Abstract
A protein-protein interaction within the helicase domain of the Tobacco mosaic virus (TMV) 126- and 183-kDa replicase proteins was previously implicated in virus replication (S. Goregaoker, D. Lewandowski, and J. Culver, Virology 282:320-328, 2001). To further characterize the interaction, polypeptides covering the interacting portions of the TMV helicase domain were expressed and purified. Biochemical characterizations demonstrated that the helicase domain polypeptides hydrolyzed ATP and bound both single-stranded and duplexed RNA in an ATP-controlled fashion. A TMV helicase polypeptide also was capable of unwinding duplexed RNA, confirming the predicted helicase function of the domain. Biochemically active helicase polypeptides were shown by gel filtration to form high-molecular-weight complexes. Electron microscopy studies revealed the presence of ring-like oligomers that displayed six-sided symmetry. Taken together, these data demonstrate that the TMV helicase domain interacts with itself to produce hexamer-like oligomers. Within the context of the full-length 126- and 183-kDa proteins, these findings suggest that the TMV replicase may form a similar oligomer.
Positive-stranded RNA viruses are a diverse group of pathogens that cause diseases in humans, plants, and animals. Although this group of pathogens is taxonomically diverse, they all encode replicase proteins involved in the synthesis of viral RNA. Enzymatic motifs within these replicase proteins can include methyltransferase (MT), helicase, and RNA-dependent RNA polymerase (POL) activities. These motifs may be present within a single multidomain protein, as found within the Tobacco mosaic virus (TMV) 183-kDa protein, or separated onto two or more virus-encoded proteins, as found in the 1a MT-HEL and 2a POL proteins of Brome mosaic virus (BMV) (2, 38). In infected cells, viral replicase proteins associate with host proteins as well as cellular membranes to produce replicase complexes that function in viral RNA synthesis. Despite the essential role of these replicase complexes in virus replication, little is known about their structure and the mechanisms that control their assembly.
TMV is a positive-stranded RNA virus that has served as a model for the study of RNA replication (3). TMV is the type member of the genus Tobamovirus and a member of the alphavirus supergroup. Its genome encodes at least four proteins (11) (Fig. 1). The 5′-proximal open reading frames (ORFs) encode 126- and 183-kDa proteins, the larger produced by the read-through of an amber stop codon (33). Both the 126- and 183-kDa proteins are necessary for efficient virus replication (17, 18, 25). Homology studies indicate that the 126-kDa-protein-encoding ORF encodes MT and helicase domains divided by an uncharacterized intervening region (IR), while the read-through portion of the 183-kDa-protein-encoding ORF encodes the POL domain (20, 21, 22) (Fig. 1). A 30-kDa cell-to-cell movement protein and a 17.5-kDa coat protein are produced from 3′ coterminal subgenomic mRNAs (6, 16, 28).
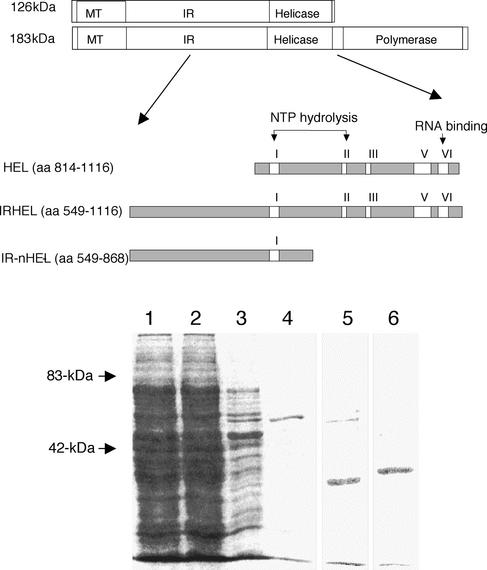
FIG. 1.
Expression and purification of 126- and 183-kDa helicase polypeptides. Shown is a schematic representation of 126- and 183-kDa helicase polypeptides expressed as hexahistidine fusion proteins from E. coli (top). Also shown are helicase polypeptides resolved by sodium dodecyl sulfate-PAGE and stained with Coomassie blue (bottom). Lane 1, total lysate from IRHEL-expressing E. coli; lane 2, flow-through fraction from Ni2+ affinity column; lane 3, 100 mM imidazole wash fraction; lanes 4, 5, and 6, 300 mM imidazole elution fractions of IRHEL (∼63 kDa), HEL (∼34 kDa), and IR-nHEL (∼36 kDa) polypeptides, respectively. Arrows mark the positions of known molecular weight markers.
Biochemical characterizations of the TMV 126-kDa protein have demonstrated the presence of MT-derived guanylyltransferase-like activity that results in the capping of viral RNAs as well as nucleotide triphosphatase activity derived from the helicase domain (7, 8, 27). Although RNA POL and helicase activities have not been demonstrated for either replicase protein in vitro, viral replicase complexes purified from infected plants have been shown to possess template-dependent and template-specific RNA POL activity (30). In addition, affinity purification of TMV replicase complexes from infected plant tissues by use of replicase-specific antibodies results in an enzyme complex that contains roughly equivalent ratios of 126- and 183-kDa proteins (39). This finding suggests that the 126- and 183-kDa proteins interact within the replicase complex.
A two-hybrid protein-protein interaction within the helicase domain of the TMV 126- and 183-kDa replicase proteins has been identified previously(12, 13). No other interactions either within or between replicase domains were found. Furthermore, mutations in the helicase domain that disrupted the two-hybrid interaction in a temperature-sensitive fashion similarly disrupted viral replication when introduced into the viral genome. Together, these findings suggest that the helicase domain interaction plays an important role in virus replication. To further examine the function and structural conformation of this domain, polypeptides representing the interacting portions of the TMV helicase domain were expressed and purified. These polypeptides displayed various abilities to hydrolyze ATP, bind RNA, and unwind duplexed RNA. Preservation of these enzymatic properties indicates that the polypeptides maintain a native structural conformation. Gel filtration and electron microscopy studies revealed that the helicase domain polypeptides self associate to form hexamer-like multimers, similar to the oligomeric structures assumed by many DNA and RNA helicases (4, 32). The implications of these findings for development of a model for the functional assembly of the TMV replicase complex are discussed.
MATERIALS AND METHODS
Expression and purification of helicase polypeptides.
The TMV amino acid numbering system used here is that of Goelet et al. (11). Three segments encompassing the helicase and IR domains of the TMV 126- and 183-kDa proteins (HEL, amino acids [aa] 814 to 1116, ∼34 kDa; IR-nHEL, aa 549 to 868, ∼36 kDa; IRHEL, aa 549 to 1116, ∼63 kDa) were PCR amplified by using TMV-specific primers containing XhoI and KpnI restriction sites (Fig. 1). PCR products were digested with the appropriate restriction enzymes and ligated into the similarly digested expression vector pTrcHis-A (Invitrogen, Carlsbad, Calif.) to produce a protein containing an N-terminal hexahistidine tag. pTrcHis constructs were transformed into Escherichia coli strain BL21, and 1-liter cultures were grown to an optical density at 600 nm of 0.6. Protein expression was induced with isopropyl-β-d-thiogalactopyranoside (1 mM), and cultures were grown for an additional 5 h. Cells were harvested by centrifugation for 10 min at 12,000 × g and resuspended on ice in 50 ml of 1× start buffer (20 mM NaHPO4, 0.5 M NaCl [pH 7.4]) containing 10 mM imidazole and 1 mg of lysozyme/ml. Cell lysates were pulse sonicated (model 500 sonic dismembrator; Fisher Scientific, Pittsburgh, Pa.) and centrifuged for 15 min at 12,000 × g. Clarified lysates were passed over HiTrap chelating Ni-affinity columns (Amersham Biosciences, Piscataway, N.J.), which were subsequently washed with 50 ml of 1× start buffer containing 100 mM imidazole, and bound polyproteins were eluted with 1× start buffer containing 300 mM imidazole. Imidazole was removed from eluted fractions via PD10 desalting columns (Amersham Biosciences), and polyproteins were concentrated in Amicon YM-30 concentrator tubes (Millipore, Bedford, Mass.). Protein concentrations were determined by Bradford assay (1), and purified protein was stored at −20°C in aliquots containing 20% glycerol.
ATPase assays.
Purified helicase polypeptides were incubated with 0.5 μCi of [γ-32P]ATP in reaction buffer (20 mM HEPES-KOH [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol [DTT], 40 μM ATP) for 30 min at 37°C (34). Reactions were terminated by the addition of EDTA to a final concentration of 0.1 M and incubation on ice. Each reaction mixture was spotted on polyethyleneimine-cellulose thin-layer chromatography plates (Fisher Scientific) and developed in buffer (150 mM formic acid, 150 mM LiCl [pH 3]). Thin-layer chromatography plates were then dried and visualized by using PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.)
RNA binding and helicase substrates.
Single-stranded RNA substrate was generated by the in vitro transcription, in the presence of [α-32P]UTP, of linearized plasmid DNA encoding TMV nucleotides (nt) 6291 to 6395. The 104-nt labeled transcripts were then separated by native polyacrylamide gel electrophoresis (PAGE), excised from the gel, and extracted by soaking overnight in buffer (80 mM KCl, 50 mM Tris-HCl [pH 8], 2 mM DTT) followed by extraction with phenol-chloroform and ethanol precipitation (36).
Duplexed RNA substrate was generated by incubating unlabeled single-stranded RNA representing TMV nt 6291 to 6395 with a complementary [γ-32P]ATP end-labeled 30-nt RNA sequence corresponding to TMV negative-stranded RNA (nt 6326 to 6355; Dharmacon Research Inc, Lafayette, Colo.). The annealing mixture was heated to 75°C for 10 min and gradually cooled to 25°C overnight. Duplexed RNA was then resolved by native PAGE, excised from the gel, and purified as described above. Both single-stranded and duplexed RNA substrates were quantified by absorbance at 260 nm.
RNA binding and helicase assays.
Purified recombinant polypeptides were added to buffer (40 mM Tris-HCl [pH 7.9], 6 mM MgCl2, 2 mM spermidine, 10 mM DTT) containing 1 pmol of either the single-stranded or the duplexed 32P-labeled RNA substrates described above (31). Binding reactions were incubated at 25°C for 30 min and resolved by native PAGE and visualized by PhosphorImager. To assess the effects of ATP on RNA binding, reactions were performed in the presence or absence of 5 mM ATP.
RNA helicase assays were carried out in buffer containing 100 μg of bovine serum albumin, 2.5 mM DTT, 5% glycerol, 2.5 U of RNasin (Promega, Madison, Wis.), and 20 mM HEPES-KOH, pH 7.4 (31, 40). Purified polypeptides were added to the mixture along with 0.25 pmol of duplexed RNA substrate as described above. Reaction mixtures were incubated at 25°C for 30 min followed by the addition of 5 mM ATP and 2.5 mM MgCl2, with a further 1-h incubation at 37°C. Reaction products were resolved by native PAGE and visualized via PhosphorImager.
Gel filtration and electron microscopy analysis.
Purified preparations of HEL and IRHEL polypeptides (75 to 150 μg) were passed over a 1- by 90-cm Superdex 200 (Amersham Biosciences) column equilibrated with 20 mM HEPES (pH 7.4) and 150 mM NaCl buffer at 0.5 ml/min. Half-milliliter fractions were collected and analyzed by sodium dodecyl sulfate-PAGE and Western immunoblotting for the detection of the hexahistidine-tagged polypeptides. Molecular weights were estimated by comparison to the markers ferritin (440 kDa) and catalase (232 kDa) (Amersham Biosciences).
Purified preparations of HEL and IRHEL polypeptides were equilibrated in buffer containing 20 mM HEPES-KOH (pH 7.4) and were spotted (3 μl) onto carbon-coated electron microscope grids and allowed to dry. The grids were then negatively stained with 1% uranyl oxalate for 3 min, wick dried with filter paper, and examined by electron microscopy. Measurements from 50 different HEL and IRHEL oligomers were averaged to determine particle diameter.
RESULTS
Expression and purification of recombinant TMV helicase polypeptides.
The three expressed histidine-tagged polypeptides (HEL, ∼34 kDa; IR-nHEL, ∼36 kDa; and IRHEL, ∼63 kDa) were designed to cover regions of the TMV 126- and 183-kDa proteins previously shown to interact in the yeast two-hybrid system (12, 13) (Fig. 1). Each polypeptide accumulated within the soluble portion of the cell lysates and was readily purified via Ni2+ affinity columns under nondenaturing conditions (Fig. 1).
TMV helicase polypeptides display ATPase activity.
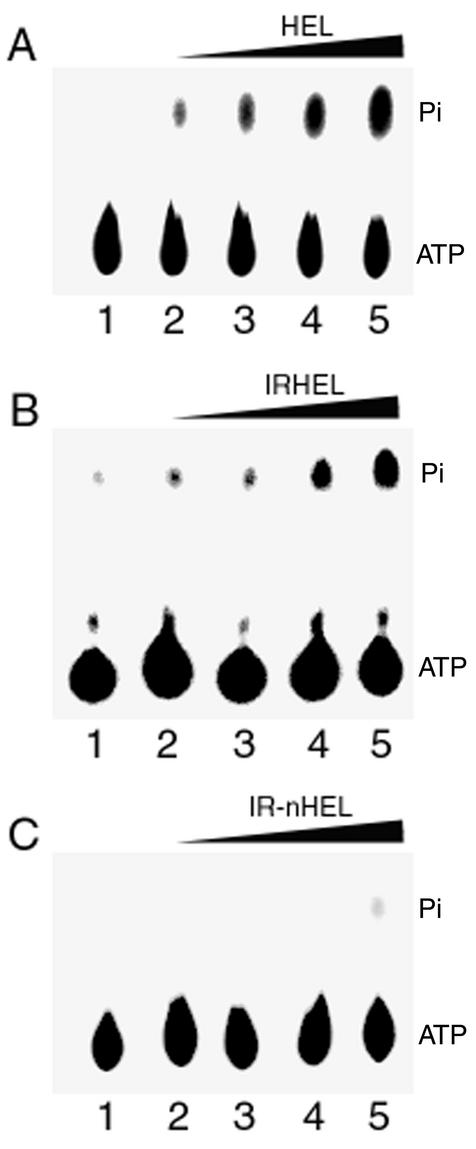
Both the IRHEL and HEL polypeptides were capable of hydrolyzing [γ-32P]ATP in a concentration-dependent fashion (Fig. 2A and B). In contrast, the IR-nHEL peptide lacks NTP hydrolysis motif II (Fig. 1) and failed to display ATPase activity (Fig. 2C). Additionally, the presence of a divalent cation was required for enzymatic activity, with Mg2+ functioning as the most efficient cofactor (Fig. 3). However, both Ca2+ and Mn2+ were able to replace Mg2+, though with a four- to sixfold reduction in efficiency (Fig. 3).
FIG. 2.
ATPase activities of 126- and 183-kDa helicase polypeptides. The panels show ATPase activity in the presence of increasing concentrations of HEL (A), IRHEL (B), or IR-nHEL (C) polypeptide. Reaction mixtures were separated by thin-layer chromatography and visualized for the release of [32Pi]. Lane 1, activity in the absence of the 126- and 183-kDa helicase polypeptides; lanes 2 to 5, activity in the presence of 25, 50, 100, and 200 ng of purified helicase polypeptides, respectively.
FIG. 3.
Effect of divalent cations on the ATPase activity of the 126- and 183-kDa helicase polypeptide IRHEL. Lane 1, activity in the absence of a cation; lanes 2 to 4, activity in the presence of 5 mM MgCl2, MnCl2, and CaCl2, respectively. Assays were done in the presence of 200 ng of purified IRHEL polypeptide.
Helicase polypeptides bind RNA in an ATP-controlled fashion.
The ability of the HEL, IR-nHEL, and IRHEL polypeptides to bind RNA was examined by electrophoretic gel mobility shift assays. The RNA substrates included a 104-nt positive-strand segment of the TMV 3′ untranslated region (nt 6291 to 6395) either single stranded or partially hybridized to a cRNA oligomer (nt 6326 to 6355). Both HEL and IRHEL polypeptides bound the single-stranded as well as partially double-stranded RNA (Fig. 4A and B). As expected, the IR-nHEL peptide did not bind RNA since it lacks RNA binding motif VI (Fig. 4C). Additional RNA binding experiments using a single-stranded RNA substrate transcribed from a plasmid DNA template produced a similar level of HEL and IRHEL binding (data not shown). Thus, HEL and IRHEL polypeptides bind RNA in a non-sequence-specific manner.
FIG. 4.
RNA binding capabilities of the TMV HEL, IRHEL, and IR-nHEL polypeptides. Electrophoretic gel mobility shift assays using native PAGE were performed to demonstrate helicase polypeptide binding to single-stranded (A and C) and partially double-stranded (B) RNAs. Lanes in panels A and B: 1, 32P-labeled single-stranded or double-stranded RNA substrate alone; 2, addition of 0.5 μg of HEL polypeptide; 3, addition of 0.5 μg of IRHEL polypeptide. Lanes in panel C: 1, 32P-labeled single-stranded RNA substrate alone; 2, addition of 0.5 μg of IR-nHEL polypeptide; 3, addition of 0.5 μg of HEL polypeptide.
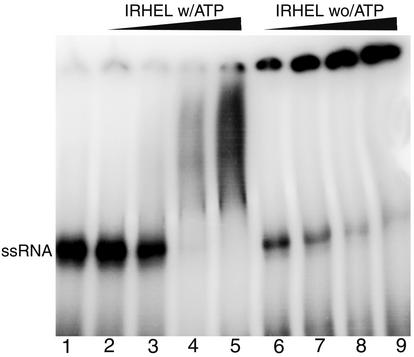
The effect of ATP on the ability of the TMV helicase domain to bind RNA was also investigated. In the absence of ATP, the IRHEL polypeptide readily bound RNA even at low protein concentrations to produce a large protein-RNA complex (Fig. 5). However, the addition of 5 mM ATP to the binding reaction completely disrupted the ability of the IRHEL polypeptide to bind RNA at low protein concentrations while producing a mixture of smaller protein-RNA complexes at high protein concentrations (Fig. 5). This finding indicates the presence of an antagonistic relationship between RNA binding and ATP binding and/or hydrolysis.
FIG. 5.
Effect of ATP on the RNA binding abilities of helicase polypeptide IRHEL. The binding reactions displayed in lanes 2 to 5 were done in the presence of 5 mM ATP, while those displayed in lanes 6 to 9 were done in the absence of ATP. Lane 1 shows the position of the unbound single-stranded RNA substrate. The remainder of the lanes display binding results in the presence of increasing concentrations of the purified IRHEL polypeptide: lanes 2 and 6, 0.05 μg; lanes 3 and 7, 0.25 μg; lanes 4 and 8, 0.5 μg; lanes 5 and 9, 1 μg.
Helicase activity of HEL and IRHEL polypeptides.
The HEL and IRHEL polypeptides were analyzed for their ability to unwind a duplexed RNA substrate containing a 35-nt 5′-end tail, a 30-nt base-paired region, and a 45-nt 3′-end tail. This RNA substrate corresponded to the TMV 3′ untranslated region (nt 6291 to 6395) and was previously used in the above-described RNA binding assays (Fig. 4). The IR-nHEL polypeptide was excluded from this analysis since it is incapable of hydrolyzing ATP or binding RNA, which are functions necessary for helicase activity. The results demonstrated that only the HEL polypeptide and not the IRHEL polypeptide was capable of releasing the labeled 30-nt segment from its complementary strand (Fig. 6). RNA release was also dependent on the presence of ATP.
FIG. 6.
Helicase activity of TMV HEL and IRHEL polypeptides. Helicase assays were done by using an RNA substrate containing a 30-base double-stranded region with 5′ (35 nt) and 3′ (40 nt) single-stranded overhangs. Similar reactions were done in the presence of purified HEL (A) or IRHEL (B) polypeptide and resolved by native PAGE. Lanes 1, double-stranded RNA substrate; lanes 2, boiled substrate showing released 32P-labeled 30-nt strand; lanes 3, helicase reaction run with 1.1 μg of either HEL or IRHEL polypeptide in the absence of ATP; lanes 4 to 8, helicase reactions run in the presence of 5 mM ATP and with increasing concentrations of either the HEL or IRHEL polypeptide (0.22, 0.44, 0.66, 0.88, and 1.1 μg, respectively).
HEL and IRHEL polypeptides associate to form oligomers.
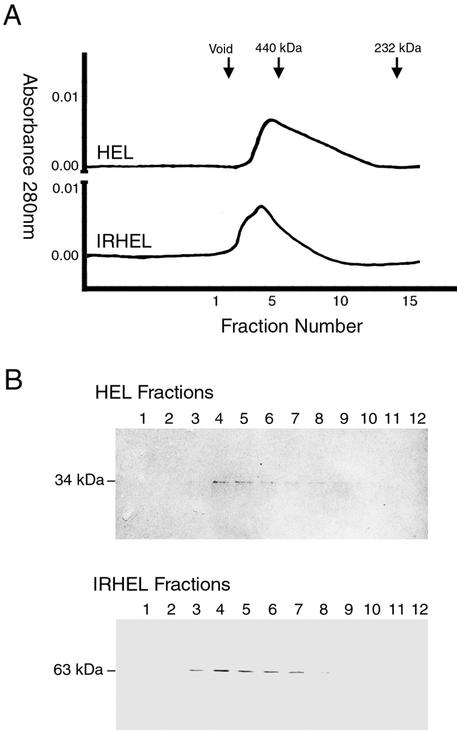
The enzymatic and RNA binding activities observed for the HEL and IRHEL polypeptides suggest that these polypeptides fold into a functional conformation and are thus suitable for additional structural studies. To assess the ability of the HEL and IRHEL polypeptides to self associate, purified HEL and IRHEL polypeptides were subjected to gel filtration chromatography. Under native conditions, both HEL and IRHEL polypeptides eluted as broad peaks with average molecular weights greater than 440 kDa, indicating that both formed large-order oligomers (Fig. 7). Consistent with their size, oligomers produced by the larger IRHEL polypeptide eluted ahead of oligomers produced by the smaller HEL polypeptide. The presence of both the 34-kDa HEL and 63-kDa IRHEL polypeptides within these high-molecular-weight peaks was confirmed by Western immunoblot detection (Fig. 7). Longer column runs failed to yield additional peaks that would indicate the presence of a monomer or smaller order oligomer.
FIG. 7.
Estimation of the native molecular mass of HEL and IRHEL oligomers. (A) Gel filtration analysis. Arrows mark the elution points of the void volume and known molecular weight markers. (B) Western immunoblot detection of HEL (∼34 kDa) and IRHEL (∼63 kDa) polypeptides present in peak fractions.
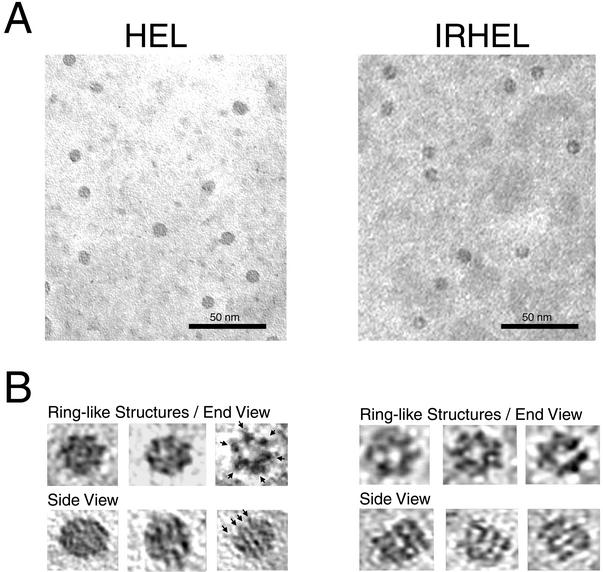
The broad, high-molecular-weight peaks produced by both HEL and IRHEL polypeptides suggest a heterogeneous mix of oligomers. Electron microscopy studies revealed that purified HEL and IRHEL solutions primarily contain a mixture of cylindrical and ring-like oligomers (Fig. 8). Additionally, what appeared to be partially formed rings or stacks of rings also were observed. These findings confirm the heterogeneous nature of the HEL and IRHEL oligomers. Higher magnifications revealed the presence of a predominant ring-like structure produced by both HEL and IRHEL polypeptides. The diameters of these ring-like structures measured 10.7 ± 0.7 nm (mean ± standard deviation) for the smaller HEL polypeptide and 12.5 ± 0.9 nm for the larger IRHEL polypeptide. The ring-like oligomers produced by both polypeptides predominantly displayed a sixfold symmetry, suggesting a hexamer-like structure, while the cylindrical oligomers appeared to be composed of stacks of the ring-like structures oriented on their sides (Fig. 8). Stacks of IRHEL ring-like structures were generally more uniform and open in structure, while the stacks produced by the HEL polypeptide were more compact, with tapered ends. This variation likely reflects the size difference between the HEL and IRHEL polypeptides. Taken together, these findings demonstrate that both HEL and IRHEL polypeptides assemble to form similar hexamer-like oligomers.
FIG. 8.
Oligomeric forms produced by HEL and IRHEL polypeptides. The samples were examined by transmission electron microscopy. (A) HEL and IRHEL oligomers viewed at low magnification (bar = 50 nm). (B) Enlarged (four times) images of individual HEL and IRHEL oligomers. Shown are end views of ring-like structures showing six-sided symmetry and side views of stacked rings showing up to four layers (arrows).
DISCUSSION
Previously, Goregaoker et al. identified a protein-protein interaction between portions of the TMV helicase domain that, when disrupted, prevented virus replication (12). In this study, we addressed the structural basis of this interaction by expressing and purifying polypeptides corresponding to the interacting portions of the TMV helicase domain from E. coli. These polypeptides were readily purified under nondenaturing conditions and displayed biochemical activities consistent with their predicted functions. Maintenance of these functions indicates that these polypeptides assume a native structural conformation and were therefore ideally suited for studies investigating the assembly of the viral replicase.
Viral-encoded helicases are likely involved in resolving double-stranded intermediates formed between the template RNA and the newly synthesized progeny strands as well as in removing RNA secondary structure. These processes require the presence of specific structural motifs (4, 32). Based on the presence of conserved motifs, the TMV helicase domain has been classified as a superfamily 1 helicase (22). Within this helicase family, motifs I and II are involved in NTP binding or hydrolysis. The HEL and IRHEL polypeptides maintain these two motifs, and both are capable of hydrolyzing ATP in the presence of a divalent cation. The helicase RNA binding site located at motif VI also is present within the HEL and IRHEL polypeptides and likely accounts for the ability of these polypeptides to bind both single-stranded and partially duplexed RNA in a non-sequence-specific manner. The importance of these motifs in helicase function is further supported by the inability of the IR-nHEL polypeptide, which lacks these motifs, to hydrolyze NTP or bind RNA. Taken together, these findings are consistent with the functions of similarly characterized viral helicases (9, 14, 19).
Interestingly, the RNA binding function of the IRHEL polypeptide was found to be ATP controlled. In the absence of ATP, the IRHEL polypeptide readily bound RNA even at low protein concentrations. In contrast, the addition of ATP to the reaction mixture disrupted RNA binding. A similar effect by ATP on RNA binding has been observed for the NS3 helicase of hepatitis C virus (10, 31). ATP binding or hydrolysis likely induces specific structural changes that allow the helicase to bind and then release the RNA during translocation. In the absence of ATP, the IRHEL polypeptide and RNA associate to form a large protein RNA complex (Fig. 5). In contrast, at high polypeptide concentrations the presence of ATP results in a mixture of smaller protein-RNA complexes. This suggests that ATP binding or hydrolysis may also affect the ability of the TMV helicase domain to form oligomers and/or maintain its quaternary structure.
The HEL polypeptide was capable of unwinding duplexed RNA in an ATP-dependent fashion, thus confirming the predicted function of this domain in virus replication. The ability of the HEL polypeptide to display helicase activity also indicates that this polypeptide maintains a structural conformation consistent with its function within the full-length 126- and 183-kDa replicase proteins. The inability of the IRHEL polypeptide to function in duplex unwinding even though it displays ATPase and RNA binding activities suggests that the truncated IR portion of this polypeptide interferes with helicase translocation. Thus, structural variations between the HEL and IRHEL polypeptides likely account for the observed differences in helicase activity.
Helicases must utilize at least two nucleic acid binding sites in order to maintain contact with the nucleic acid during translocation (23). Oligomerization of helicase subunits provides a means whereby multiple nucleic acid binding sites can interact cooperatively. In this study, gel filtration and electron microscopy results demonstrated that the HEL and IRHEL polypeptides assemble into oligomeric complexes. The molecular mass and symmetry of these complexes were consistent with the formation of hexameric ring-like structures and multimeric stacks of ring-like structures. A number of helicases, both DNA and RNA, have been shown to function as hexamers (20, 32). Furthermore, the helicase activity of the archaeal minichromosome maintenance protein is dependent upon the formation of a double hexamer that resembles a stack of ring-like structures, similar to those formed by the HEL and IRHEL polypeptides (5). Among viruses, the NS3 helicase protein of hepatitis C virus is active as an oligomer while the RNA helicase function of Simian virus 40 large tumor antigen functions as a hexamer (23, 37). Thus, the oligomeric structures produced by the TMV helicase domain are consistent with the structural conformations assumed by other characterized hexameric helicases.
Oligomeric polymerase structures produced by the poliovirus 3D-POL protein have been implicated in replication (15, 26). Additionally, the 1a MET-HEL protein of BMV has been shown to interact with itself and to accumulate within membrane-bound vesicles at levels sufficient to form a core-like inner shell, suggesting that 1a oligomerization is involved in the formation and stabilization of membrane-bound replicase complexes (29, 35). Unlike poliovirus or BMV, TMV replicase functions reside within a single multidomain protein; therefore, oligomerization of the helicase domain may dictate the structural conformation of the replicase complex. Whether or not the TMV helicase domain functions as an oligomer within the context of the full-length 126- and 183-kDa replicase proteins remains to be fully examined. However, mutations that disrupt the TMV helicase interaction within the yeast two-hybrid system also disrupt virus replication in vivo (12). Thus, the intermolecular interaction of the helicase domain appears to be essential for TMV replication. Furthermore, Watanabe et al. (39) demonstrated that within infected tissues both the 126- and 183-kDa proteins coimmunoprecipitated in a 1:1 ratio, providing additional support for the presence of an oligomeric TMV replicase structure.
A 126-kDa-183-kDa hexamer could provide a flexible platform capable of accommodating different ratios of 126-kDa proteins to 183-kDa proteins. For example, a 3:3 ratio would support the immunoprecipitation data generated by Watanabe et al. (39) (Fig. 9). However, translational read-through of the 126-kDa-protein-encoding ORF to produce the POL domain occurs only ∼10% of the time (17). Removal of the amber stop codon of the 126-kDa-protein-encoding ORF by mutation results in a TMV RNA that expresses only the 183-kDa protein and replicates at less than 20% of the efficiency of the wild-type virus (17, 18, 24, 25). Thus, although not required for RNA synthesis, excess levels of 126-kDa protein significantly enhance replication. One explanation for this phenomenon is that the excess 126-kDa protein increases the ratio of 126-kDa protein to 183-kDa protein within replicase oligomers to produce a more efficient enzyme complex (Fig. 9). In addition, excess 126-kDa protein may lead to the formation of oligomers that contain no 183-kDa protein, leading to the possibility that helicase and replicase functions reside on different oligomers. Alternatively, oligomerization of excess 126-kDa protein may provide a means of structurally encapsidating the replicase complex. The membrane-bound replicase complexes produced by BMV contain a 25:1 ratio of 1a MET-HEL protein to 2a POL protein and are similar in size and form to the virion cores produced by retroviruses (35). Based in part on these findings, Schwartz et al. (35) have suggested that the form and function of positive-stranded RNA virus replicase complexes are similar to the virion cores formed by retroviruses and double-stranded RNA viruses. What role if any a 126-kDa-126-kDa or 126-kDa-183-kDa oligomer has in the assembly of a larger core-like replicase complex remains to be determined.
FIG. 9.
Oligomeric models displaying different ratios of 126-kDa proteins to 183-kDa proteins positioned around a hexameric helicase domain.
Acknowledgments
We thank H. Shiferaw, M. Padmanabhan, and S. Golem for assistance in preparing the manuscript.
This work was supported in part by USDA-NRI grant 9902519.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 5.Chong, J. P., M. K. Hayashi, M. N. Simon, R. M. Xu, and B. Stillman. 2000. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 97:1530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deom, C. M., M. J. Oliver, and R. N. Beachy. 1987. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science 237:389-394. [DOI] [PubMed] [Google Scholar]
- 7.Dunigan, D. D., and M. Zaitlin. 1990. Capping of tobacco mosaic virus RNA. Analysis of viral-coded guanylyltransferase-like activity. J. Biol. Chem. 265:7779-7786. [PubMed] [Google Scholar]
- 8.Erickson, F. L., S. Holzberg, A. Calderon-Urrea, V. Handley, M. Axtell, C. Corr, and B. Baker. 1999. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18:67-75. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez, A., H. S. Guo, P. Saenz, L. Simon-Buela, M. Gomez de Cedron, and J. A. Garcia. 1997. The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25:4474-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallinari, P., D. Brennan, C. Nardi, M. Brunetti, L. Tomei, C. Steinkuhler, and R. De Francesco. 1998. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goelet, P., G. P. Lomonossoff, P. J. G. Butler, M. E. Akam, M. J. Gait, and J. Karn. 1982. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 79:5818-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 13.Goregaoker, S. P. 2002. Functional analysis of the tobacco mosaic virus 126/183-kDa replicase associated proteins, p. 152. Ph.D. thesis. University of Maryland, College Park.
- 14.Gross, C. H., and S. Shuman. 1995. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J. Virol. 69:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, T., T. Hunt, J. Knowland, and D. Zimmern. 1976. Messenger RNA for the coat protein of tobacco mosaic virus. Nature (London) 260:759-764. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, M., T. Meshi, F. Motoyoshi, N. Takamatsu, and Y. Okada. 1986. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 14:8291-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa, M., T. Meshi, T. Ohno, and Y. Okada. 1991. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 65:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, L., and D. L. Peterson. 1995. Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch. Biochem. Biophys. 323:47-53. [DOI] [PubMed] [Google Scholar]
- 20.Kadare, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 22.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 23.Levin, M. K., and S. S. Patel. 1999. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 274:31839-31846. [DOI] [PubMed] [Google Scholar]
- 24.Lewandowski, D. J., and W. O. Dawson. 1998. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 251:427-437. [DOI] [PubMed] [Google Scholar]
- 25.Lewandowski, D. J., and W. O. Dawson. 2000. Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90-98. [DOI] [PubMed] [Google Scholar]
- 26.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 27.Merits, A., R. Kettunen, K. Makinen, A. Lampio, P. Auvinen, L. Kaariainen, and T. Ahola. 1999. Virus-specific capping of tobacco mosaic virus RNA: methylation of GFP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 455:45-48. [DOI] [PubMed] [Google Scholar]
- 28.Meshi, T., Y. Watanabe, T. Saito, A. Sugimoto, T. Maeda, and Y. Okada. 1987. Function of the 30kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J. 6:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Reilly, E. K., Z. Wang, R. French, and C. C. Kao. 1998. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 72:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paolini, C., R. De Francesco, and P. Gallinari. 2000. Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81:1335-1345. [DOI] [PubMed] [Google Scholar]
- 32.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 33.Pelham, H. R. B. 1978. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature (London) 272:469-471. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 35.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 36.Tai, C. L., W. K. Chi, D. S. Chen, and L. H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 70:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlmann-Schiffler, H., S. Seinsoth, and H. Stahl. 2002. Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res. 30:3192-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Heijden, M. W., and J. F. Bol. 2002. Composition of alphavirus-like replication complexes: involvement of virus and host encoded proteins. Arch. Virol. 147:875-898. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, E., and G. W. Owttrim. 2000. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 28:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]