Abstract
Human papillomavirus type 16 (HPV16) is the primary etiologic agent of cervical carcinoma, whereas bovine papillomavirus type 1 (BPV1) causes benign fibropapillomas. However, the capsid proteins, L1 and L2, of these divergent papillomaviruses exhibit functional conservation. A peptide comprising residues 1 to 88 of BPV1 L2 binds to a variety of cell lines, but not to the monocyte-derived cell line D32, and blocks BPV1 infection of mouse C127 cells. Residues 13 to 31 of HPV16 L2 and BPV1 L2 residues 1 to 88 compete for binding to the cell surface, and their binding, unlike that of HPV16 L1/L2 virus-like particles, is unaffected by heparinase or trypsin pretreatment of HeLa cells. A fusion of HPV16 L2 peptide 13-31 and GFP binds (Kd, ∼1 nM) to ∼45,000 receptors per HeLa cell. Furthermore, mutation of L2 residues 18 and 19 or 21 and 22 significantly reduces both the ability of the HPV16 L2 13-31-GFP fusion protein to bind to SiHa cells and the infectivity of HPV16 pseudovirions. Antibody to BPV1 L2 peptides comprising residues 115 to 135 binds to intact BPV1 virions, but fails to neutralize at a 1:10 dilution. However, deletion of residues 91 to 129 from L2 abolishes the infectivity of BPV1, but not their binding to the cell surface. In summary, L2 residues 91 to 129 contain epitopes displayed on the virion surface and are required for infection, but not virion binding to the cell surface. Upon the binding of papillomavirus to the cell surface, residues 13 to 31 of L2 interact with a widely expressed, trypsin- and heparinase-resistant cell surface molecule and facilitate infection.
The infectious process for most viruses, including papillomavirus, is poorly understood. However, in the last decade, a plethora of primary viral receptors have been identified (reviewed in reference 1). Viruses adhere to the target cells via these primary receptors, but their uptake and transport to the site of viral replication often require interaction with other secondary viral receptors (1).
The papillomavirus capsid comprises two genetically unrelated viral proteins called L1 and L2 that surround the ∼8-kb histone-bound, closed circular viral genomic DNA (14). Expression of the major capsid protein L1 results in T=7 virus-like particles (VLPs) formed from 72 petameric L1 capsomers (21, 29). Three-dimensional image reconstructions and the ∼30:1 ratio of L1 and L2 in native bovine papillomavirus type 1 (BPV1) suggest that the L2 minor capsid protein is located in the center of the pentavalent capsomers at the virion vertices (46). Both L1 and L2 are necessary for efficient production of infectious viruses in vivo, with L2 functioning in both encapsidation and the infectious process (36, 37, 52).
Papillomaviruses bind via L1 to cells derived from a wide variety of tissues and species (34, 40). While papillomavirus pseudovirions lacking L2 are infectious (45), recent reports suggest that L2 also can bind to the cell surface, resulting in its internalization (28), and that L2 is critical to the infectious process (36, 47). Furthermore, antibody to L2 can neutralize papillomavirus without preventing virions from binding to the cell surface (41). Taken together, these findings suggest a role for L2 in facilitating infection via interaction with a secondary receptor(s) (28). Thus, we sought to define the residues of L2 that bind to the cell surface and their contribution to the infectious process of papillomavirus.
MATERIALS AND METHODS
Cell lines.
Human cervical carcinoma-derived cell lines HeLa, SiHa, and Caski; renal carcinoma cell line RCC24; ovarian carcinoma cell line OVCAR-3; colon carcinoma cell line DW-6; melanoma cell line Mel; monkey kidney epithelial cell line COS-1; mouse C127 clone C cells; and BPV1-transformed hamster fibroblast line BPHE-1 were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin G per ml, and 100 μg of streptomycin per ml. The leukemia cell lines Thomas, Salt, and K562 and the head and neck carcinoma cell line H&N013 were cultured in RPMI 1640 medium supplemented with 10% FCS, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml. The transformed monocyte cell line D-32 was grown in RPMI 1640 medium containing 10 ng of interleukin-3 per ml, 10% FCS, 100 U of penicillin G per ml, and 100 μg of streptomycin per ml.
Expression of fusion proteins.
Hexahistidine (six-His)-tagged peptides comprising residues 1 to 88, 45 to 173, 130 to 257, 216 to 340, 300 to 425, 384 to 469, and 1 to 469 of BPV1 L2 and full-length human papillomavirus 16 (HPV16) L2 were prepared as previously described (40). The glutathione S-transferase (GST)-green fluorescent protein (GFP) fusion was produced by insertion of the GFP gene of pGL-2 (Life Technologies) between the EcoRI and XhoI sites of pGEX-6P-1 (Amersham Pharmacia). HPV16 L2 gene fragments were either PCR amplified or directly synthesized and inserted between the GST and GFP genes at the EcoRI and SalI restriction sites. The fusion proteins were produced in Escherichia coli BL21 upon induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h and affinity purified with GST-trap FF columns according to the manufacturer's instructions (Amersham Pharmacia). Peptide purity and concentration, respectively, were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie blue staining and the Bradford assay with bovine serum albumin standards.
Analysis of cell surface binding by L2 and particles.
Trypsinized cells were washed and incubated in phosphate-buffered saline (PBS) with six-His-tagged BPV1 L2 fragments or whole BPV1 or HPV16 L2 protein for 1 h on ice. After washing, the cells were incubated with pentahistidine-specific monoclonal antibody (mouse immunoglobulin G1 [IgG1]; Qiagen) or an isotype control and then FITC-conjugated goat anti-mouse antibody (Kirkegaard & Perry Laboratories) for 30 min each. Finally, the cells were washed in PBS, fixed in 3.7% paraformaldehyde-PBS, and analyzed by flow cytometric analysis (FAScan; Becton Dickinson). HPV16 L2 peptides fused with GFP were bound to viable cells for an hour at 4°C in PBS, washed, and fixed. Binding was directly analyzed by flow cytometry or confocal fluorescence microscopy (UltraView Confocal Imaging System; Perkin-Elmer). The cells were mounted on a Nikon Eclipse TE 200 inverted microscope equipped with a ×40 plan fluor ×60 or ×100 plan apochromatic objective lens with a corresponding 1-, 0.8-, and 0.45-μm optical z-slice. Twelve-bit images were merged and analyzed with the Ultraview acquisition software in the RGB mode. Scatchard analysis was performed by incubation of defined concentrations of HPV16 13-31 GFP fusion protein with 106 HeLa cells for an hour at 4°C. Each supernatant was collected, and the cells were washed and harvested. The supernatants and cell lysates were separated by SDS-PAGE and analyzed by Western blotting with mouse monoclonal antibody to GFP, horseradish peroxidase-labeled anti-mouse IgG antibody, Lumiglo (Kirkegaard & Perry Laboratories), and densitometry of the bands (48). The binding of HPV16 VLPs after incubation at 4°C for an hour with HeLa cells was measured by flow cytometry with monoclonal antibody H16.V5 (1:100 dilution of ascites), and then phycoerythrin-conjugated goat anti-mouse antibody (Kirkegaard & Perry Laboratories). Heparinase treatment was performed with a combination of forms I, II, and III at 2.5 U/ml as described by Joyce et al. (23). To assess the ability of BPV1 virions to bind the cell surface, L1 was coexpressed with either L2 or L2Δ91-129 via Semliki Forest virus (SFV) in BPHE-1 cells, and the virions in sonicated cell extracts were separated from pentamers and empty VLPs by rate zonal centrifugation through a 20 to 40%(wt/vol) sucrose gradient for 90 min at 160,000 × g at 4°C (36). Equivalent quantities of virions or buffer alone were incubated with mouse C127 cells for 1 h at 4°C. The cells were washed, and bound virions were detected by indirect immunofluorescence with monoclonal antibody 5B6 to L1 (1:100 hybridoma supernatant) (12, 41). FITC-labeled anti-mouse IgG antibody was used at 5 μg/ml, and the slides were prepared with Fluoromount mounting fluid (Southern Biotechnology Associates). Fluorescence was examined with a Bio-Rad MRC 1024 laser scanning confocal system attached to a Zeiss Axioplan microscope. The images were acquired with a Zeiss ×63 N.A. 1.4 Planapo objective.
Generation of BPV1 and HPV16 pseudovirions and assay of infectivity.
To observe the blocking of BPV1 infectivity, monolayers of C127 clone C cells in 60-mm-diameter dishes were incubated with ∼200 focus-forming units (FFU) of BPV1 virions (purified from a bovine papilloma) and either 50, 25, 10, or 0 μg of L2 peptide in 1 ml of Dulbecco's PBS for 1 h at 37°C. The plates of C127 cells were washed twice with medium, maintained in culture for 3 weeks, and stained (15). To test the infectivity of BPV1 or HPV16 pseudovirions containing mutations in L2, substitutions or deletions were introduced within full-length L2 by PCR, and the mutant gene was cloned into vector pSFV4.2 and verified by sequencing. BPV1 or HPV16 pseudovirions containing wild-type or mutant L2 were prepared by infection of BPHE-1 cells with defective recombinant SFV expressing L1 and L2 or its mutants as described previously (37), and their infectivity was quantified in the focal transformation assay (15). The capacity of the BPV1 L2Δ91-129 deletion mutant to encapsidate the BPV1 genome was determined as described in reference 35. Briefly, BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 and wild-type or mutant L2. Thirty hours postinfection, the cells were harvested and lysed by sonication. The capsid proteins were immunoprecipitated with rabbit anti-BPV1 VLP, and the immunoprecipitates were treated with DNase I to eliminate nonencapsulated genomes. The amount of DNase I-resistant BPV1 genome present in the immunoprecipitates was assessed by Southern blotting (35).
ELISA.
The enzyme-linked immunosorbent assay (ELISA) was performed as follows. Peptides comprising residues 100 to 120 and 115 to 135 of BPV1 L2 were coupled to keyhole limpet hemocyanin (KLH) according to the manufacturer's instructions (Pierce), and 0.3 mg was used to immunize rabbits with Freund's complete adjuvant and incomplete adjuvant for all booster immunizations. Diluted antiserum was tested for reactivity with microtiter plates coated with (per well) 100 ng of six-His-tagged BPV1 L2 residues 45 to 173 per well or 300 ng of purified, native BPV1 virions in PBS (41). Reactivity was detected with 1:10,000 peroxidase-linked and rabbit IgG and ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)].
RESULTS
N-terminal BPV1 L2 peptide binds to the cell surface and blocks BPV1 infection in vitro.
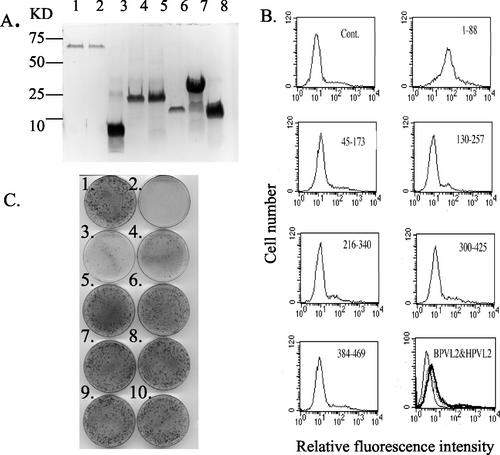
Recent genetic and biochemical studies suggest that L2 facilitates papillomavirus infectivity by binding to a secondary cell surface receptor (28, 36). Both to confirm that full-length L2 can bind to the cell surface and to identify which residues of L2 mediate this interaction, we employed full-length six-His-tagged BPV1 L2 and overlapping polypeptide fragments. These peptides span residues 1 to 88, 45 to 173, 130 to 257, 216 to 340, 300 to 425, and 384 to 469 and thus encompass the full length of the BPV1 L2 protein (41). The peptides as well as full-length BPV1 L2 and HPV16 L2 were overexpressed in E. coli and affinity purified to homogeneity (Fig. 1A). The binding of full-length HPV16 and BPV1 L2 to the surface of the human cervical carcinoma cell line HeLa was examined. The HeLa cells were incubated with six-His-tagged L2 peptides and immunofluorescently labeled with a tag-specific antibody, and the cell surface binding was analyzed by flow cytometry. Both the full-length HPV16 and BPV1 L2 proteins bound weakly to the surface of HeLa cells (Fig. 1B). Furthermore, while the BPV L2 peptide comprising residues 1 to 88 bound to HeLa cells, the other five peptides did not exhibit significant binding to HeLa cells (Fig. 1B). That the peptide encompassing residues 45 to 173 failed to bind may reflect the prior trypsin treatment of the HeLa cells (28).
FIG. 1.
A peptide comprising the first 88 amino acids of BPV1 L2 binds to HeLa cells and competes BPV1 infection. (A) Recombinant six-His-tagged HPV16 L2 (lane 1), BPV1 L2 full-length protein (lane 2), or fragments comprising residues 1 to 88 (lane 3), 45 to 173 (lane 4), 130 to 257 (lane 5), 216 to 340 (lane 6), 300 to 425 (lane 7), and 384 to 469 (lane 8) were affinity purified from E. coli, separated on an SDS-PAGE gel (15% polyacrylamide), and stained with Coomassie blue. (B) The six-His-tagged L2 polypeptides (5 μg in 0.5 ml of PBS) shown in panel A (heavy line) or buffer alone (fine line) was incubated with 2 × 106 HeLa cells for an hour on ice. After washing, bound L2 was detected in all samples by indirect immunofluorescence with a tag-specific monoclonal antibody and flow cytometric analysis. The bottom right panel shows the binding of full-length HPV16 L2 (dotted line), BPV1 L2 (heavy line), and buffer alone (fine line). Cont., control. (C) Monolayers of C127 cells, except for plate 2, were incubated with ∼200 FFU of purified BPV1, either alone (plate 1) or with 50 μg of BPV1 L2 1-88 (plate 3), 10 μg of BPV1 L2 1-88 (plate 4), 50 μg BPV1 L2 45-173 (plate 5), 10 μg of BPV1 L2 45-173 (plate 6), 50 μg of BPV1 full-length L2 (plate 7), 10 μg of BPV1 full-length L2 (plate 8), 50 μg of HPV16 full-length L2 (plate 9), or 10 μg of HPV16 full-length L2 (plate 10) in 1 ml of PBS for an hour at 37°C. The plates were washed, cultured for 3 weeks, and stained.
Infection of murine C127 cells by BPV1 is readily quantified in a focal transformation assay (15). Therefore, these recombinant L2 fragments were mixed with ∼200 FFU of purified native BPV1 virions and incubated with monolayers of mouse C127 cells. BPV L2 peptide derived from residues 1 to 88 effectively interfered with BPV1 infection of C127 cells (Fig. 1C). Consistent with the cell surface-binding data, other fragments of BPVL2, including the overlapping peptide encompassing residues 45 to 173, did not significantly affect BPV1 infection (Fig. 1C). Whereas the control plates contained over 250 foci, there were 32, 38, and 65 foci when infections were performed in the presence of (per milliliter) 50, 25, or 10 μg, respectively, of purified BPV1 L2 1-88 peptide. Full-length BPV1 L2 failed to detectably block BPV1 infection at 50 μg/ml (the molar equivalent of the BPV1 L2 1-88 peptide at 10 μg/ml). However full-length L2 also bound to HeLa cells less strongly than the 1-88 peptide fragment, and this may indicate a lower degree of correct folding of the full-length L2 compared to its N-terminal peptide when expressed in E. coli (Fig. 1B and C). It is noteworthy that of these peptides, only BPV1 L2 residues 384 to 489 bind to L1 (35), suggesting that BPV1 L2 residues 1 to 88 block BPV1 infection by binding to a cellular factor, rather than the virus itself.
BPV1 L2 residues 1 to 88 bind to a widely expressed and trypsin-resistant cell surface molecule.
Despite its limited host range and tropism for productive infection, papillomaviruses bind to and enter cells derived from a variety of tissues and species (34, 40, 48). Thus, we investigated whether the first 88 residues of BPV1 L2 can also bind to a wide range of cell lines. BPV1 L2 residues 1 to 88 strongly bound to the cervical carcinoma cell lines HeLa, SiHa, and CaSki (Fig. 2A), whereas the other 5 fragments failed to bind (not shown). Furthermore, BPV1 L2 residues 1 to 88 also bound to other human cell lines, including those derived from an ovarian carcinoma (OVCAR-3), a renal carcinoma (RCC24), a colon carcinoma (DW-6), several leukemias (Thomas, Salt, and K562), a head and neck carcinoma (H&N013), and a melanoma (Mel) (Fig. 2A). Cells of the monkey epithelial cell line COS-1 also showed an ability to bind with the N-terminal region of BPV1 L2 (Fig. 2A) (27), whereas binding to the monocyte-derived cell line D-32 was not detectable. None of the five other BPV L2 fragments detectably bound to these cell lines (data not shown).
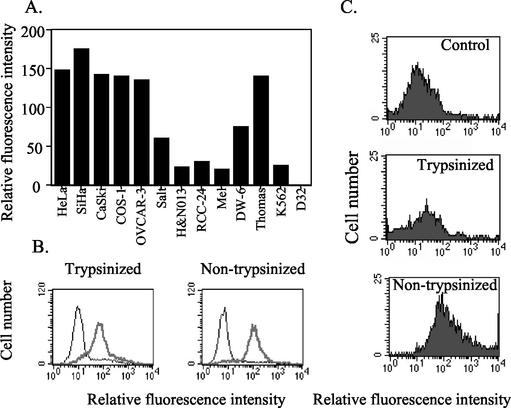
FIG. 2.
Trypsin-insensitive binding of BPV L2 residues 1 to 88 to a variety of cell lines. (A) Binding of the BPV L2 1-88 peptide at 4°C to the cell lines indicated was measured by indirect immunofluorescence and flow cytometric analysis. (B) Binding at 4°C of BPV L2 residues 1 to 88 to trypsin-treated (0.2%, 5 min at room temperature) (13) or untreated HeLa cells was measured by indirect immunofluorescence and fluorescence-activated cell sorter analysis. The heavy line indicates fluorescence in the presence of BPV L2 residues 1 to 88, and the lighter line corresponds to the fluorescence when the BPV1 L2 peptide was omitted. (C) Binding of HPV16 L1/L2 VLPs to HeLa cells to trypsin pretreated (0.2% for 5 min at room temperature) or control HeLa cells was measured by indirect immunofluorescence and flow cytometry.
Several studies show that prior treatment with trypsin prevents the binding of both VLP and L2 residues 108 to 126 to cells (17, 28, 34, 38). Prior treatment of HeLa cells with trypsin did not significantly affect the binding of BPV1 L2 residues 1 to 88 (Fig. 2B), but markedly reduced the surface binding of HPV16 L1 and L2 VLPs by HeLa cells (Fig. 2C). These data suggest both that the trypsin treatment was effective and that this peptide binds to a surface molecule distinct from the primary trypsin-sensitive receptor engaged by L1 (34).
Residues 13 to 31 of HPV16 L2 bind to the cell surface.
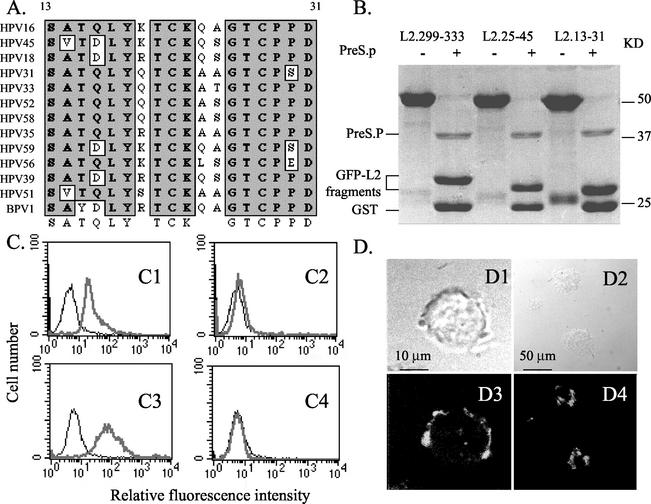
Since both of the L2 proteins of the evolutionarily distant BPV1 and HPV16 papillomaviruses bind to the surface of HeLa cells (Fig. 1B), we reasoned that the amino acid sequence of the cell surface-binding motif was likely to be well conserved. Indeed, the amino acid sequence of the N-terminal region of L2 is highly conserved between papillomavirus types (as shown on page II-L2-20 of http://www.stdgen.lanl.gov/stdgen/virus/hpv/compendium/htdocs/COMPENDIUM_PDF/97PDF/2/L2.pdf), and BLOCK and ClustalW analyses identify a putative motif (Fig. 3A). We therefore synthesized several conserved peptides from this region of HPV16 L2 and tested their ability to bind to the surface of cervical carcinoma cell lines. Conserved peptides of HPV16 L2 comprising residues 13 to 31, 25 to 45, or, as a control, 299 to 333 were expressed in E. coli as soluble fusion proteins between GST and GFP and affinity purified (Fig. 3B). Flow cytometric analysis and confocal fluorescence microscopy showed that of the fluorescent fusion proteins, only the fusion with residues 13 to 31 of HPV16 L2 clearly bound to viable SiHa cells (Fig. 3C and D), as well as HeLa and Caski cells (data not shown), whereas L2 peptides 25-45 and 299-333 failed to bind significantly (data not shown). To eliminate a possible contribution by GST to the binding to the HeLa cells, PreScission protease was used to specifically cleave GST from the L2 13-31-GFP fusion protein (49) (Fig. 3B). We observed equivalent binding of digested and undigested fusion protein to SiHa cells (Fig. 3C). Furthermore, this binding could mediate uptake of the fusion protein within 30 min at 37°C (Fig. 3D4).
FIG. 3.
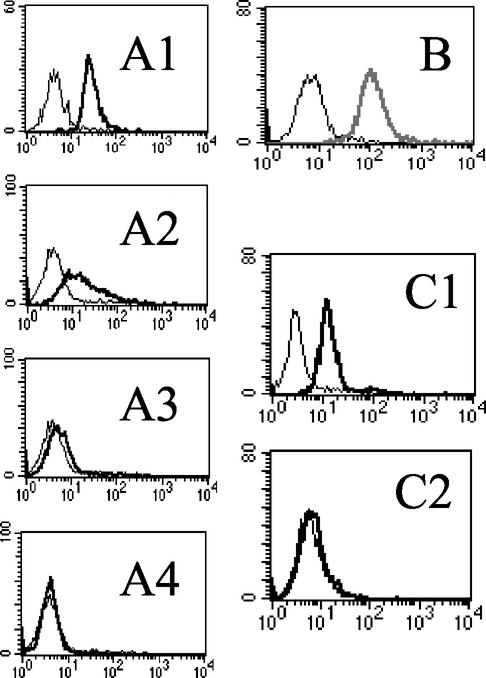
HPV16 peptide 13-31 is highly conserved and binds to the cell surface. (A) ClustalW-formatted alignment of the L2 amino acid sequence of representative high-risk HPV genotypes and BPV1 between residues 13 and 31. (B) Purified GST-GFP fusion proteins including residues 299 to 333, 25 to 45, and 13 to 31 of HPV16 L2, either with or without PreScission protease (PreS.P) digestion, were separated on an SDS-PAGE gel (15% polyacrylamide) and stained with Coomassie blue (49). The positions of the PreScission protease and GST- and GFP-L2 fragments are indicated. (C) Flow cytometric analysis of SiHa cells in the presence of buffer (fine line) or GFP fusion proteins (heavy line). (C1) PreScission protease-digested GST-HPV16 L2 13-31-GFP fusion protein. (C3) Undigested GST-HPV16 L2 13-31-GFP fusion protein. (C2) PreScission protease-digested GST-GFP. (C4) GST-GFP. (D) Analysis by confocal fluorescence microscopy of the cell surface binding and uptake of GFP fusion protein containing L2 residues 13 to 31. SiHa cells were incubated with GFP-tagged HPV16 L2 residues 13 to 31 for an hour at 4°C (D1 and D3), and then the cells were washed and shifted to 37°C for 30 min (D2 and D4). The cells were viewed by phase-contrast light microscopy (D1 and D2) or confocal fluorescence microscopy (D3 and D4).
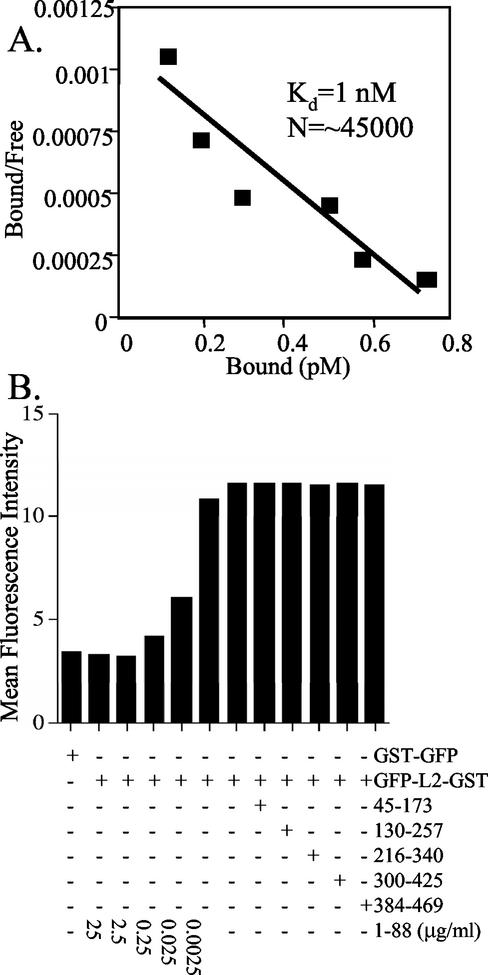
To determine the ability to saturate the putative cell surface receptor and estimate the number of receptors per HeLa cell, 106 HeLa cells were incubated for an hour at 4°C with increasing concentrations of HPV16 13-31 peptide fused with GFP. The amount of bound and free peptide was determined by Western blotting, and a Scatchard analysis was performed as described by Volpers et al. (48). A dissociation constant of 1 nM for binding of the HPV16 13-31-GFP fusion to ∼45,000 receptors per HeLa cell was estimated by linear interpolation of the data points (Fig. 4A).
FIG. 4.
Scatchard analysis and competition studies of L2 binding to the cell surface. (A) Known concentrations of the HPV16 L2 13-31-GFP fusion protein were incubated for an hour at 4°C with 106 HeLa cells. The supernatants containing free peptide and cell-bound peptide were harvested separately, and their ratio was determined by Western blotting for Scatchard analysis, as described in reference 48. (B) Binding of GST-GFP either lacking or containing residues 13-31 of HPV16 L2 to HeLa cells after incubation for 20 min at 4°C, washing, and fixation was measured directly by flow cytometry and expressed as the mean fluorescent intensity of 104 cells. The binding to HeLa cells of the HPV16 L2 13-31-GFP fusion in the presence of BPV1 L2 1-88 peptide at the concentrations indicated or control BPV1 L2 peptides at 25 μg/ml was assessed.
Since overlapping regions of BPV1 and HPV16 L2 bind to the surface of HeLa cells, we sought to determine whether the L2 peptides derived from these genotypes bind to the same surface receptors. A competition experiment was performed to determine the influence of the six overlapping BPV1 L2 peptides upon the binding of the HPV16 L2 13-31 peptide to HeLa cells. The binding to HeLa cells of GST-GFP either with or without the HPV16 L2 13-31 peptide fusion was directly measured by flow cytometry, and the mean fluorescence intensity was plotted (Fig. 4B). The presence of the BPV1 L2 1-88 peptide at 2.5 μg/ml, but not the other peptides at a 10-fold-higher concentration, reduced the binding of the HPV16 L2 peptide 13-31-GST and -GFP fusion with HeLa cells to a level equivalent to binding of nonspecific GST-GFP alone.
In order to validate the role of HPV16 L2 residues 13 to 31 in binding to the cell surface and identify residues critical for this interaction, we produced fusion proteins in which highly conserved L2 residues inserted between the GST and GFP domains were mutated and then analyzed their ability to bind to SiHa cells. While the mutation of residues 13 to 16 from ASAT to LLVV (L13L14V15V16) had a small effect upon the binding of the HPV16 L2 13-31 fusion protein to SiHa cells, the mutation of L2 residues 18 and 19 from LY to AA (A18A19) or mutation of residues 21 and 22 from TC to VV (V21V22) drastically reduced fusion protein binding to SiHa cells (Fig. 5A). Similar results were obtained when the HeLa or CaSki cell lines were substituted (data not shown).
FIG. 5.
Mutations, but not heparinase, eliminate binding of the HPV16 peptide 13-31 to cells. (A) The binding to SiHa cells of a 10-μg/ml sample of fusion protein comprising GST and GFP fused with either HPV16 L2 residues 13 to 31 (ASATQLYKTCKQAGTCPPD) or the L13L14V15V16 (LLVVQLYKTCKQAGTCPPD), A18A19 (ASATQAAKTCKQAGTCPPD), or V21V22 (ASATQLYKVVKQAGTCPPD) mutant peptides was assessed by flow cytometric analysis and plotted in panels A1 to A4, respectively. (B) The binding of a 10-μg/ml sample of fusion protein comprising GST and GFP alone (fine line) or fused with HPV16 L2 residues 13 to 31 (bold line) to SiHa cells pretreated with 2.5 U of heparinase I, II, and III per ml for 1 h at 37°C was analyzed by flow cytometry. (C) The binding of HPV16 L1/L2 VLPs to untreated (C1) or heparinase-treated (2.5 U each of forms I, II and III per ml for 1 h at 37°C) SiHa cells (C2) was detected with H16.V5 (heavy line) and FITC-conjugated anti-mouse IgG, or, as a negative control, secondary antibody alone (fine line).
Mutation of the cell surface-binding domain of L2 reduces the infectivity of HPV16 pseudovirions.
Since the mutation of residues 18 and 19 or 21 and 22 to AA or VV, respectively, reduces the ability of the cell surface interaction motif comprising residues 13 to 31 of HPV16 L2 to bind the cell surface and compete with BPV1 infection, we hypothesized that each double mutation would result in reduced infectivity when introduced within the context of an HPV16 pseudovirion. Therefore, full-length HPV16 L2 was mutated at residues 18 and 19 to AA (L2AA) or residues 21 and 22 to VV (L2VV) and cloned into the vector pSFV4.2 for the generation of defective recombinant SFV. These mutants exhibited sizes and expression levels similar to those of wild-type HPV16 L2 (data not shown). We have previously encapsidated the BPV1 genome within the HPV16 capsid to provide a simple readout of infectivity based upon focal transformation of mouse C127 cells, as was observed with native BPV1 virions (15, 37). To generate such HPV16 {BPV1} pseudovirions, the hamster fibroblast line BPHE-1, which harbors the BPV1 episome, but fails to express the BPV1 capsid genes, was coinfected with defective recombinant SFV expressing HPV16 L1 and wild-type L2 or the L2AA or L2VV mutants (37). After 30 h, the cells were harvested and sonicated, and the HPV16 pseudovirion infectivity in lysates was assayed by using the focal transformation of mouse C127 cells as a readout (15, 37). Mutation of L2 residues 18 and 19 to AA reduced the infectivity of the HPV16 pseudovirion by ∼10-fold compared to that of the wild type, whereas the mutation of residues 21 and 22 to VV resulted in only an approximately fivefold reduction in infectivity (Table 1).
TABLE 1.
Mutations within the cell surface interaction motif of L2 impair the infectivity of HPV16 pseudovirions
| Expt | No. of focia in flask
|
||||
|---|---|---|---|---|---|
| Control | L1 alone | L1+L2 | L1+L2AA | L1+L2VV | |
| 1 | 4 | 16 | 290 | 36 | 67 |
| 2 | 7 | 15 | 310 | 28 | 53 |
The control contained no virus infection. Each value represents the number of foci in a 25-cm2 flask.
Influence of heparinase pretreatment upon L2 and L1/L2 VLP binding to the cell surface.
L1 VLPs bind to heparan sulfate on the surface of cells, and heparinase pretreatment of cells inhibits both binding and infection (23, 43). To determine whether the L2 residue 13-31 motif also binds to heparan sulfate on the cell surface, HeLa cells were digested with a heparinase cocktail and incubated with GFP fusion protein either containing or lacking the HPV16 L2 13-31 motif. The binding of the L2 13-31 motif to HeLa cells was unaffected by heparinase treatment (Fig. 5B), whereas the binding of HPV16 L1 VLP was eliminated (data not shown).
Heparinase treatment dramatically reduces the binding of L1 VLPs to the cell surface, but has no effect upon L2 13-31 motif binding. Therefore, if the L2 13-31 motif is displayed upon the surface of particles, then heparinase pretreatment of HeLa cells should not eliminate the binding of HPV16 L1/L2 VLPs. Figure 5C shows that heparinase pretreatment of HeLa cells reduces the binding of HPV16 L1/L2 VLPs to background, suggesting that residues 13 to 31 are not displayed on the capsid surface. Furthermore, we compared the binding of HPV16 L1/L2 VLPs to HeLa cells in the presence or absence of HPV16 L2 13-31 peptide, both wild type and mutant, and observed no significant difference (data not shown).
Deletion of residues 91 to 128 of BPV1 L2 from the surface of BPV1 virions eliminates infectivity.
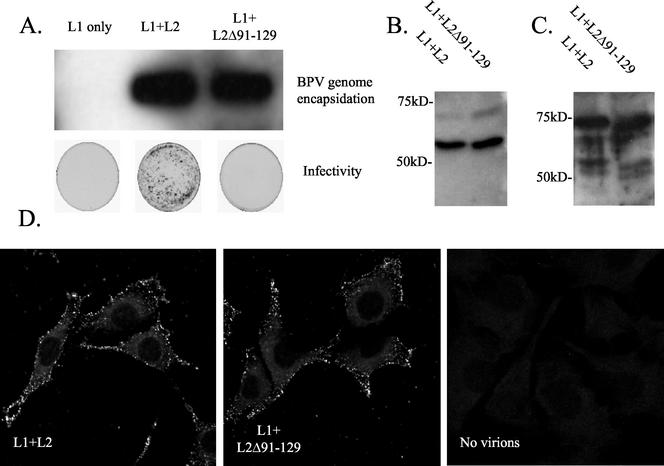
While we have demonstrated that a peptide comprising BPV1 L2 residues 1 to 88 binds to cells and blocks infection by BPV1, the BPV L2 peptide comprising residues 45 to 173 neither bound to cells nor blocked BPV1 infection of C127 cells. This result was surprising, since residues 108 to 120 of HPV16 L2 have been reported to bind to and enter cells, to contribute to infection by HPV16 pseudovirions (26, 28). It is possible that the BPV1 L2 45-173 peptide folds incorrectly when expressed in E. coli and thus fails to bind to the cell surface. Alternatively, it is possible that BPV1 L2 does not employ a domain functionally equivalent to HPV16 L2 residues 108 to 120 to facilitate BPV1 infection, particularly since residues 108 to 120 of L2 do not show the same degree of sequence conservation between the HPV16 L2 and BPV1 genotypes, as seen for residues 13 to 31. To address the latter possibility, we generated BPV1 virions containing L2 with deletion of residues 91 to 129. Because the sequence divergence between BPV1 and HPV16 in residues 108 to 120 of L2 is considerable and the alignment therefore may be inexact, we used the larger deletion in BPV1 L2 of residues 91 to 129 to ensure that this functional domain was eliminated. While a deletion in L2 of such size could potentially disrupt virion assembly, expression of L1 and wild-type L2 or L2Δ91-129 in BPHE-1 resulted in encapsidation of equivalent amounts of BPV DNA (Fig. 6A, upper panel). Furthermore, Western blot analysis of L1 and L2 in virions purified by rate zonal centrifugation demonstrates that L2Δ91-129 is incorporated into BPV1 virions at the same ratio to L1 as that seen for wild-type L2 (Fig. 6B and C). Finally, incorporation of an L2 with a deletion of this size might also perturb the entire capsid structure. Thus, we examined the ability of BPV1 virions containing wild-type L2 or L2Δ91-129 to bind to C127 cells (Fig. 6D). Purified virions were incubated with C127 cells for an hour at 4°C. Unbound material was washed away, and the virions were detected by indirect immunofluorescence with the conformationally dependent monoclonal antibody 5B6. Binding of 5B6 to L1 in the BPV1 virions containing the wild type or L2Δ91-129 was of similar intensity, consistent with normal capsid structure. Furthermore, BPV1 virions containing the wild type or L2Δ91-129 bound to C127 cells with a very similar pattern and intensity, as determined by confocal fluorescence microscopy (Fig. 6D). However, BPV1 virions containing L2 with residues 91 to 129 deleted produced no foci in the C127 focal transformation assay (Fig. 6A, lower panel). Taken together, the data suggest that deletion of residues 91 to 129 of BPV1 L2 prevents infection by BPV1 virions, but does not significantly perturb their assembly, structure, or binding to the cell surface.
FIG. 6.
Residues 91 to 129 of BPV1 L2 are not required for virion assembly or binding of virions to the cell surface, but are necessary for infectivity. (A) BPHE-1 cells were infected with recombinant SFV expressing BPV1 L1 alone or coinfected with SFV expressing BPV1 L1 and wild-type L2 or L2Δ91-129. Thirty hours postinfection, the cells were harvested and lysed by sonication. The presence of DNase I-resistant BPV1 genome that is immunoprecipitated with anti-VLP antiserum was detected by Southern blot analysis (upper panel). The lysates were also tested for the presence of infectious BPV1 virions by using the C127 focal transformation assay (see plates in lower panel). BPHE-1 cells were coinfected with BPV1 L1 and the wild type or L2Δ91-129 and harvested from the lysates 30 h postinfection by centrifugation through a 40% sucrose cushion and separated from empty VLP by further purification by rate zonal density centrifugation (10 to 40% sucrose, SW-41 rotor, 25,000 rpm, 4°C). Equivalent yields and incorporation of L2 and L2Δ91-129 into purified virions were demonstrated by Western blotting with rabbit BPV1 L1 antiserum (B) and rabbit anti-BPV1 L2 antiserum (C). Virion preparations of L1/L2 and L1/L2Δ91-129 were loaded in lanes 1 and 2, respectively (B and C). (D) Equal amounts of BPV1 L1/L2 and L1/L2Δ91-129 virions or buffer alone were incubated with C127 clone C cells at 4°C for 1 h, and surface-bound virions were detected by immunofluorescent staining with monoclonal antibody 5B6.
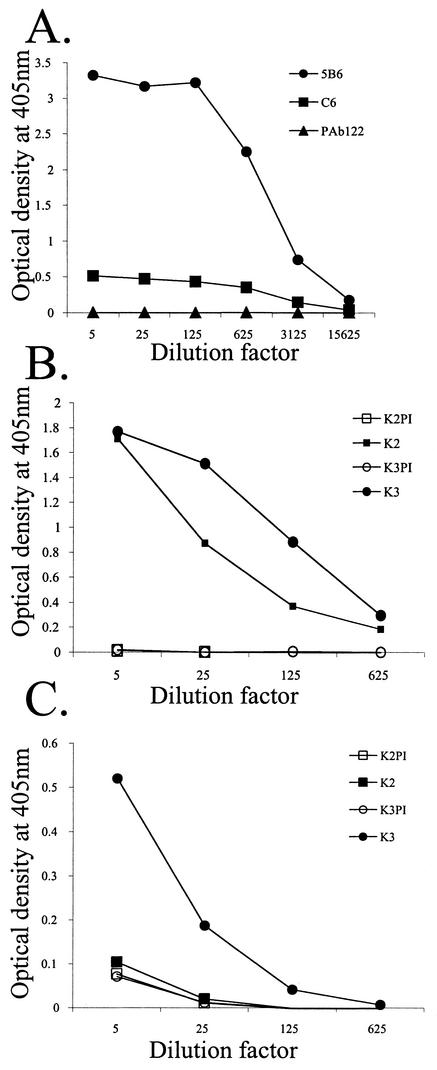
The importance of L2 residues 91 to 129 in BPV1 infection, as also described for HPV16 L2 residues 108 to 120 (28), suggests that this motif is located on the surface of BPV1 virions. Furthermore, monoclonal antibody or peptide-specific antiserum to residues 108 to 120 neutralizes HPV16 pseudovirion (26). However, Liu et al. demonstrated that monoclonal antibody C6 binds to residues 116 to 123 of L2 on the surface of BPV1 virions, yet fails to neutralize (31). Indeed, we also observed binding of C6 to the surface of BPV1 virions (Fig. 7A), but the C6 monoclonal antibody fails to neutralize BPV1 virions prepared from bovine warts (31; data not shown). The reduced binding of C6 as compared with the L1 and conformationally specific neutralizing antibody 5B6 likely reflects the stoichiometry of L2 to L1. However, it is possible that a neutralizing epitope is located elsewhere within this BPV1 L2 91-129 region. To search for such an epitope, we immunized rabbits with KLH coupled to BPV1 L2 peptides encompassing residues 85 to 105 (antiserum K1), 100 to 120 (antiserum K2), and 115 to 135 (antiserum K3). The antisera were tested by ELISA for reactivity with the purified six-His-tagged BPV1 L2 peptide comprising residues 45 to 173 as well as native BPV1 virions. The rabbit antiserum to peptide 85-105 failed to react with either BPV1 or L2 peptide 45-173 (data not shown). However, the antiserum to residues 100 to 120 and 115 to 135 of BPV1 L2 reacted strongly with the BPV1 L2 peptide 45-173 (Fig. 7B), but only the latter peptide serum showed strong reactivity with native BPV1 virions (Fig. 7C). None of the three peptide-specific antisera neutralized BPV1 virions purified from bovine warts at a dilution of 1:10 (data not shown).
FIG. 7.
L2 residues 91 to 129 are partially exposed on the surface of BPV1 virions. (A) A microtiter plate coated with 300 ng of purified BPV1 virions per well was reacted for an hour with monoclonal antibody C6 to BPV1 L2 residues 116 to 123 (31), 5B6 to a conformationally dependent and neutralizing epitope on BPV1 L1 (41), or PAb122 to an irrelevant antigen. After washing, specific binding was detected with a peroxidase-linked anti-mouse IgG antibody. The plate was developed with ABTS, and the A405 was determined. (B and C) Microtiter plates were coated with a 300-ng/well sample of either BPV1 L2 45-173 peptide that had been purified by using its hexahistidine tag (B) or BPV1 virions from a bovine wart purified on cesium chloride gradients (C). The plates were reacted with preimmune (PI) or immune sera of rabbits immunized with KLH coupled to BPV1 L2 peptides 100-120 (K2 antiserum) and 115-135 (K3 antiserum). Upon washing, specific binding was detected with a peroxidase-linked anti-rabbit IgG antibody and ABTS. The A405 was determined.
DISCUSSION
A common mechanism of viral neutralization is the binding of antibody to the cell recognition motif of viral capsid proteins. The importance of developing a vaccine that protects against infection by the plethora of oncogenic HPV genotypes has driven efforts to identify both the binding motifs of their capsid proteins and their cell surface receptors. Clearly a conserved receptor-binding motif of a capsid protein holds considerable promise as a prophylactic vaccinogen active against all oncogenic HPV genotypes (28). This is particularly attractive, since the generally papillomavirus type-specific nature of the immunodominant neutralizing epitopes on L1 renders comprehensive vaccination with L1 VLP vaccines problematic (5, 20, 37, 39). L2 has emerged as an exciting candidate prophylactic vaccine, since vaccination against L2, but not L1, induces antibodies that cross-neutralize diverse HPV genotypes (26, 42). Significantly, vaccination against L2 also protects against experimental papillomavirus infection (6, 10, 16, 30), implying that targeting of conserved epitopes in L2 with functional significance during infection could be useful in the development of a pan-HPV prophylactic vaccine.
L1 mediates the initial binding of papillomavirus to the cell surface of a wide range of cell types (34, 38, 40, 48). There are ∼26,000 receptors on the surface of HeLa cells, HPV33 L1 VLPs bind with a dissociation constant of 84 pM, and binding is sensitive to heparinase and trypsin pretreatment (23, 34, 48). Clearly the interaction of L1 VLPs with the cell surface is distinct from that of L2 residues 13 to 31, which bind to ∼45,000 receptors per HeLa cell with a Kd of ∼1 nM, and the interaction is unaffected by heparinase or trypsin pretreatment. Integrin α6 was initially identified as a receptor (17, 32) for HPV6 L1 VLPs, but it is not obligate (44). Several viruses can use alternate receptors to enter the same cell type: for example, three receptors (HveA, -B, and -C) have been described for herpes simplex virus type 1 (18, 33), and foot-and-mouth disease virus exploits αvβ6 integrin, but in certain situations can also use heparan sulfate glycosaminoglycans (GAGs) (1, 3). Similarly, papillomavirus binds via the major capsid protein L1 to heparan sulfate GAGs, and heparinase treatment of the cell surface prevents HPV33 pseudovirion infection (19, 23). Interestingly, CD16 may also contribute to immune cell surface binding by VLPs (11). However, despite the availability of the atomic structure of L1 and high-resolution structures of virions bound to two different neutralizing monoclonal antibodies, neither the residues in L1 that contribute cell surface binding nor a primary viral receptor has been defined (4, 9).
Separately from the initial binding of a primary surface receptor, other viral capsid motifs or proteins can interact with secondary cellular receptors to facilitate a viral infection: e.g., adenovirus subgroup C binds with high affinity to CAR via fiber knobs, but a second interaction between the penton base and αvβ3 or αvβ5 integrin is necessary for uptake (2, 50). Indeed, L2 plays a key role in papillomavirus infection, since deletion of the positively charged residues at either terminus of L2 renders BPV1 noninfectious (36). Kawana et al. recently suggested that L2 might bind a trypsin-sensitive secondary receptor to facilitate papillomavirus infection, because a conserved L2 motif (residues 108 to 126) can bind to and enter HeLa cells, and this process is prevented by pretreatment with trypsin (28). While L2 enhances the infectivity of in vitro-assembled HPV16 pseudovirions by only approximately twofold (27), incorporation of mutations that prevented cell surface binding of this L2 108-126 motif also eliminated L2's contribution to infectivity (28). However, infection by HPV pseudovirions prepared under more physiologic conditions is dramatically enhanced by L2 (36, 47, 51). Herein, we show that deletion of residues 91 to 129 from L2 also renders BPV1 virions noninfectious, suggesting that this region, like residues 108 to 120 of HPV16, also facilitates the infectious process and that this function is conserved even in such evolutionarily diverse papillomavirus genotypes. Thus, the failure of the BPV1 L2 peptide comprising residues 45 to 173 to bind to the cell surface most likely can be attributed to the prior trypsin treatment of the cells and/or improper folding after synthesis in E. coli.
L2-specific neutralizing antibody fails to prevent virions from binding to the cell surface, and BPV1 virions rendered noninfectious by small deletions at either terminus of L2 are still able to bind to the cell surface as for the wild type (38, 40, 41). These data suggest that L2 functions in the infectious process after initial surface binding. Consistent with this notion, we show that deletion of L2 residues 91 to 129 eliminates infection, but these virions bind to the cell surface as for wild-type BPV1. Furthermore, the HPV16 L2 13-31 peptide that interacts with the cell surface does not prevent the binding of HPV16 VLP to the cell surface. While L2 itself can bind directly to the cell surface, these data are more consistent with interaction of L2 and secondary receptor molecules after the initial L1-mediated binding of the particle to the cell surface.
We defined a cell surface-binding motif to residues 13 to 31 of HPV16 L2 and identified residues within this region that are important both for this function and infectivity. This motif is conserved in BPV1 L2, and cross-competition experiments suggest a conservation of function. Since mutations in residues 18 and 19 or 21 and 22 fail to bring infectivity to the same level as for HPV16 pseudovirions lacking L2 (Table 1), residues 13 to 31 may act in concert with the 108-126 motif, as described by Kawana et al. (28), to facilitate virion infection. However, the surface receptor for the HPV16 L2 108-126 motif is trypsin sensitive, whereas the interaction of L2 with the cell surface described herein is resistant to trypsin pretreatment, suggesting that the cellular factors bound by these two motifs are different (28). It is notable that residues 108 to 126 of HPV16 L2 are recognized by cross-neutralizing antibodies, and vaccination with this peptide induces a cross-neutralizing antibody response (24, 26). However, vaccination of rabbits with peptides comprising residues 94 to 122 of cottontail rabbit papillomavirus (CRPV) or rabbit oral papillomavirus (ROPV) protected against the homologous, but not heterologous rabbit papillomavirus (16). Embers et al. also suggest that protection via L2 peptide vaccination is mediated by neutralizing antibody papillomavirus (16). It was therefore surprising that neither the C6 monoclonal antibody (which binds with residues 116 to 123 of BPV1 L2) (31) nor peptide antiserum to BPV1 L2 residues 115 to 135 was detectably neutralizing for BPV1. Indeed, data from CRPV, ROPV, and HPV16 demonstrate a neutralizing epitope is located within residues 94 to 122 and 108 to 120, respectively (16, 24, 26). However, antiserum to BPV1 L2 residues 100 to 120 failed to bind significantly with intact BPV1 or to neutralize, while antiserum to residues 45 to 173 is neutralizing (41). Residues 101 to 120 of L2 represent an immunodominant epitope in BPV4 (8), but vaccination with this peptide alone is not protective (7).
The well-conserved motif comprising HPV16 L2 residues 13 to 31 that binds to the cell surface and facilitates infection may also represent a target for prophylaxis. Indeed the sera of many CINIII or cervical carcinoma patients are reactive with this peptide (13). The L2 13-31 peptide does not contain a heparin-binding motif, and heparinase pretreatment has no effect upon the binding of the GFP fusion protein to cells. However, heparinase treatment drastically reduced surface binding of both L1 only and L1/L2 VLPs. Furthermore, particle binding studies with monoclonal antibodies or polyclonal peptide-specific antisera suggest that, unlike L2 residues 108 to 120 (25, 26), the residue 13-31 motif is not present on the capsid surface. However an immediately adjacent region (L2 residues 32 to 81 on HPV16 VLP) is displayed on the surface of the capsid (22, 31). Taken together, the data suggest the hypothesis that particle binding to heparan sulfate causes the L2 13-31 motif to be displayed on the capsid surface for interaction with a potential secondary receptor.
Like L1 (34, 40, 48), L2 apparently binds to cells derived from multiple species and tissues (28), consistent with the ability of papillomavirus pseudovirions to infect many different cell types (45). Therefore, the exquisite host range specificity of productive papillomavirus infection is unlikely to be restricted at the level of infection, which implies that replication and/or transcriptional controls predominantly determine host range. However, this ability of papillomavirus particles to deliver encapsidated DNA to a broad range of cell types also implies that the papillomavirus pseudovirion may represent a useful vector for gene therapy and genetic immunization. Furthermore, L2's dual roles in virion assembly and infection suggest that its incorporation in papillomavirus pseudovirion used in such applications would be beneficial.
Acknowledgments
We thank N. D. Christensen (University of Pennsylvania, Hershey) for monoclonal antibody H16.V5; J. T. Schiller (NIH, Bethesda, Md.) for HeLa and C127 clone C cells; and A. Lewis (FDA, Bethesda, Md.) for BPHE-1 cells. From Johns Hopkins University, Baltimore, Md., we thank T.-C. Wu for SiHa, Caski, and COS-1 cells; X. Z. Zhu for RCC24, OVCAR-3, DW-6, and Mel cells; Q. Qia for Thomas, Salt, K562, and H&N013 cells; and Y. Yan for D-32 cells. We also thank T.-C. Wu, D. R. Lowy, and J. T. Schiller for critical review of the manuscript.
This work was funded by grants to R.B.S.R. from the Cancer Research Institute, the American Cancer Society (RSG-02-175-01-MBC), and the National Institutes of Health.
REFERENCES
- 1.Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 3.Berinstein, A., M. Roivainen, T. Hovi, P. W. Mason, and B. Baxt. 1995. Antibodies to the vitronectin receptor (integrin αVβ3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J. Virol. 69:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booy, F. P., R. B. Roden, H. L. Greenstone, J. T. Schiller, and B. L. Trus. 1998. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 Å resolution. J. Mol. Biol. 281:95-106. [DOI] [PubMed] [Google Scholar]
- 5.Breitburd, F., R. Kirnbauer, N. L. Hubbert, B. Nonnenmacher, C. Trin-Dinh-Desmarquet, G. Orth, J. T. Schiller, and D. R. Lowy. 1995. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 69:3959-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campo, M. S. 1994. Vaccination against papillomavirus in cattle. Curr. Top. Microbiol. Immunol. 186:255-266. [DOI] [PubMed] [Google Scholar]
- 7.Campo, M. S., B. W. O'Neil, G. J. Grindlay, F. Curtis, G. Knowles, and L. Chandrachud. 1997. A peptide encoding a B-cell epitope from the N-terminus of the capsid protein L2 of bovine papillomavirus-4 prevents disease. Virology 234:261-266. [DOI] [PubMed] [Google Scholar]
- 8.Chandrachud, L. M., G. J. Grindlay, G. M. McGarvie, B. W. O'Neil, E. R. Wagner, W. F. Jarrett, and M. S. Campo. 1995. Vaccination of cattle with the N-terminus of L2 is necessary and sufficient for preventing infection by bovine papillomavirus-4. Virology 211:204-208. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, N. D., J. W. Kreider, N. C. Kan, and S. L. DiAngelo. 1991. The open reading frame L2 of cottontail rabbit papillomavirus contains antibody-inducing neutralizing epitopes. Virology 181:572-579. [DOI] [PubMed] [Google Scholar]
- 11.Da Silva, D. M., M. P. Velders, J. D. Nieland, J. T. Schiller, B. J. Nickoloff, and W. M. Kast. 2001. Physical interaction of human papillomavirus virus-like particles with immune cells. Int. Immunol. 13:633-641. [DOI] [PubMed] [Google Scholar]
- 12.Day, P. M., R. B. S. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillner, J., L. Dillner, G. Utter, C. Eklund, A. Rotola, S. Costa, and D. DiLuca. 1990. Mapping of linear epitopes of human papillomavirus type 16: the L1 and L2 open reading frames. Int. J. Cancer 45:529-535. [DOI] [PubMed] [Google Scholar]
- 14.Doorbar, J., and P. H. Gallimore. 1987. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J. Virol. 61:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dvoretzky, I., R. Shober, S. K. Chattopadhyay, and D. R. Lowy. 1980. A quantitative in vitro focus assay for bovine papilloma virus. Virology 103:369-375. [DOI] [PubMed] [Google Scholar]
- 16.Embers, M. E., L. R. Budgeon, M. Pickel, and N. D. Christensen. 2002. Protective immunity to rabbit oral and cutaneous papillomaviruses by immunization with short peptides of l2, the minor capsid protein. J. Virol. 76:9798-9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evander, M., I. H. Frazer, E. Payne, Y. M. Qi, K. Hengst, and N. A. J. McMillan. 1997. Identification of the α6 integrin as a candidate receptor for papillomaviruses. J. Virol. 71:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 19.Giroglou, T., L. Florin, F. Schafer, R. E. Streeck, and M. Sapp. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giroglou, T., M. Sapp, C. Lane, C. Fligge, N. D. Christensen, R. E. Streeck, and R. C. Rose. 2001. Immunological analyses of human papillomavirus capsids. Vaccine 19:1783-1793. [DOI] [PubMed] [Google Scholar]
- 21.Hagensee, M. E., N. H. Olson, T. S. Baker, and D. A. Galloway. 1994. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J. Virol. 68:4503-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heino, P., B. Skyldberg, M. Lehtinen, I. Rantala, B. Hagmar, J. W. Kreider, R. Kirnbauer, and J. Dillner. 1995. Human papillomavirus type 16 capsids expose multiple type-restricted and type-common antigenic epitopes. J. Gen. Virol. 76:1141-1153. [DOI] [PubMed] [Google Scholar]
- 23.Joyce, J. G., J. S. Tung, C. T. Przysiecki, J. C. Cook, E. D. Lehman, J. A. Sands, K. U. Jansen, and P. M. Keller. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 274:5810-5822. [DOI] [PubMed] [Google Scholar]
- 24.Kawana, K., Y. Kawana, H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 2001. Nasal immunization of mice with peptide having a cross-neutralization epitope on minor capsid protein L2 of human papillomavirus type 16 elicit systemic and mucosal antibodies. Vaccine 19:1496-1502. [DOI] [PubMed] [Google Scholar]
- 25.Kawana, K., K. Matsumoto, H. Yoshikawa, Y. Taketani, T. Kawana, K. Yoshiike, and T. Kanda. 1998. A surface immunodeterminant of human papillomavirus type 16 minor capsid protein L2. Virology 245:353-359. [DOI] [PubMed] [Google Scholar]
- 26.Kawana, K., H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 1999. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J. Virol. 73:6188-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawana, K., H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 1998. In vitro construction of pseudovirions of human papillomavirus type 16: incorporation of plasmid DNA into reassembled L1/L2 capsids. J. Virol. 72:10298-10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawana, Y., K. Kawana, H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 2001. Human papillomavirus type 16 minor capsid protein L2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J. Virol. 75:2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, Y. L., L. A. Borenstein, R. Selvakumar, R. Ahmed, and F. O. Wettstein. 1992. Effective vaccination against papilloma development by immunization with L1 or L2 structural protein of cottontail rabbit papillomavirus. Virology 187:612-619. [DOI] [PubMed] [Google Scholar]
- 31.Liu, W. J., L. Gissmann, X. Y. Sun, A. Kanjanahaluethai, M. Muller, J. Doorbar, and J. Zhou. 1997. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 227:474-483. [DOI] [PubMed] [Google Scholar]
- 32.McMillan, N. A., E. Payne, I. H. Frazer, and M. Evander. 1999. Expression of the alpha6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virology 261:271-279. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 34.Müller, M., L. Gissmann, R. J. Cristiano, X.-Y. Sun, I. H. Frazer, A. B. Jenson, A. Alonso, H. Zentgraf, and J. Zhou. 1995. Papillomavirus capsid binding and uptake by cells from different tissues and species. J. Virol. 69:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. S. Roden. 2001. L1 interaction domains of papillomavirus l2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roden, R. B. S., P. M. Day, B. K. Bronzo, W. H. Yutzy IV, Y. Yang, D. R. Lowy, and J. T. Schiller. 2001. Positively charged termini of the L2 minor capsid protein are necessary for papillomavirus infection. J. Virol. 75:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roden, R. B. S., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roden, R. B. S., N. L. Hubbert, R. Kirnbauer, F. Breitburd, D. R. Lowy, and J. T. Schiller. 1995. Papillomavirus L1 capsids agglutinate mouse erythrocytes through a proteinaceous receptor. J. Virol. 69:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden, R. B. S., N. L. Hubbert, R. Kirnbauer, N. D. Christensen, D. R. Lowy, and J. T. Schiller. 1996. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J. Virol. 70:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roden, R. B. S., R. Kirnbauer, A. B. Jenson, D. R. Lowy, and J. T. Schiller. 1994. Interaction of papillomaviruses with the cell surface. J. Virol. 68:7260-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roden, R. B. S., E. M. Weissinger, D. W. Henderson, F. Booy, R. Kirnbauer, J. F. Mushinski, D. R. Lowy, and J. T. Schiller. 1994. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J. Virol. 68:7570-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roden, R. B. S., W. I. Yutzy, R. Fallon, S. Inglis, D. R. Lowy, and J. T. Schiller. 2000. Minor capsid protein of human genital papillomaviruses contains subdominant, cross-neutralizing epitopes. Virology 270:254-257. [DOI] [PubMed] [Google Scholar]
- 43.Selinka, H. C., T. Giroglou, and M. Sapp. 2002. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology 299:279-287. [DOI] [PubMed] [Google Scholar]
- 44.Sibbet, G., C. Romero-Graillet, G. Meneguzzi, and M. S. Campo. 2000. Alpha6 integrin is not the obligatory cell receptor for bovine papillomavirus type 4. J. Gen. Virol. 81:327-334. [DOI] [PubMed] [Google Scholar]
- 45.Touze, A., and P. Coursaget. 1998. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 26:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 47.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpers, C., F. Unckell, P. Schirmacher, R. E. Streeck, and M. Sapp. 1995. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J. Virol. 69:3258-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, P. A., L. E. Leong, P. W. Ng, S. H. Tan, S. Waller, D. Murphy, and A. G. Porter. 1994. Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Bio/Technology 12:601-605. [DOI] [PubMed] [Google Scholar]
- 50.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 51.Yeager, M. D., M. Aste-Amezaga, D. R. Brown, M. M. Martin, M. J. Shah, J. C. Cook, N. D. Christensen, C. Ackerson, R. S. Lowe, J. F. Smith, P. Keller, and K. U. Jansen. 2000. Neutralization of human papillomavirus (HPV) pseudovirions: a novel and efficient approach to detect and characterize HPV neutralizing antibodies. Virology 278:570-577. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, J., D. J. Stenzel, X. Y. Sun, and I. H. Frazer. 1993. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J. Gen. Virol. 74:763-768. [DOI] [PubMed] [Google Scholar]