Abstract
At the early stages of herpes simplex virus type 1 (HSV-1) infection, parental viral genomes have a tendency to become juxtaposed to cellular nuclear structures known as PML (promyelocytic leukemia) nuclear bodies or ND10, while the immediate-early (IE) protein ICP0 precisely colocalizes with these structures. Previous indirect-immunofluorescence studies observed that the HSV-1 transcriptional regulator ICP4 has a mainly diffuse nuclear distribution early in infection and is later recruited into viral replication compartments. We have constructed HSV-1 variants expressing ICP4 and ICP0 linked to ECFP and EYFP, respectively, both singly and in combination. Coupled with an efficient method of expressing autofluorescent PML in ND10, we have studied the dynamics of ICP0, ICP4, and ND10 in live, infected cells. The greater sensitivity and lower background signals in live cells revealed that early in infection, ICP4 forms discrete foci, some of which are juxtaposed to ND10, while ICP0 was found to colocalize precisely with PML. As expected from these results, using a double-labeled virus, we observed that foci of ICP0 and ICP4 were also juxtaposed but not colocalized early in infection. Some of the ICP4 foci must have contained parental viral genomes, because they developed into replication compartments. We propose that a proportion of the ND10-associated ICP4 foci represent ICP4 molecules being recruited onto parental viral genomes, a process likely to be a critical step early in lytic infection. These results may be analogous to the localization of IE1 and IE2 during human cytomegalovirus infection, suggesting a principle common to the alpha- and betaherpesviruses.
A crucial biological and clinical feature of infection by herpes simplex virus type 1 (HSV-1) is the ability of the virus to establish a latent state in sensory neurons after initial replication in epithelial tissues. The latent virus is carried for the lifetime of the infected individual, during which there are periodic episodes of reactivation and renewed lytic replication in the epithelia served by the latently infected ganglion. During the lytic cycle, the whole viral genome is very actively transcribed, while latency is characterized by very limited transcription restricted to the latency-associated transcripts of uncertain function. Therefore, the molecular mechanisms governing the contrast between lytic- and latent-state transcription have attracted much intensive research (for a general review, see reference 27).
The viral and cellular proteins involved in viral gene expression during the lytic cycle, and the regulatory elements within the viral genome that they act on, are now quite well understood. The lytic-cycle genes can be divided into three temporal classes, designated immediate early (IE), early, and late. The IE protein ICP4 is essential for viral infection because it is required to activate the transcription of the early and late genes (11, 37). Consistent with its ability to bind to viral DNA (3, 33), ICP4 is recruited into viral replication compartments containing large amounts of replicating viral DNA (10, 28, 38). However, prior to the onset of DNA replication, ICP4 in fixed cells has been observed to be distributed diffusely throughout the nucleus, with foci appearing as infection proceeds (10, 28, 38, 46).
In recent years, it has become increasingly clear that the environment of a gene within the nucleus is an important factor in the regulation of its transcriptional activity (8). In this regard, it is intriguing that the parental genomes of HSV-1 (and also those of several other DNA viruses that replicate in the nucleus) have been shown to have a tendency to associate with specific nuclear substructures known as ND10 or PML (promyelocytic leukemia) nuclear bodies (22, 32). We have recently developed methods to observe this association in live cells, and furthermore, we have shown that the presence of an ICP4-responsive gene within a model genome increases the probability of ND10-genome association (39). In addition to viral genomes becoming juxtaposed to ND10, another viral IE regulatory protein, ICP0, precisely colocalizes within ND10 structures and then causes their disruption (17, 31). Therefore, there appear to be intimate connections between ND10 and viral components during the early stages of infection. However, the localization of ICP4 with respect to other viral proteins and ND10 during the initial stages of infection has not been investigated in any detail. In this paper, we report the study of the localization and dynamics of ICP4 and ICP0 with respect to ND10 in live cells. We found that ICP4 forms discrete foci early in infection, some of which are associated with ND10 structures containing ICP0. We observed that a proportion of the ICP4 foci later developed into viral replication compartments, particularly those associated with ICP0 accumulated at ND10. Since replication compartment formation requires an initiating viral genome, it follows that a proportion of the ICP4 foci must contain one or more parental genomes. These data suggest that the spatial organization of ICP0, ICP4, and viral genomes plays an important role in the early stages of lytic viral infection.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 was grown and titrated in BHK cells propagated in Glasgow modified Eagle's medium containing 100 U of penicillin/ml and 100 μg of streptomycin/ml and supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. Vero cells and HEp-2 cells were grown in Glasgow modified Eagle's medium or Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal calf serum and antibiotics as described above. Human fetal foreskin fibroblast (HFFF-2) cells were grown in the same medium as HEp-2 cells.
The viruses expressing ICP4 and ICP0 linked to autofluorescent proteins (AFPs) were constructed by homologous recombination between sequences upstream and downstream of the ICP0 and ICP4 coding regions of genomes derived from HSV-1 strain 17+. Therefore, all these viruses express the AFP-linked proteins from transcription units containing their normal regulatory sequences in their proper genomic locations. The initial step was to clone the relevant IE protein coding sequences fused in frame and downstream of the ECFP (enhanced cyan fluorescent protein) or EYFP (enhanced yellow fluorescent protein) coding region in plasmid pECFP-C1 or pEYFP-C1 (Clontech). These plasmids also include the 3′ untranslated region of the ICP0 or ICP4 gene. Secondly, the cytomegalovirus (CMV) promoter region in these plasmids was replaced by the relevant IE promoter region to create plasmids of the type pIEn-AFP-ICPm, where n is IE1 or IE3, AFP is either ECFP or EYFP, and m is either 0 or 4. To isolate virus vEYFP-ICP0, the plasmid DNA was cotransfected into BHK cells with the ICP0 null mutant dl1403 DNA. To isolate viruses vECFP-ICP4 and vEYFP-ICP4, the plasmid DNA was cotransfected with DNA from HSV140, a derivative of HSV-1 17+ that contains the varicella-zoster virus gene 62 open reading frame in place of that of ICP4 (13). This virus grows less well than its parent, so replacement of the varicella-zoster virus gene 62 sequences with the AFP-linked ICP4 open reading frame leads to preferential replication of the fluorescent progeny. All viruses were plaque purified at least three times and checked for expression of full-length fluorescent fusion protein by Western blotting.
Virus AFP4/0 was isolated after a mixed infection with vECFP-ICP4 and vEYFP-ICP0. The plaques were screened for expression of both fluorescent proteins and then purified to homogeneity by successive rounds of plaque purification and Western blot screening to eliminate any IE1 or IE3 genes without the fluorescent marker from the virus isolate. Virus vEYFP-ICP4tsK contains the tsK mutation in the ICP4 coding sequence, which confers a temperature-sensitive DNA binding defect on the ICP4 protein (36, 37). It was constructed by transferring a fragment of a tsK genomic clone into pIE3-EYFP-ICP4 and then isolating the derivative virus at 31°C after cotransfection with HSV140 DNA as described above.
Baculovirus Ac.CMV.ECFP-PML.
Plasmid pECFP-PML contains the cDNA of the 633-residue isoform IV of the PML protein (24, 26) inserted in frame into plasmid pECFP-N1. This isoform of PML was chosen because its response to ICP0 has been well documented (35). The CMV promoter region of the vector plus the PML-ECFP coding region was inserted into plasmid pFastBacHTa of the Bac-to-Bac baculovirus construction system (Life Technologies), and recombinant baculovirus was isolated according to the manufacturer's protocol. The resultant baculovirus, Ac.CMV.ECFP-PML, expresses the PML-ECFP fusion protein from the CMV promoter in both insect and mammalian cells. Preliminary experiments showed that infection of HEp-2 cells with this virus at a multiplicity of 50 PFU per cell led to expression of PML located in ND10 in a high percentage of cells by 16 h postinfection. Highly efficient infection of Vero cells was achieved using a multiplicity of 5 PFU per cell.
Time lapse microscopy of live cells.
Live cells were observed with a Zeiss Axiovert S100 inverted fluorescence microscope equipped with an ×63 oil immersion objective lens and a Hamamatsu Orca 2 digital camera. The excitation wavelength was controlled by mercury lamp illumination and a motorized filter wheel equipped with filters specific for ECFP and EYFP (set 51017sbx; Chroma Technology). Dichroic and emission filter combinations (Chroma Technology) suitable for dual ECFP-EYFP and single EGFP fluorophores were housed in a static filter block. The filter wheel, shutters on fluorescence and bright-field illumination light paths, and camera image acquisition were controlled by AQM software (Imaging Associates and Kinetic Imaging). Single images or timed image sequences were exported as bitmap or TIFF files from the AQM software and then transferred to the program FLEX running in the KS300 environment (Imaging Associates) for selection of regions of interest from image sequences. Individual frames were prepared for presentation using Adobe Photoshop.
An essential part of the system was the ability to maintain the cells for long periods in a controlled environment with constant temperature, CO2, and humidity. The main body of the microscope, excluding the manual controls, was housed in a humidified gas-tight Perspex box equipped with temperature-controlled air circulation and a CO2 detection and maintenance device (supplied by EMBL Workshops, Heidelberg, Germany). The conditions within the box were maintained at 37°C, 5% CO2, and 90% humidity so that the cells remained healthy indefinitely. The cells were seeded into two-well chambered cover glass units with coverslip quality glass bottoms (Lab-Tek and Nunc) at a density of 2 × 105 per well the day before each experiment. Infections were carried out in Eagle's medium without phenol red and containing 1% calf serum. In all figures, the times given relate to the time after addition of the virus rather than the time after a period of adsorption.
Western blotting.
Whole-cell extracts of transfected cells were prepared by washing cell monolayers with phosphate-buffered saline (PBS) and adding sodium dodecyl sulfate gel boiling mix. After the cell lysates were boiled, the proteins were separated by electrophoresis on sodium dodecyl sulfate-7.5% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The filters were blocked overnight in PBS containing 0.1% Tween 20 and 5% dried milk and then incubated with primary antibodies for 2 h in the same buffer. After being extensively washed, the filters were incubated with horseradish peroxidase-conjugated secondary antibody in PBS-0.1% Tween 20-2% dried milk for 1 h and then washed again extensively before detection of bound antibody by enhanced chemiluminescence (NEN) and exposure to film. ICP0 was detected using monoclonal antibody 11060 (18), and ICP4 was detected by monoclonal antibody 10176 (15). As described previously, an antibody mixture was used to detect the proteins UL39, ICP8, and UL42 (20).
Transfection, immunofluorescence, and confocal microscopy.
HEp-2 cells were transfected using Lipofectamine Plus reagent (Gibco BRL) according to the manufacturer's instructions. For immunofluorescence of fixed cell samples, cells on glass coverslips were washed with PBS, fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose), and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The coverslips were incubated for 1 h at room temperature with primary antibodies diluted in PBS containing 1% newborn calf serum and then washed at least six times before treatment with secondary antibodies in the same manner. The primary antibodies used were anti-Sp100 rabbit serum SpGH (40), anti-PML rabbit serum r8 (5), and anti-ICP0 monoclonal antibody 11060 (18). The secondary antibodies used were fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma) and Cy5-conjugated goat anti-rabbit immunoglobulin G (Amersham). The coverslips were then mounted using Citifluor AF1 and examined using a Zeiss LSM 510 confocal microscope. The ECFP, fluorescein isothiocyanate, and Cy5 signals were excited using the 458-, 488-, and 633-nm laser lines, respectively. The data from the channels were collected sequentially using the appropriate bandpass filters built into the instrument. Data were collected with fourfold averaging at a resolution of 1,024 by 1,024 pixels using optical slices between 0.5 and 1 μm thick. The microscope was a Zeiss Axioplan utilizing an ×63 oil immersion objective lens, NA 1.4. Data sets were processed using the LSM 510 software and then exported for preparation for printing using Photoshop.
RESULTS
Characterization of viruses expressing autofluorescent ICP0 and ICP4 proteins.
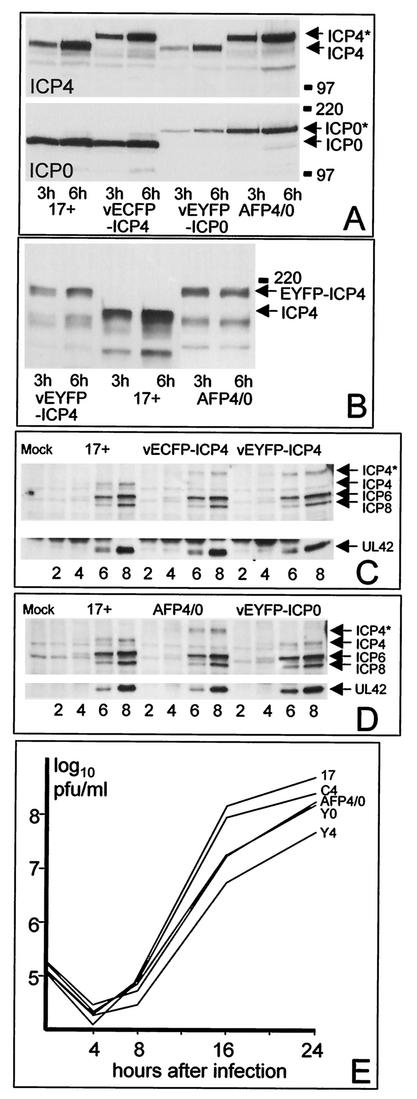
HSV-1 expressing EYFP-linked ICP0 and both ECFP and EFYP versions of ICP4 were constructed as described in Materials and Methods. In addition, virus AFP4/0, which expresses EYFP-ICP0 and ECFP-ICP4 from both copies of its IE1 and IE3 genes, was isolated after in vivo recombination and plaque purification. The expression of the AFPs by the viruses used in the present study was analyzed by Western blotting of infected whole-cell extracts. All viruses expressed fusion proteins of the expected sizes and the relevant wild-type proteins as expected (Fig. 1A and B), and they also expressed a variety of typical early viral proteins as efficiently as the wild-type 17+ control. After infection of Vero cells at a multiplicity of 2 PFU per cell, the time courses and amounts of ICP4, ICP6, ICP8, and UL42 expression by vECFP-ICP4, vEYFP-ICP4, vEYFP-ICP0, and AFP4/0 were indistinguishable from those of the wild-type strain 17+ (Fig. 1C and D). Since ICP4 is essential for the activation of early gene transcription (11, 37), these data illustrate the fact that the AFP-ICP4 fusion proteins function as efficiently as the normal protein under these conditions.
FIG. 1.
(A) BHK cells were infected with the viruses 17+, vECFP-ICP4, vEYFP-ICP4, and AFP4/0 (MOI = 10) as indicated, and whole-cell extracts were prepared 3 or 6 h after virus adsorption. The proteins were separated on 7.5% polyacrylamide gels, transferred to a nitrocellulose filter, and then detected by probing the same filter for ICP4 and ICP0 sequentially. The gel mobilities of the normal proteins are represented by the 17+ samples; the recombinant viruses express either the normal protein or the autofluorescent fusion protein of lower mobility, as expected from their genotypes. The positions of selected molecular mass markers (in kilodaltons) and the ICP4 and ICP0 proteins are indicated. The asterisks indicate the AFP versions of ICP4 and ICP0. (B) The same experiment as in panel A was conducted with viruses vEYFP-ICP4, 17+, and AFP4/0 probed for expression of ICP4. (C) Vero cells were infected with 17+, vECFP-ICP4, and vEYFP-ICP4 (MOI = 2), and whole-cell extracts were prepared 2, 4, 6, and 8 h after adsorption. The samples were analyzed by Western blotting for the expression of ICP4, ICP6, ICP8, and UL42. ICP4* represents the autofluorescent version of the protein. (D) The same experiment as in panel C was conducted using viruses 17+, AFP4/0, and vEYFP-ICP0. (E) Growth curves of the autofluorescent viruses. BHK cells in parallel 35-mm-diameter plates were infected with the indicated viruses at a multiplicity of 0.5 PFU per cell. The cells and medium were harvested 4, 8, 16, and 24 h after virus adsorption, and the samples were titrated along with the input aliquots on Vero cells. The data are derived from a single experiment but are representative of several similar experiments.
The ICP0 fusion protein expressed by vEYFP-ICP0 localized normally to ND10 at early times of infection and caused the disruption of these domains by 4 h after infection (data not shown). An EGFP-ICP0 fusion protein expressed by a recombinant virus also localized to and disrupted both ND10 and centromeres, induced the degradation of the cellular proteins PML and CENP-C at these structures, and caused infected cells to stall in mitosis in a manner similar to that of the normal ICP0 protein (29). Like the normal ICP0 protein, the EGFP fusion protein induced an accumulation of conjugated ubiquitin at ND10 and centromeres prior to their disruption (16). These data indicate that the addition of the fusion moiety does not significantly alter the targeting or biochemical activities of ICP0. Another sensitive method to test the functionality of the ICP0 fusion protein is to compare the relative plaque-forming efficiencies of vEYFP-ICP0 and AFP4/0 with those of the wild-type strain 17+ and the ICP0 null mutant dl1403 on BHK and human fibroblast cells. Whereas viruses expressing functional ICP0 plaque with similar efficiencies on both cell types, ICP0 activity is particularly important in the human fibroblasts, so viruses with decreased ICP0 activity form plaques very inefficiently on these cells (41). The ratios of the titers on HFFF-2 and BHK cells of 17+, dl1403, vEYFP-ICP0, and AFP4/0 were 0.38, 0.007, 0.35, and 0.75, respectively, indicating that the normal and ICP0 fusion proteins have similar biological activities. Finally, expression of the EYFP-ICP0 protein in Vero cells increased the plaque-forming efficiency of the ICP0 null mutant dl1403 (data not shown).
Taken together, these data show that the addition of the fusion moiety to the N-terminal ends of ICP0 and ICP4 does not significantly compromise ICP0 function or ICP4-mediated activation of gene expression during virus infection. However, growth curve experiments indicated that the addition of the fusion moieties causes a slight reduction in virus yields (Fig. 1E). Over a number of experiments, virus vECFP-ICP4 gave yields reduced by ∼2-fold, while vEYFP-ICP0 and AFP4/0 replicated to levels 3- to 4-fold lower than that of strain 17+. For reasons that are unclear, virus vEYFP-ICP4 gave yields about 10-fold smaller than those of strain 17+ (Fig. 1E). Despite this apparent difference in the growth potentials of the EYFP- and ECFP-ICP4 fusion protein viruses, no significant differences between the rates of gene expression (Fig. 1C and D) or behavior in live-cell experiments (data not shown) of these viruses were noted during the course of these studies.
ICP4 forms tiny foci at the early stages of infection, some of which later develop into replication compartments.
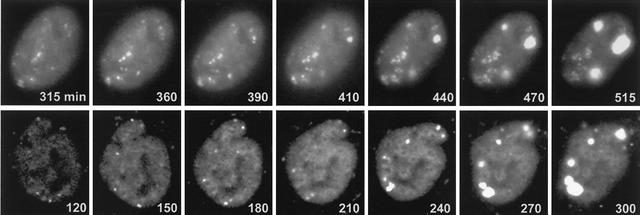
Prior to the onset of DNA replication, ICP4 in fixed cells has been observed to be mostly distributed diffusely throughout the nucleus. At later times of infection, punctate foci of ICP4 were observed, in addition to larger accumulations corresponding to replication compartments (10, 28, 38, 47). To investigate the behavior of ICP4 in live cells, HEp-2 cells were infected with virus vECFP-ICP4 and examined at intervals thereafter. Even at very early times of infection—indeed, as soon as the fluorescent ICP4 could be detected—we observed the appearance of punctate foci of the protein. As infection progressed, a subset of these foci developed into replication compartments (Fig. 2, top row). There appeared to be a relatively long lag period before this occurred in HEp-2 cells. In the example shown, the ICP4 foci remained small until at least 6 h after infection, but once initiated, the replication compartments developed very rapidly over the next 2 h (Fig. 2, top row). The development of replication compartments proceeded much more quickly in Vero cells, probably because in our hands viral gene expression was generally more efficient and developed more rapidly in Vero than in HEp2 cells. In the example shown (Fig. 2, bottom row), several of the ICP4 foci had expanded into small replication compartments by 4 h postinfection, and thereafter, the compartments developed rapidly.
FIG. 2.
Recruitment of ICP4 into foci, some of which later develop into replication compartments. HEp-2 cells (top row) or Vero cells (bottom row) were infected with vECFP-ICP4 (MOI = 10) and analyzed by live-cell microscopy. The times after addition of the virus are indicated in each panel.
The ICP4 foci formed early during HSV-1 infection are often juxtaposed to ND10 in live cells.
The formation of distinct ICP4 foci at very early times of infection in live cells was unexpected. In previous studies with fixed cells, ICP4 appeared to be mostly diffusely distributed at early times of infection (10, 28, 38, 46). The increased sensitivity, lower background signals, and absence of potential problems caused by fixation and sample preparation in the live-cell methods seem to be revealing an aspect of ICP4 dynamics that is not readily observed using fixed cells. However, the ICP4 foci were not observed in transfected cells expressing ECFP-linked ICP4 alone (see below), indicating that focus formation was not a consequence of the ECFP fusion. The distribution of ICP4 at early times of infection appears to be influenced by the fact that the cells are infected rather than simply expressing the protein.
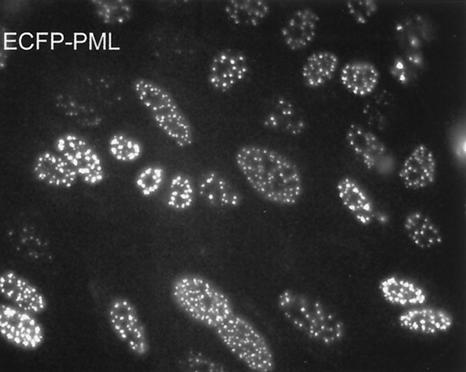
Given the precise colocalization of ICP0 at ND10 (17, 31) and the observed juxtaposition of ND10 and a proportion of parental viral genomes (22, 32), we developed methods to visualize ND10 in live cells so that the relative locations of the ICP4 foci could be established. Baculoviruses have been shown to enter mammalian cells efficiently and express proteins if they are linked into an appropriate expression cassette (6, 19), although they do not express insect proteins from the insect virus promoters (data not shown). Infection of HEp-2 cells with a baculovirus containing a human cytomegalovirus (HCMV) promoter-driven ECFP-linked PML gene cassette gave readily detectable autofluorescent PML in a high proportion of cells (Fig. 3). The expressed ECFP-PML protein was modified by conjugation to SUMO-1 (data not shown) and was recruited into punctate foci that also contained endogenous Sp100, another major ND10 constituent (Fig. 4, top row). The level of PML expression was proportional to the multiplicity of the baculovirus infection. The experiments described in this paper were conducted using multiplicities of baculovirus infection that did not result in gross overexpression of the protein and its aggregation into abnormally large foci. Only those cells that contained ECFP-PML in an apparently normal ND10-like distribution were selected for further study.
FIG. 3.
Expression of ECFP-PML. HEp-2 cells were infected with AC.CMV.ECFP-PML at a multiplicity of ∼50 PFU per cell and then examined by live-cell microscopy the following day. A very high proportion of the cells express autofluorescent PML that localizes in characteristic ND10 structures.
FIG. 4.
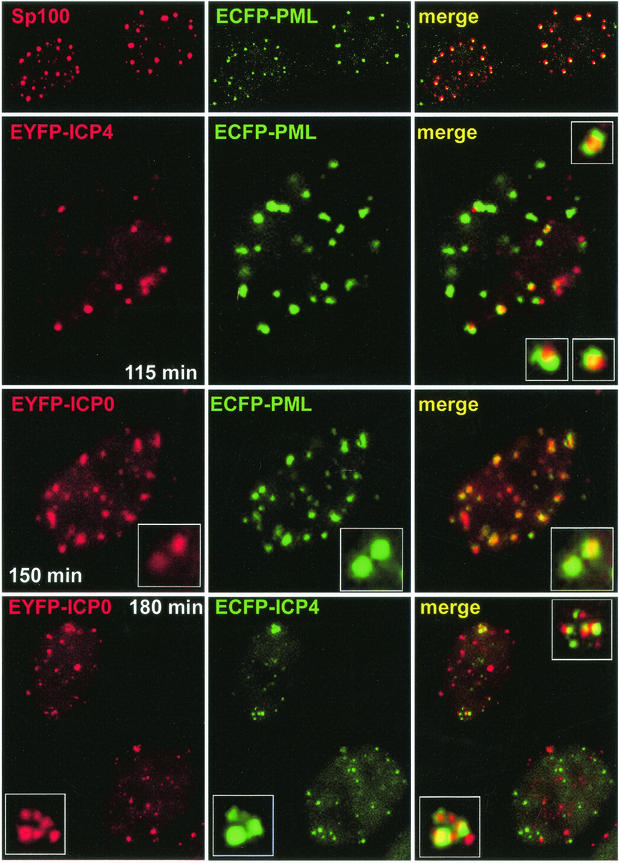
Examination of the relative locations of PML, ICP0, and ICP4 in live HEp-2 cells. Each row shows the single and merged channels of typical examples of cells from individual experiments. (Top row) ECFP-labeled PML (green) expressed from baculovirus Ac.CMV.ECFP-PML precisely colocalizes with endogenous Sp100 (red) in fixed cells as examined by confocal microscopy. (Row 2) HEp-2 cells infected the day before with Ac.CMV.ECFP-PML were infected with vEYFP-ICP4 (MOI = 10). The image was captured 115 min after addition of the virus (i.e., 55 min after a 60-min adsorption period). The inset images in the merged image show at higher magnification examples of ICP4 foci associated but not precisely colocalizing with PML. (Row 3) HEp-2 cells infected the day before with Ac.CMV.ECFP-PML were infected with vEYFP-ICP0 (MOI = 10). The image was captured 150 min after addition of the virus. Note that the colocalization of ICP0 and PML, indicated by the expanded insets, is far greater than that exhibited by ICP4 and PML. (Bottom row) HEp-2 cells were infected with virus vAFP4/0 (MOI = 10). The image was captured 180 min after addition of the virus. The inset images show examples of association but not precise colocalization of the ICP0 and ICP4 foci.
Subsequent infection of HEp-2 cells expressing ECFP-linked PML with vEYFP-ICP4, an HSV-1 derivative expressing EYFP-linked ICP4, showed that a proportion of the ICP4 foci were juxtaposed to but not precisely colocalized with PML in ND10 (Fig. 4). These images are remarkably similar to the juxtaposition of ND10 to both HSV-1 and HCMV genomes (22, 32) and the localization of the HCMV IE2 transcriptional regulator protein in foci associated with ND10 (23).
Fluorescently labeled PML and ICP0 colocalize, and ICP4 and ICP0 form juxtaposed foci in live, infected cells.
Fixed-cell studies have shown that ICP0 precisely colocalizes with PML in ND10 at early times of infection (17, 31). This behavior was reproduced in live HEp-2 cells that had first been infected with Ac.CMV.ECFP-PML and subsequently with vEYFP-ICP0 (Fig. 4). A prediction from this and the previous experiment is that the foci formed by ICP0 and ICP4 at the early stages of infection should be juxtaposed or associated rather than precisely colocalized. To test this hypothesis, a virus, vAFP4/0, was constructed that expresses ECFP-ICP4 from both copies of the IE3 gene and EYFP-ICP0 from both copies of the IE1 gene. At early times of infection of HEp-2 cells with vAFP4/0, both ICP0 and ICP4 formed punctate foci (Fig. 4). Although many of the foci were not associated, several of the ICP4 foci were indeed juxtaposed to ICP0 accumulations. Even in cases where the ICP4 and ICP0 signals appeared to be colocalizing, detailed analysis at high magnification revealed that the signals from the two proteins were distinct (Fig. 4, lower set of insets). From these data, it can be concluded that ICP0 precisely colocalizes with PML in ND10 while ICP4 forms punctate foci corresponding to separate bodies, some of which are associated with but not identical to the ICP0-ND10 structures. The juxtaposition of the ICP0 and ICP4 foci is again remarkably reminiscent of the relative locations of IE1 and IE2, their functional homologues in HCMV (23). It is also reminiscent of the formation of foci of ICP8 and other replication proteins in close association with ND10 (30, 44). The observation that two viral tegument proteins can also form foci that are associated with ICP0 structures (21) implies that several viral proteins may behave in a similar manner. However, our data indicate that ICP4 forms such foci rapidly, at times expected to precede abundant expression of early or late viral proteins.
Development of replication compartments from ICP4 foci juxtaposed to ICP0.
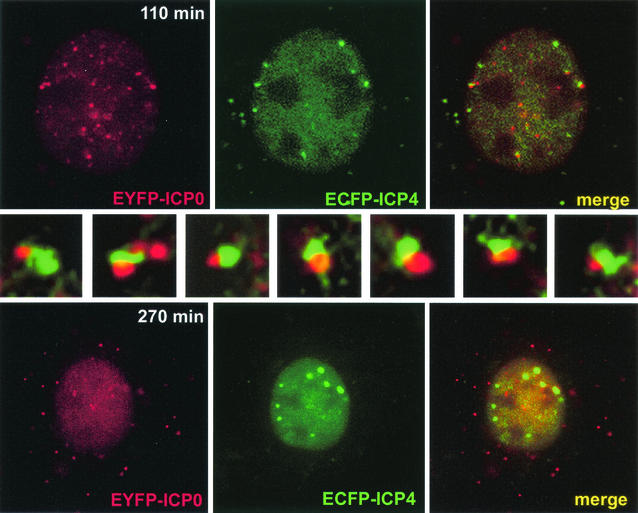
Vero cells are the preferred cell type to study the development of HSV-1 replication compartments, since this process takes place more rapidly and with greater efficiency in Vero than in HEp-2 cells (Fig. 2). A series of images was taken at 5-min intervals starting 110 min after the infection of Vero cells with AFP4/0. Figure 5 shows a cell from the first image and the same cell 160 min later when replication compartments had started to develop. These enlarged ICP4 foci continued to expand into typical large replication compartments at later times of infection (not shown). In the selected cell, there were large numbers of ICP0 foci early in infection and a smaller number of ICP4 foci, but in this case, most of the ICP4 foci were initially associated with an ICP0 structure. The insets in the center of Fig. 5 show expanded portions of the main 110-min image, illustrating the seven ICP4 foci that later develop into replication compartments. It appears that replication compartments develop from foci of ICP4 that at early times of infection are associated with ICP0 structures. Note that at later times of infection, ICP0 is diffusely distributed in the nucleus and has also migrated to form punctate foci in the cytoplasm (Fig. 5, 270 min).
FIG. 5.
Development of replication compartments from ICP4 foci that are initially juxtaposed to ICP0. Vero cells were infected with vAFP4/0 (MOI = 10). (Top row) Cell 110 min after addition of the virus, with multiple ICP0 foci (red) and a smaller number of ICP4 foci (green), many of which were juxtaposed to an accumulation of ICP0. (Middle row) Magnifications of seven examples of this juxtaposition (starting with the example at the top left of the cell and proceeding in a clockwise direction around the cell; the magnified image at the right end of the row corresponds to the ICP4 at the top right and was taken from the previous image in the time sequence, when the associated ICP0 dot was more prominent). (Bottom row) The same cell 270 min after addition of the virus, when the seven original foci had developed into small replication compartments (there is an additional developing compartment below and to the left of the right topmost original focus). At later times of infection, all these foci expanded further into substantial replication compartments (not shown).
Virus infection causes ICP4 expressed from a transfected plasmid to be recruited into foci.
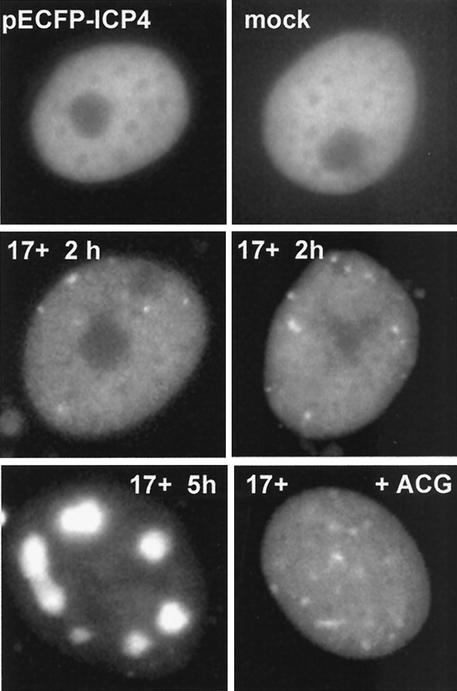
We next investigated whether virus infection is required for the formation of the ICP4 foci. Vero cells were transfected with plasmid pECFP-ICP4, and the localization of the fluorescent ICP4 protein was studied in cells that were either not infected or subsequently infected with HSV-1 strain 17+ under a variety of experimental conditions. It has previously been reported that ICP4 expressed from a transfected plasmid was distributed evenly in the nucleoplasm (46, 47). Similarly, we found that the ECFP-ICP4 protein by itself was diffusely distributed, and there was no appearance of distinct nuclear foci (Fig. 6, top). For the purposes of this analysis, cells expressing very high levels of ICP4, some of which could be in large aggregates, were not considered. In cells expressing low to medium levels of otherwise diffuse ICP4, distinct foci appeared as quickly as 40 min after addition of the virus, and by 2 h postinfection, the majority of cells contained ICP4 foci (Fig. 6, middle). By 5 h postinfection, many cells contained the fluorescent ICP4 protein in replication compartments (Fig. 6, bottom left). The formation of the replication compartments, but not the initial ICP4 foci, was abrogated by the DNA replication inhibitor acycloguanosine (Fig. 6, bottom right). Repetition of the experiment in the presence of cycloheximide or actinomycin D indicated that focus formation could not be attributed solely to recruitment of preexpressed ICP4, since both drugs greatly reduced the effect (data not shown). Therefore, it is likely that other viral proteins are also present in and required for the proper formation of the ICP4 foci. This question can be addressed in the future by construction of viruses expressing combinations of autofluorescently labeled wild-type and mutant viral proteins.
FIG. 6.
Formation of ICP4 foci requires virus infection. Vero cells were transfected with plasmid pECFP-ICP4, and individual cells were examined by live-cell microscopy the following day. (Top) In the absence of further treatment, ICP4 gave a diffuse nuclear distribution. (Middle) Two hours after infection of the transfected cells with HSV-1 17+ (MOI = 10), foci of ICP4 appeared. (Bottom) By 5 h after infection, ICP4 had been recruited into replication compartments in a number of cells (left), but this did not occur in the presence of 5 μM acycloguanosine (+ACG; right).
The ICP4 foci are not formed at the nonpermissive temperature by tsK, a virus with a lesion in the DNA binding domain of ICP4.
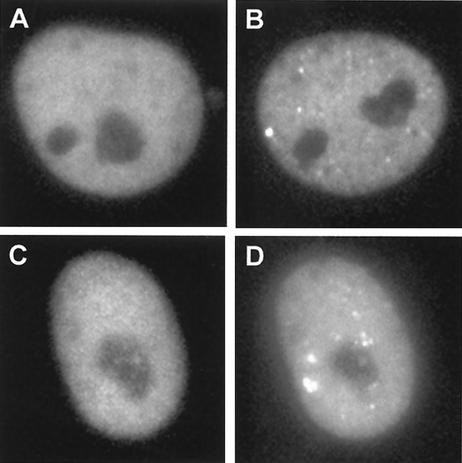
We next tested the role of DNA binding by ICP4 in the formation of the foci. The ICP4 tsK mutation was transferred into plasmid pIE3-EYFP-ICP4, and then the mutant plasmid was used to generate virus vEYFP-ICP4tsK. The presence of the mutation was confirmed by DNA sequencing of the transfer plasmid and by the temperature sensitivity of the recombinant virus (not shown). The tsK mutation produces a reversible thermolabile defect (37), and since it lies in the DNA binding domain of ICP4, it confers a temperature-sensitive DNA binding phenotype (36). Vero cells infected with this virus at the nonpermissive temperature displayed a diffuse nuclear localization of the mutant ICP4 protein, and this pattern did not change over several hours. When the temperature of the microscope incubation chamber was reduced to the permissive temperature, nuclear foci appeared within 15 min (Fig. 7). Therefore, correct folding of the DNA binding domain of ICP4 is required for the formation of the foci. This result does not exclude the role of other viral proteins in this effect, however, since the tsK lesion could also affect the interaction of ICP4 with other proteins.
FIG. 7.
Formation of ICP4 foci is dependent on correct folding of its DNA binding domain. Vero cells were infected with virus vEYFP-ICP4tsK (MOI = 10) at either the permissive or nonpermissive temperature (33 and 38.5°C, respectively). (A and B) At 38.5°C, the protein gave a diffuse nuclear distribution (A; 6 h postinfection) in contrast to the appearance at 33°C of nuclear foci that were similar to those in a wild-type virus infection (B; 4 h postinfection) (compare with Fig. 2). (C) Cells that had been held at nonpermissive temperature for 6 h and then shifted down to the permissive temperature exhibited a rapid appearance of ICP4 foci. (D) The same cell 15 min after temperature downshift.
DISCUSSION
This study demonstrates that the dynamics of ICP0 and ICP4 with respect to ND10 can be studied in live cells. Most significantly, ICP4 was observed to form ND10- and ICP0-associated foci in the early stages of infection, and a proportion of these foci later developed into replication compartments. These results give a graphic illustration of the general principle that the locations of viral proteins and genomes with respect to the cellular architecture are likely to be crucial factors in the progress of viral infection. Thus, the biochemical properties of a protein in functional or in vitro assays must be considered in conjunction with its localization in the infected cell. Indeed, the proper functioning of regulatory proteins that act on the viral genome can only occur when they are assembled with other relevant proteins into nucleoprotein complexes on parental viral genomes. The development of these complexes into productive replication compartments is likely to be influenced by their functional interactions and associations with structural components of the nucleus, and therefore their locations within the nucleus.
Taken together with observations of the localization of parental viral genomes in fixed and live cells (22, 32, 39), it is likely that a proportion of the ICP4 foci that are observed early after infection contain one or more parental viral genomes. Indeed, those that develop into replication compartments must contain at least one parental genome. Other viral and cellular proteins that bind to functional elements on the viral DNA, or interact with the DNA-bound proteins, must also be located in these structures. Thus, from its earliest stages, virus infection must involve a structured assembly of viral proteins and DNA, implying that intracellularly replicating virus has a more discrete structural aspect than might be implied from the apparently rather general localization of many of its regulatory proteins throughout the nucleus. The results presented in this paper, taken together with our recent observation that replication compartments develop preferentially from parental viral genomes that are associated with ND10 (39), strongly suggest that the viral nucleoprotein complexes that are associated with ND10 (and hence also with accumulations of ICP0) have an increased probability of giving rise to a replication compartment and progeny viral genomes.
While the ICP4 foci that later develop into replication compartments must contain at least one parental viral genome, it is uncertain whether all such foci contain viral DNA. The situation is likely to be complex. For example, the tendency for parental genomes to migrate to ND10 implies that at a high multiplicity of infection more than one parental genome may congregate in one place, so the number of foci may be less than the applied number of PFU. On the other hand, the normally high particle/PFU ratio of viral stocks means that there are far more viral genomes in the inoculum than PFU, which could in principle increase the number of foci at low multiplicities of infection (MOI). Other complicating factors include likely cell-to-cell variability in the efficiencies of particle adsorption, transport, and uncoating and even the presence of aggregates of input viral particles. Although this analysis is difficult (or even intractable, given the variations in dot intensity and detection), those foci that develop into replication compartments must contain at least one viral genome. Given the number of AFP molecules required to give a detectable signal, there must also be large amounts of ICP4 present in such structures. It is interesting that matrix mean analysis has identified as many as 500 potential ICP4 binding sites scattered throughout the HSV-1 genome (12). If ICP4 molecules accumulate on viral genomes in such numbers, they could have a significant effect on the assembly of the viral DNA into a chromatin-like structure, and it is possible that this assembly contributes, at least in part, to the transcriptional properties of ICP4 and the distinction between viral and cellular chromatin.
The relative localization of ICP0 in ND10 and ICP4 adjacent to ND10 early during HSV-1 infection has many parallels to the situation of HCMV infection. The IE1 protein of HCMV, which has some functional similarities to ICP0, precisely colocalizes with PML in ND10 during the early stages of infection, while IE2 (the transcriptional regulator that is functionally analogous to ICP4) forms foci that are associated (but not colocalized) with ND10 (23). Since HCMV parental genomes, like those of HSV-1, have a tendency to become associated with ND10, it is likely that the situations of HSV-1 and HCMV infection are analogous: one regulatory protein (IE1 or ICP0) precisely colocalizes with ND10, while the transcriptional regulator (IE2 or ICP4) forms adjacent foci that may also include viral genomes. This proposal would partly explain why the HCMV IE2 protein precisely colocalizes with ND10 in transfected cells but forms juxtaposed foci during infection (23); in transfected cells, IE2 may be aberrantly located because viral genomes are absent, so the protein is unable to assemble into the macromolecular complexes characteristic of a viral infection. We note that simian virus 40 (SV40) large T antigen, a transcriptional regulator with many functional similarities to IE2 and ICP4, also forms foci adjacent to ND10 (7, 14, 25), as do model SV40 genomes (43). Therefore, the related but different localizations of ICP0 and ICP4 may reflect a situation that is common to many different viral infections. The significance of the juxtaposition or association, rather than precise colocalization, is that ICP0 and ICP4 are present in separate structural entities (ND10 and viral nucleoprotein complexes, respectively) and therefore cannot be precisely colocalized but can be in close proximity.
The observations of the localization of ICP4, ICP0, parental viral genomes, and ND10, and the preferential development of replication compartments from the ND10-associated genomes (39), raise several questions concerning the basis of protein and genome localization juxtaposed to ND10 and whether this has a positive or negative effect on the efficiency of viral infection. The kernel of the argument is whether ND10 structures are sites of repression or whether they are depots of nuclear factors that may benefit viral infection (34). There is accumulating evidence that viral-genome association with ND10 is increased when virus-induced nucleoprotein complexes, such as those involved in transcription or DNA replication, assemble on the viral chromatin (4, 39, 42, 43). In the case of HSV-1, because the disruption of ND10 by ICP0 correlates with an increased probability of productive infection, it is possible that the ND10 environment has an initial repressive effect on viral gene expression. At later times of infection, generally after the original structures have been disrupted, the development of replication compartments from genomes that earlier were associated with ND10 suggests a more positive role for these bodies (or at least for their environment once modification by ICP0 has taken place). The results presented in this paper are consistent with those of previous studies of SV40, human papillomavirus, HSV-1, HCMV, Epstein-Barr virus, and human herpesvirus 8, which, taken together, suggest that association of viral genomes or replicating episomes with ND10 greatly increases the probability of the onset of productive DNA replication (1, 4, 22, 23, 32, 39, 42, 43, 45). We note that latent Epstein-Barr virus genomes are not associated with ND10 (4), and indirect evidence indicates that latent human herpesvirus 8 genomes may not be either (2, 9, 45). It will be very interesting to determine the site of quiescent HSV-1 genomes and to follow their location during the reactivation process.
Acknowledgments
This work was supported by the Medical Research Council. G.S. is a postdoctoral researcher funded by a Marie Curie Fellowship of the European Community Framework 5 program (contract number HPMF-CT-2000-01078).
We thank Duncan McGeoch for encouragement and helpful comments on the manuscript, Patrick Lomonte for development of the EGFP-linked ICP0 HSV-1 that led to the start of this work, Melanie Lambotin for investigating the expression of baculovirus proteins in HEp-2 cells, Dagmar Knebel-Morsdorf for anti-baculovirus protein antibodies, Alexandros Zafiropoulos for the image of endogenous Sp100 colocalizing with ECFP-PML, and Chris Boutell for help with the initial characterization of virus Ac.CMV.ECFP-PML. We also thank Jens Rietdorf and Andreas Girod (EMBL) for help with the live-cell management system and Paul Sheppard (Imaging Associates) for help with the automated microscopy.
REFERENCES
- 1.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Beard, P., S. Faber, K. W. Wilcox, and L. I. Pizer. 1986. Herpes simplex virus immediate early infected-cell polypeptide 4 binds to DNA and promotes transcription. Proc. Natl. Acad. Sci. USA 83:4016-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, P., P. M. Lieberman, and G. G. Maul. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800-11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 6.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 93:2348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockell, M., and S. M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9:199-205. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, M. A., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162-178. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiDonato, J. A., J. R. Spitzner, and M. T. Muller. 1991. A predictive model for DNA recognition by the herpes simplex virus protein ICP4. J. Mol. Biol. 219:451-470. [DOI] [PubMed] [Google Scholar]
- 13.Disney, G. H., and R. D. Everett. 1990. A herpes simplex virus type 1 recombinant with both copies of the Vmw175 coding sequences replaced by the homologous varicella-zoster virus open reading frame. J. Gen. Virol. 71:2681-2689. [DOI] [PubMed] [Google Scholar]
- 14.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R., A. Cross, J. Tyler, and A. Orr. 1993. An epitope within the DNA-binding domain of the herpes simplex virus immediate early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J. Gen. Virol. 74:1955-1958. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 19:6155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann, C., V. Sandig, G. Jennings, M. Rudolph, P. Schlag, and M. Strauss. 1995. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. USA 92:10099-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, W. L., and R. D. Everett. 2001. Human neuron-committed teratocarcinoma NT2 cell line has abnormal ND10 structures and is poorly infected by herpes simplex virus type 1. J. Virol. 75:3819-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, W. Q., L. Szekely, G. Klein, and N. Ringertz. 1996. Intranuclear redistribution of SV40T, p53, and PML in a conditionally SV40T-immortalized cell line. Exp. Cell Res. 229:289-300. [DOI] [PubMed] [Google Scholar]
- 26.Kastner, P., A. Perez, Y. Lutz, C. Rochette-Egly, M. P. Gaub, B. Durand, M. Lanotte, R. Berger, and P. Chambon. 1992. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 11:629-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knipe, D. M., P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). 2001. Fields virology, 4th ed. Lippincott-Williams and Wilkins, Philadelphia, Pa.
- 28.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G(1) into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 32.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 33.Muller, M. T. 1987. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J. Virol. 61:858-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, T., V. G. Preston, and R. D. Everett. 1990. A mutant of herpes simplex virus type 1 immediate early polypeptide Vmw175 binds to the cap site of its own promoter in vitro but fails to autoregulate in vivo. J. Gen. Virol. 71:851-861. [DOI] [PubMed] [Google Scholar]
- 37.Preston, C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 39.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sternsdorf, T., H. H. Guldner, C. Szostecki, T. Grotzinger, and H. Will. 1995. Two nuclear dot-associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand. J. Immunol. 42:257-268. [DOI] [PubMed] [Google Scholar]
- 41.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 42.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 45.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Z., and P. A. Schaffer. 1995. Intracellular localization of the herpes simplex virus type 1 major transcriptional regulatory protein, ICP4, is affected by ICP27. J. Virol. 69:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]