Abstract
Viruses utilize numerous mechanisms to counteract the host's immune response. Interferon production is a major component of the host antiviral response. Many viruses, therefore, produce proteins or RNA molecules that inhibit interferon-induced signal transduction pathways and their associated antiviral effects. Surprisingly, some viruses directly induce expression of interferon-induced genes. SM, an early lytic Epstein-Barr virus (EBV) nuclear protein, was found to specifically increase the expression of several genes (interferon-stimulated genes) that are known to be strongly induced by alpha/beta interferons. SM does not directly stimulate alpha/beta interferon secretion but instead induces STAT1, an intermediate step in the interferon signaling pathway. SM is a posttranscriptional activator of gene expression and increases STAT1 mRNA accumulation, particularly that of the functionally distinct STAT1β splice variant. SM expression in B lymphocytes is associated with decreased cell proliferation but does not decrease cell viability or induce cell cycle arrest. These results indicate that EBV can specifically induce cellular genes that are normally physiological targets of interferon by inducing components of cytokine signaling pathways. Our findings therefore suggest that some aspects of the interferon response may be positively modulated by infecting viruses.
Epstein-Barr virus (EBV), a human gammaherpesvirus, is the agent of infectious mononucleosis and is associated with Burkitt lymphoma, nasopharyngeal carcinoma, and lymphomas in immunosuppressed hosts (for a review, see reference 32). Infection by human herpesviruses of all classes specifically modulates cellular-gene expression. Because herpesviruses establish lifelong infections in the face of a competent immune system, many of the cellular genes affected are components of the innate or adaptive immune response. For example, an EBV immediate-early gene product inhibits gamma interferon (IFN-γ) signaling and down-regulates expression of the IFN-γ receptor (42).
The EBV SM protein is a posttranscriptional gene regulatory protein expressed early during lytic replication (9, 12, 14, 53, 66). Homologues of SM are found in herpes simplex virus (HSV), human cytomegalovirus (CMV), varicella-zoster virus, and Kaposi's sarcoma-associated virus (human herpesvirus 8) and act as transcriptional and posttranscriptional regulators (2, 10, 17, 26, 29, 33, 40). During lytic EBV replication, SM is expressed prior to other early genes but after the immediate-early genes BRLF1 and BZLF1. SM enhances the expression of several EBV genes and heterologous genes in cotransfection assays (30, 31, 39, 52, 55). Its ability to activate expression of cotransfected genes in a promoter-independent fashion has led to it being described as a promiscuous transactivator. Further studies demonstrated that several genes containing introns were inhibited by SM, whereas intronless genes were activated by SM (52). The majority of cellular genes and latent EBV genes are spliced, whereas most lytic EBV genes are not spliced, consisting of single open reading frames (21). Moreover, intronless genes are generally inefficiently expressed (27, 38), suggesting that SM could preferentially activate lytic EBV genes. SM binds mRNA, shuttles from nucleus to cytoplasm, interacts with components of nuclear export pathways, and enhances both nuclear and cytoplasmic accumulation of target gene RNA transcripts (6, 8, 20, 51, 55).
The global effect of SM on host cell gene expression and phenotype is unknown. The HSV ICP27 gene product, which is homologous to SM, has a global inhibitory effect on host cell splicing but nevertheless activates the expression of α-globin, an intron-containing gene (11). Although it appears likely that one function of SM is to facilitate the expression of lytic EBV genes at the expense of spliced genes at the appropriate point in the replicative cycle, SM does not enhance expression of all intronless genes equally. SM increases cytoplasmic accumulation of chloramphenicol acetyltransferase (CAT) and EBV BMRF1, BALF2, BSLF1, and DNA polymerase mRNAs, but not firefly luciferase, growth hormone, or EBV BBLF2/3 cDNA-derived transcripts (51, 52, 55). Although SM binds mRNA in vivo, its discriminatory effect is not based simply on a differential ability to bind various mRNAs, and a specific RNA sequence motif necessary for binding by SM has not been identified (51). Since the molecular basis of SM's gene-specific activity remains to be determined, it is difficult to predict a priori the effect of SM on a given gene. Thus, the net effect of SM on an individual cellular gene could be negative or positive.
Since modulation of host cell gene expression by EBV is likely to be important both in altering the cellular environment and in affecting EBV replication, we wished to identify cellular genes that are specifically regulated by SM. We therefore devised a system in which SM expression could be synchronously induced in EBV-negative B-lymphoma cells and compared to cells not expressing SM, thereby allowing an analysis without the confounding effects of other EBV genes and lytic replication as a whole. The effect of SM on the cellular transcriptional profile was analyzed by hybridizing mRNA from these two populations of cells to microarrays representing known cellular genes. Of ∼1,700 human mRNA transcripts represented on the oligonucleotide arrays used for this study, SM specifically induced STAT1 and several members of a family of genes known to be induced by IFN-α/β (IFN-stimulated genes [ISGs]) via the JAK/STAT pathway.
MATERIALS AND METHODS
Cell lines and plasmids.
BJAB is an EBV-negative B-lymphoma cell line (41). CMV-SM, CMV-CAT, PSP65GC, Gal4-ER-VP16, and pJ6Ω-puro have been described previously (7, 52). SM and EBNA3C cDNAs were cloned into pSP65GC, which contains a synthetic Gal4-responsive promoter, to yield pSP65-SM and pSP65-EBNA3C. These were used to generate SM-BJAB or EBNA3C-BJAB cell lines in which SM or EBNA3C expression, respectively, is inducible with 4-hydroxytamoxifen. SM-BJAB, EBNA3C-BJAB, and VP16-BJAB cells were maintained in 800 μg of neomycin/ml and 0.25 μg of puromycin/ml.
Construction of inducible cell lines and transfections.
To generate VP16-BJAB cells, BJAB cells were transfected with 10 μg of Gal4-ER-VP16 DNA by electroporation and placed under selection with 800 μg of neomycin/ml until resistant clones were obtained. VP16-BJAB cells were transfected with 2 μg of pJ6Ω-puro and 10 μg of either pSP65-SM or pSP65-EBNA3C and selected with 800 μg of neomycin/ml and 0.25 μg of puromycin/ml. For reporter assays, BJAB cells were electroporated with 5 μg of CMV-CAT and treated with 100 nM 4-hydroxytamoxifen or mock treated. CAT assays were performed 48 h after transfection as previously described (52). HeLa cells were transfected with Lipofectamine (Invitrogen Corp.) in accordance with the manufacturer's protocols 48 h prior to RNA isolation.
RNA analysis.
RNA was isolated with RNAzol B (Teltest, Friendswood, Tex.), and Northern blotting was performed as previously described (51, 52). To isolate nuclear and cytoplasmic RNA fractions, cells were first lysed in 0.5% Nonidet P-40 buffer, and the nuclei were separated by centrifugation prior to RNA isolation as previously described (52). Probes for ISG and STAT1 genes were generated by reverse transcription and PCR amplification using gene-specific primers. RNA for microarray analysis was isolated using RNAzol B and further purified by passage over RNAeasy columns (Qiagen). cRNA synthesis, labeling, and hybridization to microarrays (Human Cancer G110 array; Affymetrix, Santa Clara, Calif.) were performed in accordance with the manufacturer's protocols.
Tamoxifen treatment, IFN assays, and cocultivation experiments.
SM-BJAB, EBNA3C-BJAB, and VP16-BJAB cells were induced by treatment with 100 nM 4-hydroxytamoxifen (Sigma). For IFN assays, the supernatants were harvested prior to induction, 12 h postinduction, and every 4 h thereafter for 48 h. Assays were performed with enzyme-linked immunoassay (ELISA) kits for IFN-α and IFN-β, and sensitivity was validated by simultaneous measurement of standardized IFN preparations (Research Diagnostics Inc., Flanders, N.J.). Growth curves were performed by withdrawal of cells at each time point, addition of trypan blue, and direct counting of viable cells in a hemacytometer. Cells were analyzed by flow cytometry after propidium iodide staining according to published protocols (16). Cocultivation experiments were performed by placing 4 × 106 SM-BJAB cells in the bottom chamber of a 75-mm-diameter Transwell cell culture apparatus (Corning Costar) and placing 4 × 106 BJAB cells in the upper chamber. Tamoxifen (100 nM) or IFN-β (1,000 U/ml) was added to the growth medium, and the cells were incubated for an additional 48 h.
Immunofluorescence and immunoblotting.
Immunofluorescence microscopy of SM-BJAB cells and immunoblotting for SM were performed with polyclonal anti-SM or anti-IRF-3 antibodies as described previously (6). Monoclonal anti-phospho-STAT1 antibodies were purchased from BD Transduction Laboratories (Lexington, Ky.). Polyclonal anti-STAT1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Polyclonal anti-IRF-3 antibodies have been described previously (47).
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed using a 32P end-labeled probe corresponding to the IFN-stimulated response element (ISRE) of the ISG15 promoter (5′-GATCCATGCCTCGGGAAAGGGAAACCGAAACTGAAGCC-3′). Equal amounts of protein were incubated with poly(dI-dC) and the labeled oligonucleotides in ISRE binding buffer (40 mM KCl, 20 mM HEPES [pH 7.0], 1 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol, 4% Ficoll, 0.02% Nonidet P-40). Electrophoresis was performed on 6% nondenaturing polyacrylamide gels, and the gels were dried and subjected to autoradiography. For supershift experiments, nuclear extracts were incubated on ice with anti-STAT1 antiserum for 1 h at 4°C prior to the addition of the labeled oligonucleotide as previously described (47).
RESULTS
Construction of an inducible SM-expressing cell line.
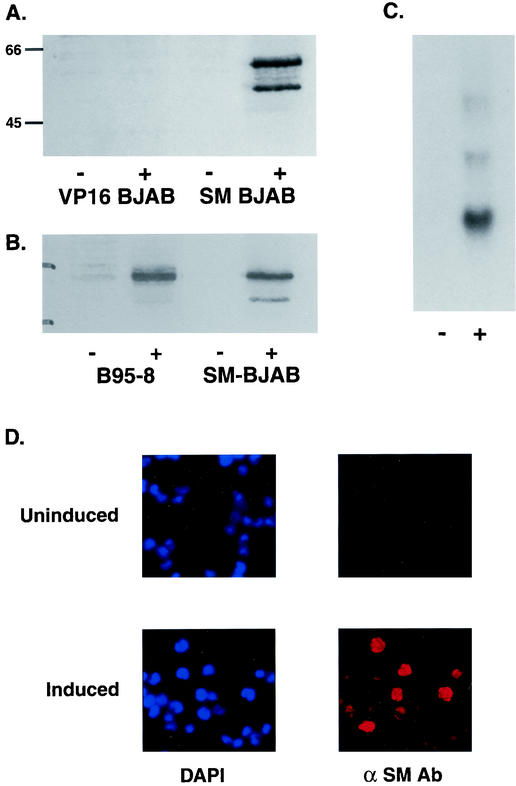
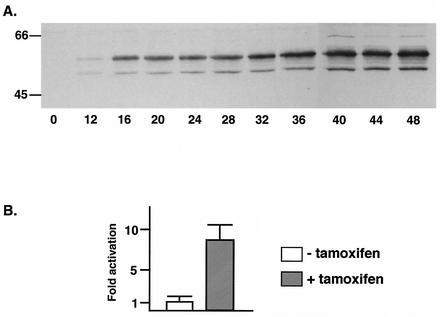
To study the effects of SM on host cell gene expression, we generated a cell line that inducibly expresses SM upon treatment with 4-hydroxytamoxifen using the EBV-negative B-lymphoma cell line BJAB (41). BJAB cells were transfected with Gal4-ER-VP16, which expresses a fusion between the DNA binding domain of the yeast transcriptional activator Gal4, the hormone binding domain of the estrogen receptor (ER), and the acidic activation domain of the HSV transcriptional activator VP16. This chimeric transcriptional activator binds only to sequences that are not present in mammalian DNA but functions well in a variety of cell types when bound to its cognate promoter. Furthermore, transcription is activated only in the presence of estrogen or the ER agonist 4-hydroxytamoxifen. Cell lines stably expressing Gal4-ER-VP16 (VP16-BJAB cells) were established by drug selection. We placed SM transcription under the control of Gal4-ER-VP16 by cloning SM cDNA downstream of a synthetic promoter with four Gal4 binding sites that is minimally active in the absence of Gal4-ER-VP16. VP16-BJAB cells were transfected with the SM plasmid, and drug-resistant clones were selected. Individual clones were tested for reproducibly inducible SM expression by immunoblotting tamoxifen-treated and mock-treated cell lysates. An immunoblot of lysate from a representative clone in which SM expression is tightly regulated by tamoxifen is shown in Fig. 1A. The amount of SM protein expressed in these cells was similar to that produced by EBV in B95-8 cells which had been induced to undergo lytic replication by tetradecanoyl phorbol acetate treatment (Fig. 1B). RNA isolated from induced and mock-induced cells was analyzed by Northern blotting, which confirmed that SM expression was stringently dependent on tamoxifen for induction (Fig. 1C). Immunofluorescence microscopy revealed that >90% of induced SM-BJAB cells express SM but that SM expression could not be detected in uninduced SM-BJAB cells (Fig. 1D). To determine the kinetics of SM protein accumulation, lysates of SM-BJAB cells harvested at various times after induction were analyzed by immunoblotting. SM was detectable as early as 12 h postinduction and reached maximal levels ∼40 h postinduction (Fig. 2A). In order to confirm that the SM protein being produced was functional, we transfected the SM-responsive reporter plasmid CMV-CAT into SM-BJAB cells and measured CAT activity in the presence and absence of SM induction. CAT activity was increased approximately sevenfold in SM-BJAB cells treated with tamoxifen compared to mock-induced cells, demonstrating that SM produced by SM-BJAB cells was capable of regulating gene expression (Fig. 2B).
FIG. 1.
Induction of SM expression by tamoxifen in SM-BJAB cells. (A) Immunoblotting of lysates from VP-16 BJAB or SM-BJAB cells was performed with cells harvested 48 h after induction (+) or mock induction (−). Molecular mass (in kilodaltons) is shown at left. (B) RNA from SM-BJAB cells induced (+) or mock induced (−) with tamoxifen was probed with SM cDNA. (C) Lysates from equal numbers of EBV-infected B95-8 cells induced (+) with tetradecanoyl phorbol acetate or SM-BJAB cells induced with tamoxifen were immunoblotted with anti-SM antibodies. Lysates were also prepared from mock-induced (−) B95-8 and SM-BJAB cells. (D) Induced and uninduced SM-BJAB cells were fixed 48 h after induction and examined by immunofluorescence microscopy for SM expression. DAPI staining was performed to visualize nuclei.
FIG. 2.
Kinetics of expression and function of SM in SM-BJAB cells. (A) SM-BJAB cells were treated with tamoxifen at time zero, and aliquots were withdrawn for immunoblotting with anti-SM antibodies at the times shown (in hours) below the blot. Molecular mass (in kilodaltons) is shown at left. (B) SM-BJAB cells were transfected with CMV-CAT reporter plasmid and either induced with tamoxifen (+) or mock induced (−). CAT activity was measured in the cell lysates 48 h after induction. The error bars indicate standard errors of the means.
SM inhibits cell proliferation.
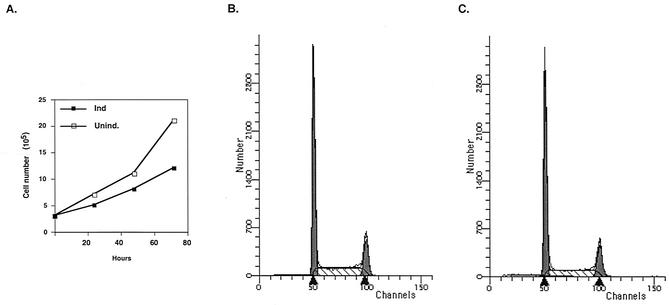
After induction, SM-expressing cells did not appear morphologically different from mock-induced cells under the light microscope (data not shown). However, by 48 h, the growth medium in flasks containing SM-expressing cells was invariably less acidic than that in flasks containing control cells, suggesting that the former were metabolically less active or growing more slowly. Trypan blue staining indicated that SM-expressing cells were as viable as uninduced cells, with viability of >98% in both cases. Comparison of growth curves revealed that SM-expressing cells had an increased doubling time of ∼36 h versus 26 h for SM-negative cells (Fig. 3A). We performed cell cycle analysis of SM-expressing and non-SM-expressing cells at 48 h postinduction, at which time control cells had doubled twice. The profiles of the two cell populations were similar, indicating that SM did not lead to cell cycle arrest (Fig. 3B and C). These data therefore indicated that SM led to a decrease in the growth rate but not to increased cell death or cell cycle arrest over a period of 48 h.
FIG. 3.
Effects of SM on cell growth. (A) Induced (Ind.) or uninduced (Unind.) SM-BJAB cells were directly counted by addition of trypan blue and microscopy at times shown. (B and C) SM-BJAB cells were fixed 48 h after induction with tamoxifen (B) or mock induction (C) and stained with propidium iodide. The cells were analyzed by flow cytometry.
SM expression leads to induction of IFN-stimulated genes.
Having confirmed that the inducible SM-expressing cells could be compared to completely SM-negative control cells and that adequate amounts of SM were being produced to have effects on the host cell phenotype, we analyzed the cell RNA by hybridization to oligonucleotide microarrays (Affymetrix). RNAs harvested 48 h after induction from tamoxifen-treated SM-expressing cells and from control cells that contained VP16-ER only (VP16-BJAB cells) were compared. Experiments were conducted with arrays that contained oligonucleotides representing 1,700 individual human genes. Interestingly, the RNA from SM-expressing cells revealed only eight genes consistently up-regulated above the cutoff level of a twofold increase. In contrast, ∼25 genes appeared to be down-regulated at least twofold. Expression of the vast majority of the remaining genes either was decreased moderately or remained unchanged. These findings are consistent with SM having a globally negative effect on the expression of intron-containing host cell genes, since the majority of host cell genes contain introns.
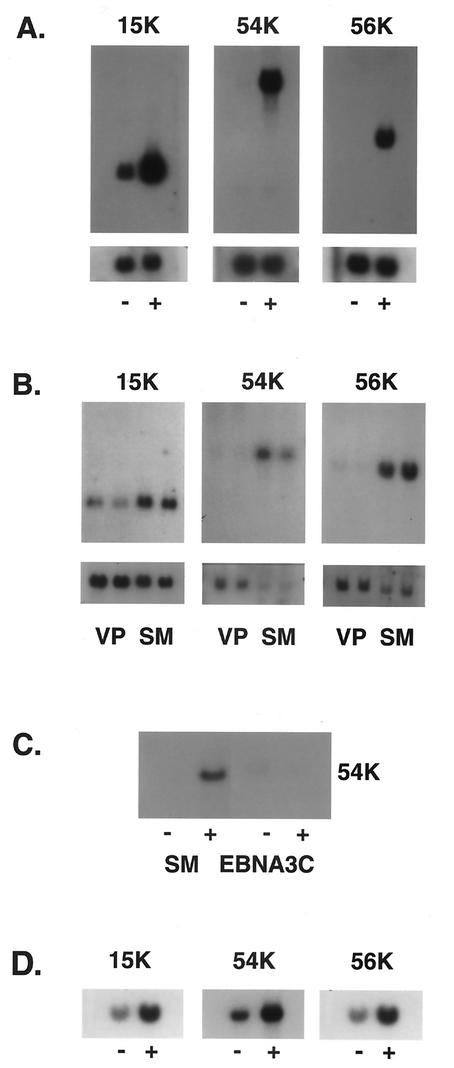
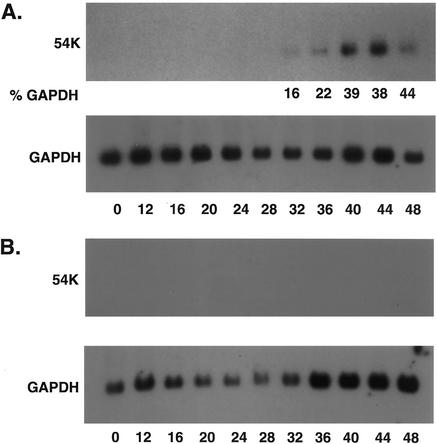
Surprisingly, the three most highly induced genes (ISG15K, ISG54K, and ISG56K) had originally been identified as ISGs (22, 37, 64). The promoter of each of these genes contains a sequence referred to as an ISRE that mediates IFN responsiveness (for a review, see reference 59). In addition, STAT1, a component of ISGF3, the transcription factor that binds and activates ISREs, was also induced by SM expression. The functions of many of the ISGs remain unclear despite their early discovery as IFN-inducible genes. In order to verify the array data, we analyzed ISG expression by Northern blotting of RNA from induced and uninduced SM-BJAB cells. As shown in Fig. 4A, SM expression led to a dramatic increase in ISG15K, ISG54K, and ISG56K expression. ISG54K and ISG56K were not constitutively expressed in the absence of SM, whereas ISG15K expression was increased 7.2-fold by SM. Tamoxifen-treated VP16-BJAB cells (SM negative) did not express increased amounts of ISGs compared to tamoxifen-treated SM-BJAB cells, demonstrating that the effect was not tamoxifen mediated (Fig. 4B). In order to confirm that ISG stimulation was a specific SM-mediated effect and not a nonspecific consequence of high-level synthesis of an exogenous viral mRNA, we measured the level of ISG54K mRNA in BJAB cells in which an EBV latent nuclear protein, EBNA3C, was expressed from an inducible promoter (EBNA3C-BJAB cells). As shown in Fig. 4C, tamoxifen treatment of EBNA3C-BJAB cells did not lead to ISG54K expression. To verify that SM-mediated induction of ISGs was not restricted to the tamoxifen-regulated system, we asked whether SM expression in HeLa cells could also induce ISGs. HeLa cells were transfected with an SM expression plasmid or control plasmid, and ISG expression was measured 48 h after transfection. SM-transfected HeLa cells expressed significantly greater amounts of ISG15K, ISG54K, and ISG56K mRNAs than controls (Fig. 4D). These data therefore confirm that SM specifically induces ISGs and also demonstrate that ISG induction by SM is not confined to B lymphocytes.
FIG. 4.
ISG expression is specifically induced by SM. (A) RNA from induced (+) or mock-induced (−) SM BJAB cells was analyzed by Northern blotting using ISG-specific 32P-labeled probes. The identity of the ISG detected is shown above each blot. The blots were also probed for glyceraldehyde-3-phosphate dehydrogenase (shown below the corresponding ISG blots). (B) RNA from tamoxifen-induced SM-BJAB (SM) or VP16-BJAB (VP) cells was analyzed as for panel A. (C) RNA from induced (+) or uninduced (−) EBNA3C-BJAB or SM-BJAB cells was probed for ISG54K expression. (D) RNA from HeLa cells transfected with control plasmid (−) or SM (+) was analyzed for ISG expression.
We next measured the kinetics of ISG induction to correlate it with the expression of SM protein. RNA was isolated from parallel aliquots of tamoxifen-treated VP16-BJAB or SM-BJAB cells withdrawn at various times after induction and was analyzed by Northern blotting. ISG54K mRNA first became detectable at 32 h and continued to increase during the course of the experiment (Fig. 5A). As expected, there was no ISG54K mRNA detectable in VP16-BJAB cells after tamoxifen treatment (Fig. 5B). The RNA used in this experiment was derived from aliquots of cells harvested in parallel with those used to measure SM protein expression shown previously (Fig. 2A). Thus, ISG mRNA expression began ∼20 h after SM protein expression was first detectable. The delay between SM expression and the onset of ISG mRNA accumulation suggested that the effect of SM may not be direct but that the synthesis or activation of an intermediary factor might be required for the induction of ISG mRNA.
FIG. 5.
Kinetics of ISG induction. RNA from SM-BJAB (A) or VP16-BJAB (B) cells was prepared before induction and every 4 h thereafter for 48 h. ISG54K expression was measured by Northern blotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was measured in the same blots and is shown below each ISG blot. In panel A, the amount of signal in each ISG band is shown as a percentage of the GAPDH signal (%GAPDH) to normalize for loading variation.
SM increases STAT1 mRNA levels.
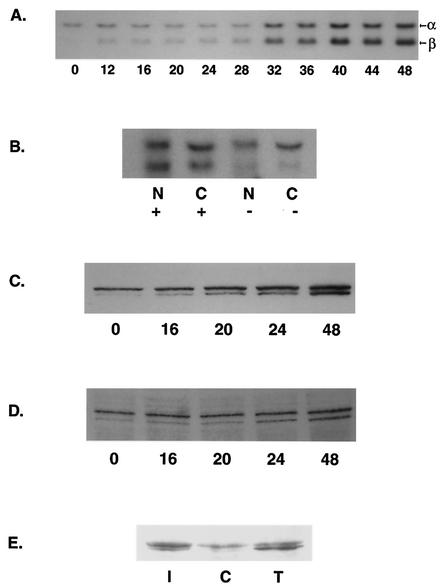
In order to confirm that SM also had effects on STAT1 mRNA as indicated by the array data, we measured the amounts of STAT1 mRNA at various times after SM induction. As shown in Fig. 6A, SM expression led to an increase in STAT1 mRNA levels. The increase began relatively soon after SM expression but increased significantly after 32 h, consistent with positive autoregulation of STAT1 transcription. STAT1 is expressed as two isoforms, STAT1α and STAT1β, which are generated by alternative splicing (4). The increase in STAT1 mRNA was accompanied by a progressive increase in the amount of the alternatively spliced STAT1β mRNA. Thus, SM led to an alteration in the splicing pattern and the amounts of STAT1 mRNA, suggesting posttranscriptional effects on STAT1 gene expression. These data indicate that the net effect of SM includes not only induction of STAT1 mRNA but also alteration of the STAT1β/STAT1α ratio. SM increases nuclear and cytoplasmic accumulation of target mRNAs and enhances their nuclear export. To examine the effect of SM on nuclear and cytoplasmic accumulation of STAT1 mRNA, we performed Northern blotting on fractionated RNAs from cells expressing or not expressing SM. SM led to an increase in steady-state levels of both nuclear and cytoplasmic STAT1 mRNA (Fig. 6B). The increase was approximately threefoldin both nucleus and cytoplasm, suggesting that the SM effect was primarily on mRNA accumulation as opposed to export. SM has not been found to enhance transcription from promoters examined previously. However, particularly as the STAT1 promoter is itself responsive to STAT1, we cannot rule out transcriptional activation as a contributory mechanism for the induction of STAT1 mRNA.
FIG. 6.
Kinetics of STAT1 mRNA and protein induction by SM. (A) RNA from SM-BJAB cells harvested at the times shown from 0 to 48 h after induction was analyzed by Northern blotting for STAT 1 mRNA. The upper band (α) corresponds to STAT1α (p91), and the lower band (β) corresponds to STAT1β (p84). (B) SM-BJAB cells were treated (+) or mock treated (−) with tamoxifen; 48 h after induction, the cells were lysed and separated into nuclear (N) and cytoplasmic (C) fractions prior to Northern blotting for STAT 1 mRNA. (C) Lysates of SM-BJAB cells harvested 0 to 48 h after induction were analyzed by immunoblotting them with antibodies specific for STAT1. (D) Lysates of SM-BJAB cells harvested 0 to 48 h after induction were analyzed by immunoblotting them with antibodies specific for Y701-phosphorylated (activated) STAT1 (phospho-STAT1). (E) Activation of STAT1 by SM was compared to that by IFN by treating SM-BJAB cells with either tamoxifen (T), medium (C), or IFN-β (I) and immunoblotting them with antibodies specific for Y701 phospho-STAT1.
Although receptor-mediated STAT1 activation occurs by phosphorylation of preexisting STAT1 protein, there are many different mechanisms by which STAT1 protein levels and activity are regulated (4, 44, 45). In addition, there are functional differences between the two STAT1 isoforms, as the β form is thought to be ineffective in mediating activation of IFN-γ-responsive promoters (46). Measurement of total STAT1 levels in tamoxifen-induced SM-BJAB cells demonstrated increasing levels of STAT1 protein, particularly of the STAT1β isoform, consistent with the previously demonstrated induction of STAT1 mRNA (Fig. 6C). STAT1 levels began to increase by 20 h, shortly after the time that SM protein became clearly detectable (cf. Fig. 2A), and continued to increase at later times. Since STAT1 phosphorylation is essential for STAT1 activation, we examined whether STAT1 was phosphorylated in SM-BJAB cells by immunoblotting the same lysates from tamoxifen-induced cells with antibodies specific for the tyrosine 701-phosphorylated form of STAT1 (Fig. 6D). The levels of phosphorylated STAT1 increased after SM induction, paralleling the increase in the total levels of STAT1. Comparison with the effect of IFN-β on SM-BJAB cells demonstrated a difference between SM- and IFN-β-mediated STAT1 activation. IFN-β primarily increased activated STAT1α levels, whereas SM led to increases in both phosphorylated STAT1α and STAT1β (Fig. 6E), consistent with the effects on STAT1 mRNA noted previously.
SM expression induces ISRE binding activity.
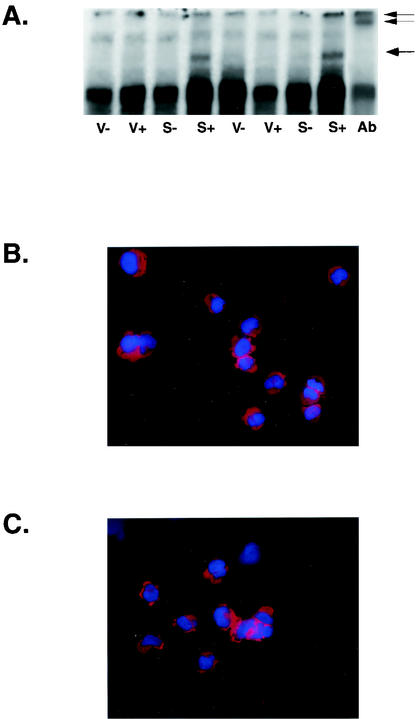
Since the previous experiments indicated that SM expression induced STAT1, a component of the signal transduction pathway that induces ISGs, it appeared likely that ISG induction by SM was mediated by a STAT1-dependent pathway. Binding of IFN-α/β to the IFN receptor leads to a cascade of signal transduction resulting in transcription of ISGs. Phosphorylation of JAK1 and TYK2 kinases ensues and ultimately leads to phosphorylation of STAT1 and STAT2 on specific tyrosine residues (15, 28). Activated STAT1 and STAT2 heterodimerize, associate with the DNA binding protein IRF-9 (p48), and translocate to the nucleus, forming a complex known as ISGF3 (23). ISGF3 binds to ISREs in the promoters of ISGs and activates transcription. To examine whether SM led to activation of ISGF3, EMSAs were performed using nuclear extracts from induced and uninduced SM-BJAB and VP16-BJAB cells and an oligonucleotide corresponding to the ISRE from the ISG15K promoter. Only extracts from SM-expressing cells contained an ISRE binding activity resulting in a complex with altered mobility (Fig. 7A). This complex contained STAT1, as its mobility was further shifted by incubation with anti-STAT1 antibody (Fig. 7A), suggesting that the ISRE-binding complex was ISGF3.
FIG. 7.
ISRE binding activity and IRF-3 localization in SM-expressing cells. (A) Nuclear extracts were prepared in duplicate from induced (+) or mock-induced (−) VP16-BJAB cells (V) or SM-BJAB cells (S) and used in EMSA with a labeled ISRE oligonucleotide. A band with altered mobility when incubated with SM-BJAB lysates is indicated with a single arrow. Anti-STAT1 antibody was added to induced SM-BJAB nuclear extract prior to incubation with a 32P-labeled ISRE fragment (Ab). The resulting supershifted band is indicated with a double arrow. (B and C) SM-BJAB cells were mock induced (B) or induced (C) with tamoxifen, fixed, and stained for immunofluorescence microscopy with anti-IRF3 antibodies (red). The nuclei were counterstained with DAPI (blue).
Virus infection may also lead to phosphorylation and nuclear translocation of IRF-3, which can independently activate ISRE-containing promoters (1, 61). To determine whether IRF-3 also occurred during SM-mediated ISG induction, we examined the intracellular location of IRF-3 in induced and uninduced SM-BJAB cells by immunofluorescence microscopy. In both cases, IRF-3 remained localized to the cytoplasm, suggesting that SM did not also activate IRF-3 (Fig. 7B and C). We also examined nuclear and cytoplasmic fractions of SM-expressing and non-SM-expressing cells by immunoblotting them with anti-IRF3 antibodies and were unable to detect nuclear translocation of IRF-3 in response to SM induction (data not shown).
ISG induction by SM is not mediated by a diffusible factor.
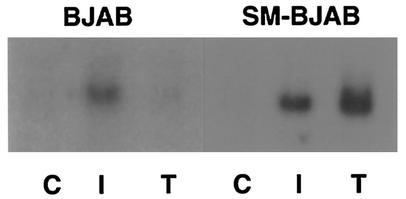
Human IFN-α/β genes consist of single unspliced open reading frames (35, 36, 62). Since SM activates intronless genes (51), it was possible that SM could stimulate ISG expression by enhancing IFN-α/β production. We therefore measured IFN-α in the supernatants of induced and uninduced SM-BJAB and control VP16-BJAB cells. Supernatants were collected from induced cells every 4 h between 12 and 48 h postinduction and analyzed by an ELISA which detects all known isoforms of human IFN-α except IFN-α F, with a limit of detection of 10 pg/ml, which corresponds to ∼30 to 50 U/ml. No IFN-α was detected in any of the supernatants, suggesting that stimulation of IFN-α synthesis was not the mechanism by which SM led to ISG expression. Identical experiments using an ELISA for IFN-β yielded similar results. These results nevertheless left open the possibility that small amounts of IFN, not detectable by the ELISA but sufficient to trigger ISG expression, were being produced and secreted by SM-expressing cells. To directly address this possibility, the following cocultivation experiment was performed. Parent BJAB cells were incubated in the upper chamber of a Transwell apparatus, in which the upper and lower chambers are separated by a membrane with a pore size of 0.4 μM, which permits diffusion of soluble molecules but not cells. SM-BJAB cells were placed in the lower chamber, and SM expression was induced with tamoxifen. An identical dual-chamber apparatus was prepared in which the SM-BJAB cells were mock induced. Forty-eight hours after induction, RNA was prepared from cells harvested from each chamber and analyzed by Northern blotting. As expected, mock-induced SM-BJAB cells expressed no detectable ISG54K, whereas induced SM-BJAB cells expressed easily detectable amounts of ISG54K mRNA (Fig. 8). Significantly, BJAB cells grown in the same medium as induced SM-BJAB cells expressed no ISG54K mRNA. As a control, another dual-chamber dish containing SM-BJAB and BJAB cells was treated with 1,000 U of IFN-β/ml. Both BJAB and SM-BJAB cells responded to IFN-β treatment, demonstrating that BJAB cells can respond to IFN. Interestingly, induction of SM in SM-BJAB cells actually led to greater expression of ISG54K than treatment with 1,000 U of IFN-β/ml. These results therefore indicated that a diffusible factor was not responsible for the stimulation of ISG expression by SM.
FIG. 8.
ISG induction by SM is limited to SM-expressing cells. Equal numbers of BJAB and SM-BJAB cells were placed in the upper and lower chambers, respectively, of Transwell dishes. Tamoxifen (T), IFN-β (I), or medium (C) was added to each chamber. RNA was prepared from cells in each compartment 48 h after treatment and analyzed by Northern blotting for ISG54K expression.
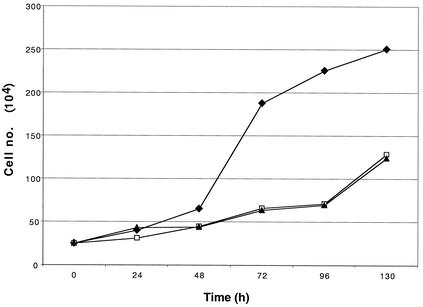
SM expression had a pronounced antiproliferative effect on SM-BJAB cells. Because SM led to decreased expression of cellular genes, as measured by the microarray as well as ISG induction, we asked whether SM had a greater antiproliferative effect than did IFN. We therefore compared the growth of SM-induced, IFN-treated, and untreated SM-BJAB cells. The antiproliferative effect of SM was essentially identical to that of IFN-β, as shown in Fig. 9. These data therefore suggest that ISG expression may be sufficient to explain the antiproliferative effect of SM, although the relative effects of SM and IFN-α/β on B lymphocytes will require further analysis and comparison of the complete transcriptional profiles of IFN-treated and SM-expressing cells.
FIG. 9.
Comparison of antiproliferative effects of SM and IFN-β. SM-BJAB cells were either mock treated (diamonds) or treated with 1,000 U of IFN-β (triangles) or tamoxifen (squares)/ml, and incubated at 37°C. Aliquots of each culture were withdrawn and counted at the times shown.
DISCUSSION
In this study, we demonstrate that an EBV protein expressed early during the lytic cycle of replication inhibits cell growth and induces components of signal transduction pathways involved in cytokine signaling. Specifically, EBV SM, a posttranscriptional regulator of EBV gene expression, leads to induction of STAT1 and several ISGs in EBV-negative B cells and epithelial cells. SM is known to act posttranscriptionally to both activate and inhibit gene expression (52). While the presence of introns can lead to inhibition of gene expression by SM, the data presented here show that SM can nevertheless induce some spliced cellular genes, since STAT1 and the ISGs are spliced. SM increases accumulation of its target RNAs and enhances their nuclear export (51, 52, 55). Consistent with its known mechanism of action, our data suggest that SM acts to increase the accumulation of STAT1 mRNAs. Further, SM alters the splicing pattern of STAT1, leading to increased production of the STAT1β isoform. Activated STAT1 has the capacity to form several different complexes involved in gene regulation. When complexed to STAT2 and IRF-9 (p48) to form ISGF3, STAT1 binds and activates ISREs found in the promoters of IFN α/β-responsive genes (23, 24, 54, 63). We have shown that STAT1 induction by SM is associated with increased expression of several ISGs, most likely via ISGF3 formation. The ISGs shown to be induced by SM all have ISRE-containing promoters. These ISGs, particularly ISG54K and -56K, are essentially not transcribed in the absence of ISGF3 and are thus extremely differentially expressed when induced, making them most likely to be detected by microarray technology. We therefore consider it most likely that their induction is due to increased transcription as a consequence of increased STAT1 levels. Such an interpretation is consistent with the delayed onset of ISG accumulation compared to earlier increases in STAT1 levels. However, it should be noted that there may also be posttranscriptional effects of SM on ISG mRNAs. Although we have previously shown that SM does not increase transcription from various promoters, we have not directly ruled out a transcriptional effect of SM on the ISG or STAT1 promoters. Such analyses are part ongoing investigations.
IFN expression by the host cell is viewed as a defensive host response against viral infection. IFNs stimulate the expression of many genes with antiviral and antiproliferative effects, producing an antiviral state. It is therefore somewhat surprising that the EBV protein SM itself induces genes normally expressed in response to IFN. Several aspects of SM-mediated ISG induction are notable. First, ISG induction by SM is not dependent on EBV infection or the interaction of virus glycoproteins with cell receptors, known triggers for the cellular antiviral response (5, 60, 65). Second, SM-mediated induction of ISGs does not require any other EBV proteins or EBV DNA replication, as SM expression alone was sufficient to induce ISGs in B lymphocytes and other cell types. Finally, STAT1 induction by SM does not appear to involve IFN or other diffusible cytokines.
Is STAT1 induction a defensive cellular response to SM, or is it advantageous to viral replication? Although several of the ISGs, particularly those belonging to the ISG56K family, are among the genes most highly induced by IFN, little is known about their roles in cellular physiology. Both ISG54K and ISG56K mRNAs are essentially undetectable prior to IFN treatment but are induced to very high levels by IFN-α/β. SM is a potent inducer of ISGs, leading to higher levels of ISG expression than 1,000 U of IFN-β/ml. Based on the limited present knowledge of the functions of these ISGs, it is possible that their induction is advantageous to EBV during the stage of its lytic replication when SM is normally expressed, immediately preceding and during EBV DNA replication. Recently, the protein encoded by ISG56K was shown to interact with translation initiation factor eIF-3 and inhibit translation (25). Inhibition of cell protein synthesis and an antiproliferative effect may facilitate rapid synthesis and accumulation of early lytic EBV mRNAs that encode proteins required for subsequent EBV DNA replication. SM-mediated slowing of cell metabolism might then be relieved as SM levels drop during the later stages of EBV replication. Alternatively, other subsequently synthesized EBV proteins might serve to counteract the SM-mediated effect and the host antiviral response. In this context, it is relevant that another IFN-induced gene, viperin, is directly induced by CMV infection, specifically by binding of the CMV glycoprotein gB to the cell surface (13). Paradoxically, viperin has antiviral effects and inhibits CMV replication. CMV is able to modulate viperin effects, and it has been proposed that CMV may use viperin to facilitate virion production.
Although the ISGs may play an important role in the antiproliferative effects of SM, it is likely that there are other SM-mediated, STAT1-dependent effects on host cell gene expression. In addition to their role in cytokine signaling, STAT1 molecules are thought to be involved in the basal transcription of several genes, including caspases (34). It has recently been shown that specific caspases are required for replication of Aleutian mink disease parvovirus (3). It should also be noted that BZLF1, an immediate-early protein expressed prior to SM during lytic replication, inhibits STAT1 activation (42). Thus, SM may be required to restore levels of STAT1 activity necessary for optimal EBV virion production.
STAT1 activation may also be required for expression of non-ISRE-dependent genes that are potentially important for virus replication. STAT1 has been shown to negatively regulate c-myc expression, as well as that of other genes involved in cell proliferation (50). STAT1 is known to interact with BRCA1, which acts synergistically to activate transcription of IFN-γ target genes involved in growth inhibition, such as p21/WAF1 (48). IFN-γ induction of some genes in STAT1-null cells, but not in wild-type cells, suggests that STAT1-dependent repression also occurs (49).
We have shown that ISGs are induced by SM, but the net transcriptional effect of SM on STAT1-regulated genes is predicted to be complex, as SM not only increases STAT1 mRNA levels but alters the relative amounts of its two different functional isoforms in a manner distinct from that of IFN-α. STAT1α and -β (p91 and p84) are generated by alternative splicing (54). STAT1α homodimers can bind and activate IFN-γ-activated sequences found in promoters of genes activated by IFN-γ (56-58), whereas STAT1β homodimers are ineffective in stimulating IFN-γ-activated sequences (46). Thus, SM could interfere with IFN-γ-mediated antiviral effects by increasing the levels of STAT1β. Whether STAT1β is capable of mediating STAT1-dependent repression remains to be established. The net effect of the SM-mediated increase in both STAT1α and STAT1β is therefore difficult to predict, but modulation of STAT1 levels and function by SM is likely to have extensive effects on the resistance of EBV-infected cells to innate and adaptive immune responses.
Other herpesviruses also induce ISGs by IFN-dependent and -independent pathways. HSV infection triggers expression of ∼20 IFN-α-stimulated genes (43). Human CMV infection or binding of CMV gB to the cell surface induces expression of a large number of IFN-responsive genes in addition to viperin, and human CMV infection leads to the formation of an ISRE binding complex containing IRF-3 (47). Induction of ISGs by various herpesviruses indicates that STAT-dependent and -independent transcriptional regulatory pathways may be activated as well as inhibited by infecting viruses to maximize their survival. There are at least 300 cell genes that are induced to various levels and in different patterns by IFN-α/β and IFN-γ (18, 19). Depending on the specific kinases that are activated and the natures of the transcriptionally active complexes that are formed, the net effect on cell physiology of the alteration of IFN signal transduction pathways by viral proteins is likely to be extremely complex. Comparing the effect of SM on the large number of STAT1-responsive genes to infection by EBV and to the effects of IFN will require extensive use of transcriptional profiling and is the subject of investigation.
Acknowledgments
This work was supported by NIH grants CA81133 (S.S.), CA80105 (M.D.), and CA56645 (C.E.S.).
REFERENCES
- 1.Au, W. C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bello, L. J., A. J. Davison, M. A. Glenn, A. Whitehouse, N. Rethmeier, T. F. Schulz, and J. Barklie Clements. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 3.Best, S., J. B. Wolfinbarger, and M. E. Bloom. 2002. Caspase activation is required for permissive replication of Aleutian mink disease parvovirus in vitro. Virology 292:224-234. [DOI] [PubMed] [Google Scholar]
- 4.Blesofsky, W. A., K. Mowen, R. M. Arduini, D. P. Baker, M. A. Murphy, D. D. Bowtell, and M. David. 2001. Regulation of STAT protein synthesis by c-Cbl. Oncogene 20:7326-7333. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, S. M., V. Ruvolo, A. K. Gupta, and S. Swaminathan. 1999. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J. Virol. 73:6872-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braselmann, S., P. Graninger, and M. Busslinger. 1993. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl. Acad. Sci. USA 90:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buisson, M., F. Hans, I. Kusters, N. Duran, and A. Sergeant. 1999. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J. Virol. 73:4090-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buisson, M., E. Manet, M. C. Trescol-Biemont, H. Gruffat, B. Durand, and A. Sergeant. 1989. The Epstein-Barr Virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J. Virol. 63:5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M., and B. Barrell. 1990. Herpesviruses: a study of parts. Trends Genet. 6:86-91. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, P., K. S. Ellison, R. Verity, and J. R. Smiley. 2000. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated alpha-globin pre-mRNA in infected HeLa cells. J. Virol. 74:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevalier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho, M. S., K. T. Jeang, and S. D. Hayward. 1985. Localization of the coding region for an Epstein-Barr virus early antigen and inducible expression of this 60-kilodalton nuclear protein in transfected fibroblast cell lines. J. Virol. 56:852-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 16.Darzynkiewicz, Z., and G. Juan. 1997. Nucleic acid analysis, p. 7.5.2. In P. Robinson (ed.), Current protocols in molecular cytometry, vol. 1. John Wiley and Sons, New York, N.Y. [Google Scholar]
- 17.Davison, A. J., and P. Taylor. 1987. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J. Gen. Virol. 68:1067-1079. [DOI] [PubMed] [Google Scholar]
- 18.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 20.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell, P. 1989. Epstein-Barr virus genome. Adv. Viral Oncol. 8:103-132. [Google Scholar]
- 22.Farrell, P. J., R. J. Broeze, and P. Lengyel. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279:523-525. [DOI] [PubMed] [Google Scholar]
- 23.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 87:8555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu, X. Y., C. Schindler, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl. Acad. Sci. USA 89:7840-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, M. T. F., and C. M. Gorman. 1990. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Improta, T., C. Schindler, C. M. Horvath, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1994. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl. Acad. Sci. USA 91:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inchauspe, G., S. Nagpal, and J. M. Ostrove. 1989. Mapping of two varicella-zoster virus-encoded genes that activate the expression of viral early and late genes. Virology 173:700-709. [DOI] [PubMed] [Google Scholar]
- 30.Kenney, S., J. Kamine, E. Holley-Guthrie, E. C. Mar, J. C. Lin, D. Markovitz, and J. Pagano. 1989. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J. Virol. 63:3870-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenney, S., J. Kamine, D. Markovitz, R. Fenrick, and J. Pagano. 1988. An Epstein-Barr virus immediate-early gene product trans-activates gene expression from the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. USA 85:1652-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lipincott-Raven, Philadelphia, Pa.
- 33.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, A., M. Commane, T. W. Flickinger, C. M. Horvath, and G. R. Stark. 1997. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science 278:1630-1632. [DOI] [PubMed] [Google Scholar]
- 35.Lawn, R. M., J. Adelman, T. J. Dull, M. Gross, D. Goeddel, and A. Ullrich. 1981. DNA sequence of two closely linked human leukocyte interferon genes. Science 212:1159-1162. [DOI] [PubMed] [Google Scholar]
- 36.Lawn, R. M., J. Adelman, A. E. Franke, C. M. Houck, M. Gross, R. Najarian, and D. V. Goeddel. 1981. Human fibroblast interferon gene lacks introns. Nucleic Acids Res. 9:1045-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy, D., A. Larner, A. Chaudhuri, L. E. Babiss, and J. E. Darnell, Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA 83:8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markovitz, D. M., S. Kenney, J. Kamine, M. S. Smith, M. Davis, and E.-S. Huang. 1989. Disparate effects of two herpesvirus immediate-early gene trans-activators on the HIV LTR. Virology 173:750-754. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menezes, J., W. Liebold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 42.Morrison, T. E., A. Mauser, A. Wong, J. P. Ting, and S. C. Kenney. 2001. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity 15:787-799. [DOI] [PubMed] [Google Scholar]
- 43.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mowen, K., and M. David. 2000. Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol. 20:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mowen, K. A., J. Tang, W. Zhu, B. T. Schurter, K. Shuai, H. R. Herschman, and M. David. 2001. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104:731-741. [DOI] [PubMed] [Google Scholar]
- 46.Muller, M., C. Laxton, J. Briscoe, C. Schindler, T. Improta, J. E. Darnell, Jr., G. R. Stark, and I. M. Kerr. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouchi, T., S. W. Lee, M. Ouchi, S. A. Aaronson, and C. M. Horvath. 2000. Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-gamma target genes. Proc. Natl. Acad. Sci. USA 97:5208-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramana, C. V., M. P. Gil, Y. Han, R. M. Ransohoff, R. D. Schreiber, and G. R. Stark. 2001. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 98:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramana, C. V., N. Grammatikakis, M. Chernov, H. Nguyen, K. C. Goh, B. R. Williams, and G. R. Stark. 2000. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J. 19:263-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruvolo, V., A. K. Gupta, and S. Swaminathan. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 75:6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruvolo, V., E. Wang, S. Boyle, and S. Swaminathan. 1998. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. USA 95:8852-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sample, J., G. Lancz, and M. Nonoyama. 1986. Mapping of genes in BamHI fragment M of Epstein-Barr virus DNA that may determine the fate of viral infection. J. Virol. 57:145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindler, C., X. Y. Fu, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA 89:7836-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shuai, K., C. M. Horvath, L. H. Huang, S. A. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 57.Shuai, K., C. Schindler, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258:1808-1812. [DOI] [PubMed] [Google Scholar]
- 58.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 59.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 60.Swaminathan, S., R. Hesselton, J. Sullivan, and E. Kieff. 1993. Epstein-Barr virus recombinants with specifically mutated BCRF1 genes. J. Virol. 67:7406-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 62.Tavernier, J., R. Derynck, and W. Fiers. 1981. Evidence for a unique human fibroblast interferon (IFN-beta 1) chromosomal gene, devoid of intervening sequences. Nucleic Acids Res. 9:461-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veals, S. A., C. Schindler, D. Leonard, X. Y. Fu, R. Aebersold, J. E. Darnell, Jr., and D. E. Levy. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wathelet, M., S. Moutschen, P. Defilippi, A. Cravador, M. Collet, G. Huez, and J. Content. 1986. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur. J. Biochem. 155:11-17. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, A. D., and A. J. Morgan. 2002. Primary immune responses by cord blood CD4+ T cells and NK cells inhibit Epstein-Barr virus B-cell transformation in vitro. J. Virol. 76:5071-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong, K.-M., and A. J. Levine. 1986. Identification and mapping of Epstein-Barr virus early antigens and demonstration of a viral gene activator that functions in trans. J. Virol. 60:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]