the established cell lines were treated with IFN-α (1,000 U/ml) for 0 h (A and B), 1 h (A), or 16 h (B). Extracts from the cells were subjected to SDS-6.5% PAGE, followed by Western blot analyses with anti-pY-STAT1 (no. 9171) or anti-STAT1 (sc-464) antibody.
The ability to inhibit STAT2 activation correlates well with the inhibitory effect on the IFN-α response.
A current model for the STAT activation in IFN-α signaling proposes sequential activation of STAT2 and STAT1 in this order (28). Effect of the C protein on STAT2 activation would therefore be expected to be similar to the effect on STAT1 activation. At the early phase of infection, STAT2 activation in response to short-term IFN-α stimulation is indeed inhibited (24). In contrast to the partial inhibition of STAT1 activation in the C-expressing cells (Fig. 4, lanes 3 and 4), STAT2 activation was, unexpectedly, completely inhibited by the C protein irrespective of duration of IFN-α stimulation (Fig. 5, lanes 3 and 4). This complete inhibition was likewise observed in the cells expressing either Y1or Y2 (Fig. 5, lanes 5 to 8), whereas STAT2 was normally activated in the cells expressing either D2, D3, or C/FS that lost the STAT1-binding capacity (Fig. 5A, lanes 1, 2, and 11 to 16). Although the D1 strongly suppressed tyrosine phosphorylation of STAT2, minor phosphorylation leak was observed (Fig. 5A, lanes 9 and 10). The leak of STAT1 and STAT2 phosphorylation in the D1-expressing cells could account for its incompleteness in inhibition of the IFN-α response. It should be noted that the C, which exhibited incomplete suppression of STAT1 phosphorylation and complete inhibition of STAT2 phosphorylation, effectively inhibited the IFN-α response. The capacity to inhibit STAT2 activation thus correlated well with the inhibitory effect on the IFN-α response. These results strongly suggested that the target of the C protein was the activation process of STAT2 rather than STAT1. The pY-STAT2 level reaches a peak 1 to 2 h after IFN-α stimulation and gradually decreases to the basal level (see Fig. 6A). At 16 h after IFN-α stimulation, pY-STAT2 was undetectable in all of the cell lines under the usual exposure condition (data not shown). Even longer exposure hardly detected pY-STAT2 bands in the C, Y1, Y2, and D1 (Fig. 5B, lanes 3 to 10), whereas faint bands of pY-STAT2 were often seen in HeLa cells and cells expressing either D2, D3, or C/FS (Fig. 5B, lanes 2, 12, 14, and 16). The pY-STAT2 band in C/FS was obscure in this figure (Fig. 5B, lane 16), but a similar faint band was observed in another experiment.
FIG. 5.
Effect of the C mutant proteins on tyrosine phosphorylation of STAT2 in response to short (A)- or long (B)-term stimulation with IFN-α. Confluent monolayers of the established cell lines were treated with IFN-α (1,000 U/ml) for 0 h (A and B), 1 h (A), or 16 h (B). Extracts from the cells were subjected to SDS-6.5% PAGE, followed by Western blot analyses with anti-pY-STAT2 (no. 07-224) or anti-STAT2 (sc-476) antibody.
FIG. 6.
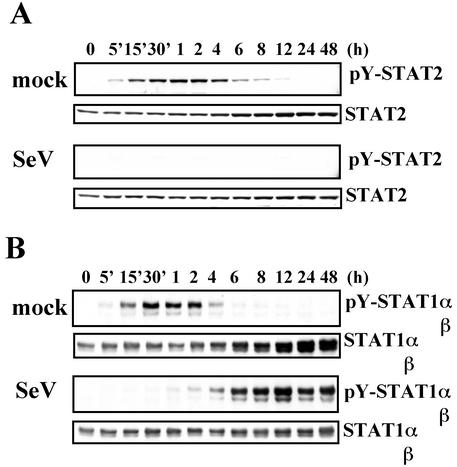
Levels of pY-STAT2 (A) and pY-STAT1 (B) at various intervals after IFN-α stimulation in SeV-infected cells. HeLa cells were mock infected or infected with SeV at an MOI of 10. The media were replaced with media containing IFN-α (1,000 IU/ml) at 2 h p.i. The cells were harvested at the indicated times. Extracts from the cells were subjected to SDS-6.5% PAGE, followed by Western blot analyses with anti-pY-STAT2 (no. 07-224) or anti-STAT2 (sc-476) (A) or anti-pY-STAT1 (no.9171) or anti-STAT1 (sc-346) (B) antibody.
To confirm that the STAT2 activation process is a crucial target for the blockade of IFN-α signaling, we next looked for IFN-α-stimulated STAT2 activation in infected cells. Cells were infected or mock infected with SeV at a multiplicity of infection (MOI) of 10. At 2 h p.i., the cells were treated with IFN-α for various time periods, and their intracellular pY-STAT2 and pY-STAT1 levels were estimated by Western blot analyses (Fig. 6). The levels of both pY-STAT2 and pY-STAT1 in mock-infected cells reached a peak ca. 1 h after IFN-α stimulation and gradually decreased to the basal level (Fig. 6, mock). In contrast, the pY-STAT2 level in infected cells retained below the detection level irrespective of duration of IFN-α stimulation, as expected (Fig. 6A, SeV). On the other hand, tyrosine phosphorylation of STAT1 was suppressed for short-term stimulation but long-term stimulation caused marked elevation of the pY-STAT1 level (Fig. 6B, SeV).
Involvement of the C-STAT1 interaction in the inhibition of STAT2 activation.
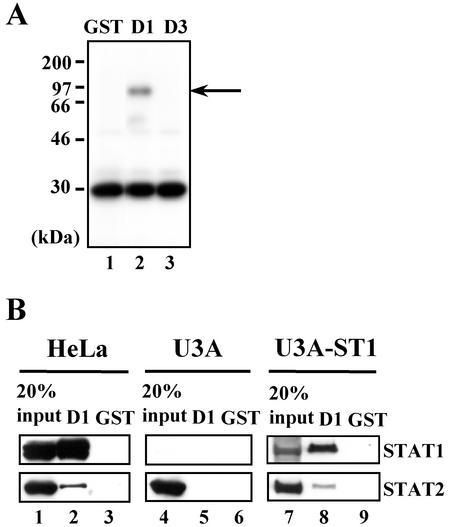
Since the STAT1-binding capacity correlated with the inhibitory effect on STAT2 activation, it seemed very likely that the C protein would inhibit STAT2 activation through the STAT1 interaction. Indeed, Western blot analysis with anti-STAT2 antibody on the membrane used in Fig. 1C could not provide evidence for binding of the C, Y1, Y2, or D1 to STAT2 (data not shown), suggesting that the C-STAT2 binding, even if present, would be very weak. To check the possibility of the direct C-STAT2 interaction, the binding assay was performed by using GST-D1 and [35S]methionine-labeled HeLa cell extracts. GST-D1 was used instead of GST-C, GST-Y1, or GST-Y2 in this experiment, as well as the following experiments, because it was difficult to prepare large amounts of the latter three proteins, due to their lability. No specific band was observed except for a band, which migrated at the same position as STAT1 did (Fig. 7A, lane 2). Since STAT2 (∼113 kDa) was not detected by this method, large amounts (∼1 mg) of unlabeled HeLa, (STAT1-deficient) U3A, and (HA-STAT1 α-expressing) U3A-ST1 cytoplasmic extracts were subjected to the binding assay with GST-D1 under the isotonic conditions and analyzed by Western blotting. In this experiment, the concentration of sodium chloride in the washing and binding buffers was reduced from 300 to 100 mM to detect weak interactions. Not only STAT1 but also STAT2 was coprecipitated with GST-D1 from HeLa cell extracts (Fig. 7B, lane 2), whereas the IFN receptor subunits (IFNAR1 and IFNAR2) and the receptor-associated kinases (JAK1 and TYK2) were not pulled down (data not shown). STAT2 was, however, present in extremely low amounts compared to STAT1 (Fig. 7B, lane 2). Furthermore, no STAT2 was pulled down from STAT1-deficient U3A cell extracts (Fig. 7B, lane 5), whereas expression of HA-STAT1α in U3A cells restored the interaction of D1 with STAT2 (Fig. 7B, lane 8). These results indicated that the binding of D1 to STAT2 was indirect and dependent on the presence of STAT1. Of the major factors associating with the IFN-α/β receptor complex, only STAT1 seemed to be a component that directly interacted with D1.
FIG. 7.
STAT1-dependent interaction of the C protein with STAT2. (A) HeLa cells (∼107 cells) were labeled with Redivue l-[35S]methionine (1.8 MBq/ml; 37 TBq/mmol; Amersham Pharmacia Biotech) for 4 h. The radioisotope-labeled cell extract (∼100 μg) was subjected to binding assay with GST, GST-D1, or GST-D3. Bound products were separated by SDS-10% PAGE and visualized with the aid of Amplify (Amersham Pharmacia Biotech) according to the manufacturer's instructions. An arrow indicates the position of STAT1. (B) Cytoplasmic extracts were prepared by lysis of HeLa, U3A, or U3A-ST1 cells with a hypotonic buffer (10 mM HEPES-KOH [pH 7.9], 0.25% NP-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 1 mM DTT). After centrifugation, NaCl concentration of the clarified lysate was adjusted to 100 mM. Large amounts (∼1 mg) of HeLa, U3A, and U3A-ST1 extracts were subjected to binding assay with GST-D1 or GST. The hypotonic buffer containing 100 mM NaCl was used as a washing buffer. Bound proteins were separated by SDS-6.5% PAGE, followed by Western blot analyses with anti-STAT1 (sc-346) or anti-STAT2 (sc-476) rabbit antibody.
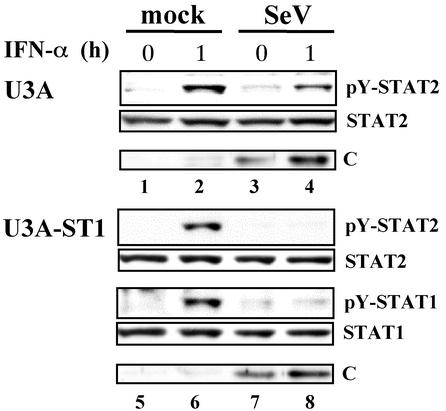
To obtain more definite evidence for the involvement of STAT1 in the inhibitory process for STAT2 activation, we studied the effects of C protein on STAT2 activation in the absence of STAT1 by using U3A cells. U3A and U3A-ST1 cells were infected with SeV at an MOI of 10. At 6 h p.i., the cells were treated with IFN-α for 1 h. As shown in Fig. 8, STAT2 was tyrosine phosphorylated in response to IFN-α stimulation in U3A irrespective of infection (Fig. 8, lanes 1 to 4), whereas tyrosine phosphorylation of not only STAT2 but also STAT1 was strikingly inhibited in infected U3A-ST1 cells (Fig. 8, lanes 7 and 8). The C protein expressed in infected U3A cells was detected in almost the same amount as that observed in infected U3A-ST1 cells (Fig. 8, lanes 3, 4, 7, and 8). Thus, STAT1 had the ability to potentiate the inhibitory effect of the C protein on STAT2 activation.
FIG. 8.
STAT1 enhanced the inhibitory effect of the C protein on STAT2 activation. U3A or U3A-ST1 cells were infected or mock infected with SeV at an MOI of 10. At 6 h p.i., the cells were treated with IFN-α (1,000 IU/ml) for 0 or 1 h. Extracts from the cells were subjected to SDS-6.5% PAGE (or SDS-11% PAGE for analysis of the C protein), followed by Western blot analysis with anti-pY-STAT2 (no. 07-224), anti-STAT2 (sc-476), anti-pY-STAT1 (no. 9171), anti-STAT1 (sc-346), or anti-C antibody.
DISCUSSION
Long-term IFN-α stimulation causes marked elevation of the pY-STAT1 level in SeV-infected cells (Fig. 6B) (23). Furthermore, the suppressive effect on STAT1 activation was not observed when HeLa cells constitutively expressing the C protein were treated with a relatively high dose of IFN-α (3,000 IU/ml) (36). These results had suggested that the SeV C protein targeted the other process except for the STAT1 activation for the blockade of IFN-α signaling. The present study addresses this issue and reveals that the inhibition of STAT2 activation but not STAT1 activation is essential for the blockade of IFN-α signaling. The effects of the C mutant proteins on IFN-α signaling are summarized in Table 1. There exists a definite positive correlation between the ability to inhibit STAT2 activation and the inhibitory effect on the IFN-α response. It should be noted that all of the original C proteins (C, Y1, and Y2) that are expressed in SeV-infected cells exhibit not partial but complete inhibition of STAT2 activation (Fig. 5). Furthermore, this complete inhibition was also observed in infected cells (Table 1 and Fig. 6A). These results strongly suggest that the C protein targets the activation process of STAT2 rather than STAT1. Nuclear transport of pY-STAT1 generated by IFN-α stimulation was impaired in the presence of the C protein (36). This impairment may be explainable by this complete inhibition of STAT2 activation, which is indispensable for STAT1-STAT2 heterodimer formation. The complete inhibition of IFN-α stimulated tyrosine phosphorylation of STAT2 in the presence of the C protein was not affected by treatment with a general phosphatase inhibitor, vanadate (unpublished result). Accordingly, it seems unlikely that the C protein activates a cellular phosphatase that may dephosphorylate STAT2.
TABLE 1.
Effects of the C mutant proteins on IFN-α signaling
| Protein or cell group | Binding to STAT1 | Inhibition of IFN-α response | Inhibition of tyrosine phosphorylation of
|
|
|---|---|---|---|---|
| STAT1 | STAT2 | |||
| C | + | + | (+) | + |
| Y1 | + | + | + | + |
| Y2 | + | + | + | + |
| D1 | + | (+)a | (+) | (+) |
| D2 | − | − | − | − |
| D3 | − | − | − | − |
| C/FS | − | − | − | − |
| SeV-infected cells | + | (+) | + | |
(+), Partial inhibition.
The reporter gene assay showed that the ability of D1 to inhibit the IFN-α response was comparable to the abilities of C, Y1, and Y2 (Fig. 3A). Consistent with this finding, the elevation of the STAT1 level as an ISG product (27) was indeed suppressed in C, Y1, Y2, and D1 (Fig. 3C, lanes 3 to 10). Nevertheless, a slight but definite induction of STAT1 was reproducibly observed in cells expressing D1 after long-term (16 h) IFN-α treatment (Fig. 3C, lanes 9 and 10). This discrepancy presumably resulted from a difference in the duration of IFN-α stimulation (6 h for the former and 16 h for the latter) in these two experiments and/or from a difference in sensitivity of these two methods. This intermediate induction of STAT1 is explainable by the generation of small amounts of the pY-STAT2 and pY-STAT1 as a signaling leak in the D1-expressing cells (Fig. 4A and 5A, lanes 9 and 10). ISGF3 complex, which must be present in the extract from the D1-expressing cells, appears to be present in too small an amount to be detected by EMSA (Fig. 3B). The incomplete inhibition by D1 was unlikely to be due to its low expression level because the C protein, which was present in a lower amount (Fig. 2A), completely inhibited the IFN-α response.
A single point mutation from Phe170 to Ser eliminated STAT1-binding capacity from the C protein and simultaneously causes abrogation of the inhibitory effect on STAT2 and STAT1 activation. This suggests that STAT1-binding capacity is essential for the inhibition of activation of not only STAT1 but also STAT2. Our binding assay could not detect direct interaction between D1 and STAT2 in the absence of STAT1 (Fig. 7B). Instead, a small amount of STAT2 was found to interact with D1 in the presence of STAT1. These results indicate that the C protein is accessible to STAT2 via the interaction with STAT1. Stancato et al. observed preassociation of STAT1 with STAT2 and STAT3 in hypotonic (10 mM HEPES [pH 7.35], 1 mM EDTA) extracts from HeLa cells but not in an extract prepared under harsh conditions (10 mM HEPES [pH 7.35], 1 mM EDTA, 1% Triton X-100, 150 mM NaCl) (40). Although the conditions in the present study were similar to the harsh conditions used by Stancato et al., the results presented here may represent a preassociation of STAT1 with STAT2. Enhancement of the inhibitory effect on STAT2 activation by STAT1 (Fig. 8) and this STAT1-dependent STAT2 interaction strongly suggest that the C protein inhibits STAT2 activation via the interaction with STAT1. Nevertheless, the possibility cannot be ruled out that other cellular molecules besides STAT1 participate in this inhibition process because, at the late phase of infection, IFN-α-stimulated tyrosine phosphorylation of STAT2 was completely inhibited even in U3A cells (unpublished results). This suggests that STAT2 activation is inhibited by the STAT1-independent process in the presence of a large amount of the C protein. On the other hand, STAT2 activation was completely inhibited even at 2 h p.i. (Fig. 6A). At this very early stage, the C protein is produced in very small amounts. Therefore, the C protein may favor preassociated STAT1-STAT2-IFNAR1 complexes to free STAT1 for the interaction. Thus, the other unidentified molecules, if present, might play an important role in the recognition of preassociated complexes and/or STAT1-independent inhibition. More refined experiments are, however, needed to prove these possibilities because changes of intracellular milieu, as well as large amounts of other viral proteins produced in infected cells, might strikingly affect STAT activation process.
STAT2 and STAT1 preassociate with the cytoplasmic tail of IFNAR2 (Fig. 9). Preassociation of STAT1 with IFNAR2 depends on the presence of STAT2 but not vise versa (28). IFN-α-mediated aggregation of the IFN receptor subunits, IFNAR1 and IFNAR2, results in activation of TYK2 and JAK1. The activated TYK2 phosphorylates IFNAR1 on Tyr466, creating a docking site for the SH2 domain of STAT2. Interaction of the STAT2 SH2 domain with the docking site results in phosphorylation of STAT2 on Tyr690, providing the docking site for the SH2 domain of STAT1. STAT1 is then phosphorylated on Tyr701. In the presence of the C protein, STAT1 can be partially activated without STAT2 activation (Fig. 4A, lanes 3 and 4; Fig. 5A, lanes 3 and 4). The present study thus demonstrates that the C protein not only inhibits STAT2 activation but also disrupts this sequential activation process. How is STAT1 tyrosine-phosphorylated without the aid of pY-STAT2? The answer to this question may be linked to the elucidation of a molecular basis for the complete inhibition of STAT2 activation. Since STAT1, STAT2, and IFNAR2 preassociate in IFN-untreated cells, binding of the C protein to STAT1 results in C-STAT1-STAT2-IFNAR2 complex formation. This complex formation possibly results in impairment of activation process of not only STAT1 but also STAT2 (Fig. 9). At the early phase of infection, activation of TYK2 is partially inhibited (24). Since the C protein did not interact with TYK2 directly, this inhibition may also result from the C-STAT1 interaction.
FIG. 9.
Schematic diagram for the proposed molecular mechanism by which SeV blocks IFN-α signaling. Black circles indicate tyrosine residues phosphorylated. STAT2 preassociates with not only the cytoplasmic tail of IFNAR2 but also STAT1 in IFN-untreated cells. In infected cells, the C protein binds to the STAT1-STAT2-IFNAR2 complex. Formation of the C-STAT1-STAT2-IFNAR2 complex results in the complete inhibition of STAT2 activation, as well as the partial inhibition of STAT1 activation. This complex formation may also affect activation of TYK2 and JAK1.
Garcin et al. initially underscored the specific importance of the AUG114-initiated C protein of the four C proteins (C′, C, Y1, and Y2) for blocking IFN-α signaling (12). Later, these authors suggested that not only the larger forms (C and C′) but also the shorter forms (Y1 and Y2) can inhibit IFN-α signaling (10, 11); however, only the larger proteins prevent establishment of the IFN-mediated anti-vesicular stomatitis virus (VSV) state (10). In contrast, Kato et al. have presented evidence for the equal activities of the longer and shorter C proteins for the inhibition of IFN-mediated anti-VSV state, as well as IFN signaling (22). In our established cell lines—His-C-HeLa, His-Y1-HeLa, and His-Y2-HeLa—IFN-α-mediated induction of the anti-VSV state was strikingly inhibited (unpublished results), thus supporting the latter conclusion. Accordingly, conflicting views exist regarding the effect of the shorter proteins, Y1 and Y2, on the IFN-mediated anti-VSV state. Although it is unclear at the moment how the discrepancy has taken place, there is a difference between the results of Garcin et al. and the results of Kato et al. in the experimental strategy used. The former conclusion was drawn from experimental results achieved mainly with mutated recombinant SeV-infected cells, in which the anti-IFN ability of the mutant C proteins would be affected by many factors, including other viral proteins, autocrine IFN associated with infection, and the expression level of the mutant C protein at the time of IFN treatment. It is possible that these factors might have led to the discrepancy (16, 17). In contrast to the STAT degradation-independent mechanism presented here, Garcin et al. emphasized the degradation of STAT1 in SeV-infected MEF cells as an SeV blocking mechanism for IFN signaling and attempted to find the common denominator between functions of SeV C protein and rubulavirus V proteins (10, 11, 13, 30). Longer C protein-specific degradation of STAT1 in NIH 3T3 mouse MEF cells, however, appears to be a special case. Because the expression of the C protein did not cause STAT1 degradation in most cell lines, including four human cell lines (HeLa, U118, 2fTGH, and HEC1B) and one mouse cell line (BF) (12, 18, 23, 24, 42, 49).
During the progress of our research, two related studies were published. Kato et al. reported that a C deletion mutant that lacked N-terminal region (98 aa) retains the abilities to counteract the antiviral action of IFNs and to downregulate viral RNA synthesis (21). Another study about the binding abilities of several mutant C proteins (13) suggested that the STAT1-binding region is mapped to the Y1 region and that the amino acid Phe170 is important for the binding capacity. These results are basically consistent with the results presented here.
The C protein inhibits transcriptional activation of ISGs in response not only to IFN-α/β but also to IFN-γ. When the C protein was expressed sufficient in amount, IFN-γ-stimulated tyrosine phosphorylation of STAT1 was not inhibited at all (23, 42). This finding and the conclusions in the present study reveal that the blocking mechanism for IFN-γ is totally different from that for IFN-α/β. Further analyses of the effects of the C mutant proteins on IFN-γ signaling will provide important information regarding the molecular basis for the inhibition of the IFN-γ response.
Acknowledgments
We thank S. Ishida and S. Kitagawa (Kubo) for excellent technical assistance and Y. Ohnishi and Y. Nagai for constant encouragement of our research. We are grateful to G. Stark for permission to use U3A cells, T. Hirano for permission to use pEF-BOS-HA-STAT1, H. Asao for providing the pEFneo expression vector and sending U3A cells, T. Shioda for providing the cDNA clone H-33, M. Hibi for sending pEF-BOS-HA-STAT1, and A. Kato and Y. Nagai for providing anti-C rabbit serum. We also thank Y. Kimura for helpful discussions.
This work was supported in part by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science and by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asao, H., and X. Y. Fu. 2000. Interferon-gamma has dual potentials in inhibiting or promoting cell proliferation. J. Biol. Chem. 275:867-874. [DOI] [PubMed] [Google Scholar]
- 3.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 4.Colamonici, O., H. Yan, P. Domanski, R. Handa, D. Smalley, J. Mullersman, M. Witte, K. Krishnan, and J. Krolewski. 1994. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol. Cell. Biol. 14:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran, J., and D. Kolakofsky. 1989. Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J. 8:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii, N., N. Yokosawa, and S. Shirakawa. 1999. Suppression of interferon response gene expression in cells persistently infected with mumps virus, and restoration from its suppression by treatment with ribavirin. Virus Res. 65:175-185. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 10.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind STAT1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 14.Giorgi, C., B. M. Blumberg, and D. Kolakofsky. 1983. Sendai virus contains overlapping genes expressed from a single mRNA. Cell 35:829-836. [DOI] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response, and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2001. Paramyxovirus accessory proteins as interferon antagonists. Microbiol. Immunol. 45:787-800. [DOI] [PubMed] [Google Scholar]
- 17.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 18.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, A., Y. Ohnishi, M. Hishiyama, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2002. The amino-terminal half of Sendai virus C protein is not responsible for either counteracting the antiviral action of interferons or down-regulating viral RNA synthesis. J. Virol. 76:7114-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511:139-144. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 26.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 27.Lehtonen, A., S. Matikainen, and I. Julkunen. 1997. Interferons upregulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 159:794-803. [PubMed] [Google Scholar]
- 28.Li, X., S. Leung, I. M. Kerr, and G. R. Stark. 1997. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol. Cell. Biol. 17:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajima, K., Y. Yamanaka, K. Nakae, H. Kojima, M. Ichiba, N. Kiuchi, T. Kitaoka, T. Fukada, M. Hibi, and T. Hirano. 1996. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15:3651-3658. [PMC free article] [PubMed] [Google Scholar]
- 30.Nishio, M., D. Garcin, V. Simonet, and D. Kolakofsky. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 300:92-99. [DOI] [PubMed] [Google Scholar]
- 31.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick, D., B. Cohen, and M. Rubinstein. 1994. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77:391-400. [DOI] [PubMed] [Google Scholar]
- 33.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 Acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 35.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito, S., T. Ogino, N. Miyajima, A. Kato, and M. Kohase. 2002. Dephosphorylation failure of tyrosine-phosphorylated STAT1 in IFN-stimulated Sendai virus C protein-expressing cells. Virology 293:205-209. [DOI] [PubMed] [Google Scholar]
- 37.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809-813. [DOI] [PubMed] [Google Scholar]
- 38.Shioda, T., Y. Hidaka, T. Kanda, H. Shibuta, A. Nomoto, and K. Iwasaki. 1983. Sequence of 3,687 nucleotides from the 3′ end of Sendai virus genome RNA and the predicted amino acid sequences of viral NP, P and C proteins. Nucleic Acids Res. 11:7317-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 40.Stancato, L. F., M. David, C. Carter-Su, A. C. Larner, and W. B. Pratt. 1996. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J. Biol. Chem. 271:4134-4137. [DOI] [PubMed] [Google Scholar]
- 41.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 43.Veals, S. A., C. Schindler, D. Leonard, X. Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 45.Yan, H., K. Krishnan, A. C. Greenlund, S. Gupta, J. T. Lim, R. D. Schreiber, C. W. Schindler, and J. J. Krolewski. 1996. Phosphorylated interferon-alpha receptor 1 subunit (IFNαR1) acts as a docking site for the latent form of the 113-kDa STAT2 protein. EMBO J. 15:1064-1074. [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoo, J., B. Gotoh, T. Komatsu, K. Takeuchi, and T. Miyadai. 1999. Replication-incompetent Sendai virus can suppress the antiviral action of type I interferon. Arch. Virol. 144:1043-1055. [DOI] [PubMed] [Google Scholar]
- 47.Yokosawa, N., T. Kubota, and N. Fujii. 1998. Poor induction of interferon-induced 2′,5′-oligoadenylate synthetase (2-5 AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1 alpha. Arch. Virol. 143:1985-1992. [DOI] [PubMed] [Google Scholar]
- 48.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]