Abstract
The vaccinia virus A30L protein is required for the association of electron-dense, granular, proteinaceous material with the concave surfaces of crescent membranes, an early step in viral morphogenesis. For the identification of additional proteins involved in this process, we used an antibody to the A30L protein, or to an epitope appended to its C terminus, to capture complexes from infected cells. A prominent 42-kDa protein was resolved and identified by mass spectrometry as the vaccinia virus G7L protein. This previously uncharacterized protein was expressed late in infection and was associated with immature virions and the cores of mature particles. In order to study the role of the G7L protein, a conditional lethal mutant was made by replacing the G7L gene with an inducible copy. Expression of G7L and formation of infectious virus was dependent on the addition of inducer. Under nonpermissive conditions, morphogenesis was blocked and viral crescent membranes and immature virions containing tubular elements were separated from the electron-dense granular viroplasm, which accumulated in large spherical masses. This phenotype was identical to that previously obtained with an inducible, conditional lethal A30L mutant. Additional in vivo and in vitro experiments provided evidence for the direct interaction of the A30L and G7L proteins and demonstrated that the stability of each one was dependent on its association with the other.
Vaccinia virus (VV), the prototypical member of the Poxviridae, has a double-stranded DNA genome of approximately 190 kbp which encodes nearly 200 polypeptides. Viral transcription, DNA replication, and progeny assembly occur entirely in the cytoplasm of the infected cells within discrete areas known as viral factories. VV morphogenesis is a complex process, occurring through a series of intermediate structures that can be visualized by electron microscopy (10, 13, 17, 24). By conventional techniques, the first discernible structures appear as rigid, crescent-shaped membranes with a brush-like border of spicules on their convex surfaces and electron-dense granular material called viroplasm adjacent to their concave sides. These structures evolve into spherical, immature virions (IV), which enclose a nucleoprotein mass. The IV undergo additional maturational events, including proteolytic processing of viral core proteins (19), creating the brick-shaped intracellular mature virions (IMV). The IMV constitute the majority of the infectious progeny in infected cells. Some IMV become wrapped by a double membrane, derived from the trans-Golgi network or endosomal cisternae (14, 27, 32), forming intracellular enveloped virions. Intracellular enveloped virions are transported to the periphery on microtubules (12, 15, 23, 36, 37) and exit cells by fusing their outermost membranes with plasma membranes, leaving the resulting extracellular cell-associated enveloped virions (CEV) still associated with cell surfaces (4). Actin polymerization occurs in the cytoplasm below the CEV and projects the CEV at the tips of long, motile microvilli toward neighboring cells (15, 23, 28, 36). Some extracellular enveloped virions are released into the medium (2). CEV and extracellular enveloped virions are thought to be responsible for cell-to-cell and long-range virus spread, respectively (2, 5, 6, 22).
Electron microscopic studies of conditionally lethal mutant viruses have provided insights into the formation of the IV. Few recognizable viral structures are formed in cells infected with a virus containing a temperature-sensitive mutation in the F10L protein kinase (33, 35). Repression of A17L expression arrests VV morphogenesis at an early stage, characterized by the absence of typical viral membranes and the accumulation of spherical masses of electron-dense material that are surrounded by numerous membrane vesicles and tubules (24, 25, 39). Inhibition of A14L expression also leads to the accumulation of membrane vesicles and tubules, but aberrant crescent-like membranes that are detached from the granular masses also appear (26, 34). Inhibition of D13L expression or addition of the drug rifampin results in the accumulation of spherical masses of electron-dense material surrounded by irregularly shaped viral membranes and tubules that lack spicules (13, 21, 24, 40). Taken together, these data suggest that viral membranes are formed from vesicular and tubular elements derived from a cellular compartment, through an unknown mechanism that requires the kinase activity of the F10L protein and the participation of the A17L, the A14L, and probably other viral proteins. The D13L protein is also targeted to the viral membrane, providing the characteristic rigid convex shape of the crescent and IV membranes.
Recently, we constructed and characterized a conditionally lethal recombinant virus, vA30Li, that has an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible A30L gene (30). Repression of the expression of A30L resulted in the accumulation of normal-looking crescent-shaped and circular viral membranes that were separated from large masses of electron-dense granular material, suggesting that the A30L protein has a role in the association of the dense viroplasm with viral membranes. In addition, a temperature-sensitive mutant with a similar phenotype was mapped to the A30L open reading frame (ORF) (29). Because the A30L protein lacks any enzyme motifs, we considered that it might act through association with other viral proteins. Here, we describe the identification and characterization of the G7L protein, a VV core protein that interacts with and stabilizes the A30L protein and is also required for the association of dense viroplasm with viral membranes.
MATERIALS AND METHODS
Cell and viruses.
BS-C-1 (ATCC CCL6) and HeLa S3 (ATCC CCL2.2) cells were grown in Eagle's minimal essential medium (MEM) and Dulbecco's MEM, respectively, each of which was obtained from Quality Biologicals Inc. and supplemented with 10% fetal bovine serum (FBS). Unmodified VV (WR strain) and the recombinant VVs vT7LacOI (1) and vTF7-3 (11) were propagated in HeLa cells as previously described. The recombinant viruses vA30Li (30), vA30LiHA (30), vA17Li (39), and vG7Li (described below) were replicated in the continuous presence of 50 μM IPTG and 2.5% FBS. All virus stocks were stored at −70°C. Plaque assays and one-step virus growth experiments were carried out with BS-C-1 cells.
Plasmid construction and generation of recombinant virus.
To construct the pVOTE.1G7L plasmid, a complete copy of the G7L ORF flanked by the NcoI and BamHI restriction sites at the 5′ and 3′ ends, respectively, was generated by PCR using VV genomic DNA as the template and the oligonucleotide primers 5′-CGATCCATGGCTGCAGAACAGCGTCGTTC-3′ and 5′-TGCGGATCCTTAACATTTTGGCAAATTGATTACC-3′ (NcoI and BamHI restriction sites underlined). The PCR product was digested with BamHI and NcoI and inserted into pVOTE.1 (38).
To construct the G7L knockout plasmid, for removal of the endogenous copy of the G7L gene, standard overlap PCR methodology was used to generate DNA containing (i) a complete copy of the β-glucuronidase (gus) gene under the control of the G7L gene promoter and (ii) flanking sequences of the G7L ORF, comprising a portion of the G8R gene (right flank), and a complete copy of the G6R ORF together with portion of the G5R gene (left flank). An 808-bp DNA segment corresponding to the flanking region to the right of the G7L ORF was generated by PCR using VV genomic DNA as the template and the oligonucleotide primers 5′-CTACAGGACGTAACATTTAACTTTAAATAATTTACAAAAATTTAAAATGAGCATCC-3′ and 5′-GATGTTGATCTTAAAGGATTGAACTCTATCC-3′. A PCR product of 1,835 bp containing the complete bacterial gus gene and part of the putative G7L promoter was generated using the pZippy-NEO/GUS plasmid (provided by T. Shors) as the template and the oligonucleotide primers 5′-TTAAAGTTAAATGTTACGTCCTGTAGAAACCCCAACC-3′ and 5′-CCTTAAAGGGTGATCATTGTTTGCCTCCCTGCTGC-3′. A 977-bp DNA segment corresponding to the flanking region to the left of the G7L ORF was generated by PCR using VV genomic DNA as the template and the oligonucleotide primers 5′-GCGTTAGAACCACGCAAGGAGATTGATG-3′ and 5′-GGAGGCAAACAATGATCACCCTTTAAGGTAATCAATTTGC-3′. The final PCR product of 3,566 bp was inserted into the pCR2.1-TOPO vector (Invitrogen) to produce the G7L knockout plasmid.
The recombinant VV vG7Li was constructed in two steps essentially as previously described for vA30Li (30). First, plasmid pVOTE.1G7L containing the inducible copy of the G7L gene was inserted into the hemagglutinin (HA) locus by homologous recombination to generate vG7L/G7Li. Then, the original copy of the G7L gene was deleted and replaced with the gus reporter gene to produce vG7Li.
Antibodies.
Rabbit anti-G7L protein sera were produced by immunizing rabbits with synthetic peptides corresponding to amino acids 285 to 301 (RMLDTSEKYSKGYKTDGC) or to amino acids 26 to 38 (ISINSEYIESKAKC) of the predicted G7L ORF with an additional cysteine residue (Covance Research Products Inc., Denver, Pa.). The C- and N-terminal-peptide antisera were used for immunoblotting and immunoelectron microscopy, respectively. The antipeptide sera to the A30L, A14L, and H3L proteins have been previously described (3, 8, 30). The murine monoclonal antibody (MAb) MHA.11 (Covance Research Products Inc.) recognizes the 9-amino-acid influenza HA epitope tag (YPYDVPDYA). The immobilized anti-HA high-affinity rat MAb covalently linked to agarose beads (anti-HA affinity matrix) was purchased from Roche Molecular Biochemicals.
Radioimmunoprecipitation analysis.
BS-C-1 cells were infected with the wild type or recombinant viruses at a multiplicity of infection of 10 PFU per cell for 1 h at 37°C except where indicated in the figures. After adsorption, the cells were washed twice and incubated for 5 h with complete Eagle's MEM containing 2.5% FBS in the absence or presence of 50 μM IPTG. At 6 h after infection, the medium was removed and replaced with methionine- and cysteine-free medium containing 2.5% dialyzed FBS (Invitrogen) and labeled with 50 μCi of a [35S]methionine and [35S]cysteine mixture (NEN-Perkin-Elmer) per ml. After 18 h the cells were harvested, washed with cold Tris-buffered saline, and lysed with 50 mM Tris (pH 8.0)-150 mM NaCl-1% (vol/vol) Nonidet P-40 (NP-40) (lysis buffer) containing protease inhibitor cocktail tablets (Roche Molecular Biochemicals) for 30 min on ice. Lysates were clarified by centrifugation at 20,000 × g for 30 min at 4°C. These extracts were rotated with 50 μl of 20% protein A-Sepharose beads for 1 to 2 h at 4°C. The lysate was then centrifuged at 1,000 × g for 1 min, and the supernatant was incubated overnight at 4°C with a 1:250 dilution of either A30L or G7L protein antiserum. The antigen-antibody complexes were then rotated with protein A-Sepharose beads for 1 to 2 h at 4°C. When the anti-HA affinity matrix was used, the cell lysates were rotated with the resin for 4 h at 4°C. The immune complexes were washed three times with lysis buffer and resuspended in sample buffer (0.125 M Tris HCl [pH. 6.8], 4% sodium dodecyl sulfate [SDS], 20% [vol/vol] glycerol, 10% [vol/vol] β-mercaptoethanol, and 0.004% bromophenol blue). Samples were incubated at 95°C for 3 min and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the proteins were visualized after autoradiography.
Affinity purification.
Three roller bottles containing 4.5 × 108 BS-C-1 cells were infected with the vA30LiHA virus at a multiplicity of infection of 1 PFU per cell for 2 h and incubated at 37°C in the presence of 500 μM IPTG. At 48 h after infection, the infected cells were harvested, washed with cold Tris-buffered saline, and lysed with lysis buffer in the presence of protease inhibitor cocktail tablets for 1 h on ice. The cell lysate was clarified by centrifugation at 20,000 × g for 1 h at 4°C, and the supernatant was rotated with 1 ml of a 50% (vol/vol) slurry of the anti-HA affinity matrix for 4 h. The resin was washed five times with 10 ml of lysis buffer and once with 10 ml of equilibration buffer (20 mM Tris [pH 7.5], 100 mM NaCl, and 0.1 mM EDTA). To elute proteins, the resin was washed three times with 0.5 ml of equilibration buffer containing 0.5 mg of HA peptide and four times with 0.5 ml of 100 mM glycine (pH 2.0). The eluted proteins were precipitated with 20% trichloroacetic acid and resuspended in 1 M Tris (pH 9.0) plus 2× sample buffer. Immunoprecipitated proteins were analyzed by SDS-PAGE and visualized by either silver or Coomassie blue staining. The Coomassie blue-stained band migrating with an apparent mass of 42 kDa was excised from the gel and identified by mass spectrometry (MS).
MS.
Coomassie blue-stained bands were excised and subjected to in-gel tryptic digestion as previously described (18). The resulting digests were concentrated in a Speed Vac (Savant, Farmingdale, N.Y.) to 10 to 20 μl, and their volumes were adjusted to 125 μl with 0.1% trifluoroacetic acid. Heptafluorobutyric acid (1.5 μl) was added to each sample prior to analysis as previously described (16), with the Magic 2002 model capillary high-pressure liquid chromatograph (Michrom BioResources, Auburn, Calif.) coupled to a model LCQ ion trap mass spectrometer (ThermoFinnigan, San Jose, Calif.) equipped with an electrospray interface. The high-pressure liquid chromatography was carried out with a 0.3- by 150-mm Magic MS C18 column (Michrom BioResources), and samples were eluted at a rate of 8 μl/min with a linear gradient of 2 to 65% CH3CN-H2O-HCOOH-heptafluorobutyric acid (900/100/2.2/0.005) at 40°C for 30 min. Column effluent was monitored at a wavelength of 215 nm. The mass spectrometer was operated in the “top-five” mode to automatically acquire a full scan between m/z 300 to 1,300 and MS/MS spectra (relative collision energy, 35%) of the five most intense ion peaks in the full scan. Uninterpreted MS/MS spectra were searched against the nonredundant database utilizing BioWorks and SEQUEST software (ThermoFinnigan).
Western blotting.
The procedures for Western blotting were similar to those previously described (30) with the following additions. The membranes were incubated for 1 h with the anti-G7L protein polyclonal antibody at a 1:1,000 dilution or the anti-H3L protein polyclonal antibody at a 1:1,000 dilution.
SDS-PAGE analysis of metabolically labeled proteins.
The procedures for labeling and analyzing the proteins were similar to those previously described (30).
Virion fractionation.
Purified VV particles (108 PFU) were incubated in a reaction mixture containing 50 mM Tris-HCl (pH 7.5) and 1% (vol/vol) NP-40, with or without 50 mM dithiothreitol for 1 h at 37°C. The insoluble and soluble materials were separated by centrifugation at 20,000 × g for 30 min at 4°C.
Electron microscopy.
BS-C-1 cells were grown in 60-mm-diameter dishes and infected with vG7Li at a multiplicity of infection of 5 PFU per cell in the presence or absence of 50 μM IPTG. At 20 h after infection, cells were prepared for transmission electron microscopy as previously described (9). For immunoelectron microscopy, the cells were fixed and prepared for freezing as previously described (8) with the anti-G7L protein antibody at a 1:10 dilution.
Transient expression of the G7L and A30L proteins.
BS-C-1 cells in a six-well plate were infected with the vTF7-3 virus at a multiplicity of infection of 10 PFU per cell at 37°C in the absence or presence of 40 μg of cytosine arabinoside (AraC) per ml. After 1 h, the inocula were removed and the cells were washed three times with Opti-MeM I (Invitrogen) with or without AraC. Cells infected with vTF7-3 in the presence of AraC were then transfected with 1 μg of different combinations of the pVOTE.1G7L, pVOTE.1A30L, and pVOTE.1 plasmids per well using 8 μl of Lipofectamine 2000; the AraC concentration was maintained. At 6 h after infection, cells were labeled with 50 μCi of a mixture of [35S]methionine and [35S]cysteine and incubated at 37°C for 18 h. Cells were then harvested, and immunoprecipitation was carried out as described above.
In vitro transcription and translation of the G7L and A30L ORF.
The reticulocyte lysate-based TNT quick coupled transcription and translation system (Promega) was used as directed by the manufacturer. To each 40 μl of TNT mixture, 20 μCi of [35S]methionine and 1 μg of total DNA (pVOTE.1A30L, pVOTE.1G7L, and pVOTE.1) were added. The samples were incubated at 30°C for 90 min and immunoprecipitated with the anti-A30L protein antibody as described above.
RESULTS
Interaction of a 42-kDa protein with the A30L protein.
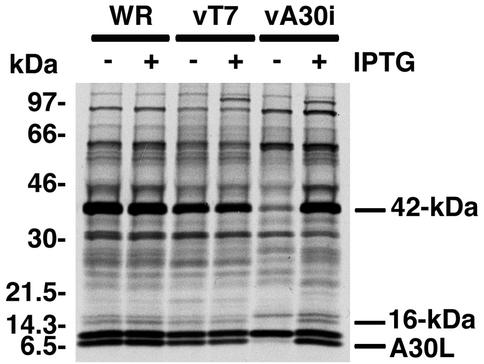
Our previous studies showed that the A30L protein is required for the association of VV membranes with the viroplasm. To investigate whether this process is mediated through interactions with other proteins, VV-infected cells were metabolically labeled and the extracts were incubated with an antibody to the A30L protein. Upon analysis of the immune complexes by SDS-PAGE and autoradiography, we detected a prominent band of approximately 42 kDa in addition to the A30L protein. To determine whether the 42-kDa and A30L proteins were specifically associated, the experiment was repeated employing recombinant vA30Li, which expresses the A30L protein only in the presence of IPTG. A strong band of 42 kDa and a weaker one of 16 kDa were detected in immune complexes from cells infected with control viruses WR and vT7lacOI in the presence or absence of IPTG but from cells infected with vA30Li only in the presence of the inducer (Fig. 1). The induced band of approximately 100 kDa, not seen in cells infected with VV WR, is T7 RNA polymerase, which was nonspecifically immunoprecipitated. Similar experiments were carried out using vA30LiHA, a recombinant virus that inducibly expresses an HA epitope-tagged version of the A30L protein, and an HA-specific MAb. Again, a prominent 42-kDa protein was detected only in immune complexes from cells infected in the presence of IPTG (data not shown).
FIG. 1.
Coimmunoprecipitation of a 42-kDa protein with the A30L protein from VV-infected cells. BS-C-1 cells were infected with wild-type VV (WR), vT7LacOI (vT7), or vA30Li (vA30i) in the presence (+) or absence (−) of 50 μM IPTG as indicated. At 6 h after infection, the cells were labeled with a mixture of [35S]methionine and [35S]cysteine and incubated for 18 h at 37°C. Cell extracts were prepared and incubated with antiserum directed to the C-terminal 11 amino acids of the A30L protein. The immunoprecipitated products were resolved by SDS-PAGE and visualized by autoradiography. Bands corresponding to the A30L protein (which is the lower band of a doublet) and proteins with masses of 42 and 16 kDa are indicated on the right. Numbers on the left correspond to molecular masses of the marker proteins.
Identification of the 42-kDa protein.
To identify the 42-kDa protein, extracts from cells infected with vA30LiHA were incubated with a high-affinity anti-HA antibody covalently linked to agarose beads. After extensive washing, the bound proteins were eluted with HA peptide and then low pH. The affinity-purified proteins were separated by SDS-PAGE and visualized either by silver (Fig. 2A) or Coomassie blue (not shown) staining. The band corresponding to the 42-kDa protein was excised from the Coomassie blue-stained polyacrylamide gel and subjected to in-gel tryptic digestion. The resulting digest was analyzed by liquid chromatography-MS-MS, and seven tryptic peptides were identified as peptides encoded by the G7L ORF of VV (Fig. 2B). Analysis of the 16-kDa band revealed peptides corresponding to the J1R protein ORF (to be described elsewhere).
FIG. 2.
Identification of the 42-kDa protein as the product of the G7L ORF. (A) Affinity purification of the A30L protein complex. Proteins extracted from cells infected with vA30LiHA in the presence of IPTG were purified using a high-affinity anti-HA affinity matrix. The F.T (Flowthrough) lane contains material that did not bind to the HA matrix. The HA matrix was washed with lysis buffer (wash lanes, fractions 1 to 3), and the proteins were eluted with the HA peptide (HApep lanes, fractions 1 to 3) followed by glycine (glycine lanes, fractions 1 to 4). Proteins from each fraction were analyzed by electrophoresis on an SDS-10 to 20% polyacrylamide gel in Tricine buffer followed by silver staining. Bands corresponding to the A30L and 42-kDa proteins are indicated. Numbers on the left indicate the positions and molecular masses in kilodaltons of marker proteins. (B) Predicted amino acid sequence of the VV G7L ORF. Peptides identified by MS are underlined. Amino acids comprising the peptide used to produce polyclonal antibody are in italics, and consensus proteolysis cleavage sites are indicated by arrows. (C) Reactivity of the 42-kDa protein with G7L protein antiserum. (Left) Lysates of uninfected (lane U) and VV WR-infected BS-C-1 cells were resolved by SDS-PAGE, reacted with G7L peptide antiserum by Western blotting, and detected by chemiluminescence. (Right) Extracts of cells infected with the vA30LiHA virus in the presence (+) or absence (−) of 100 μM IPTG were incubated with the high-affinity anti-HA affinity matrix, and bound proteins were analyzed by Western blotting with G7L protein antiserum. The positions of migrations and molecular masses of marker proteins are indicated on the left.
A rabbit polyclonal antibody to a peptide derived from amino acids 285 to 301 of the G7L protein (Fig. 2B) was prepared and found by Western blotting to interact with a band of approximately 42 kDa from cells infected with VV but not from uninfected cells (Fig. 2C, left gel). To confirm the interaction of the G7L and A30L proteins, extracts of cells infected with the vA30LiHA virus in the presence or absence of IPTG were incubated with the anti-HA affinity matrix and the bound proteins were analyzed by SDS-PAGE followed by Western blotting with the G7L protein antiserum. The 42-kDa band was detected only in the complex that was affinity purified from cells infected with the vA30LiHA virus in the presence of IPTG (Fig. 2C, right gel), further demonstrating that the A30L and G7L proteins interact during VV infection. Additional experiments indicated that the A30L protein was specifically immunoprecipitated with G7L protein antiserum (data not shown).
Temporal expression of the G7L ORF.
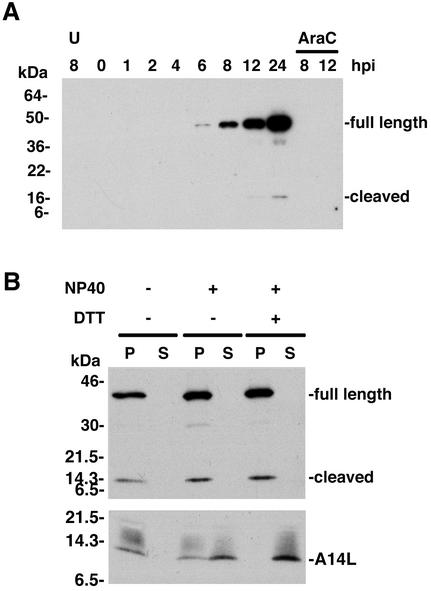
The only available information regarding the G7L protein was a report describing the detection of a C-terminal fragment during an N-terminal sequence analysis of core proteins (31). Homologs of the G7L protein were found in the genomes of other poxviruses but not in unrelated viruses or organisms. Inspection of the predicted amino acid sequence of the G7L ORF revealed two putative viral protease cleavage sites (Ala-Gly-X) (Fig. 2B) but no membrane localization or other motifs that would suggest a function. Late expression of the G7L ORF was predicted from the presence of a consensus late promoter sequence upstream of the G7L ORF (data not shown). To confirm this prediction, whole-cell lysates were prepared at various times after VV infection or in the presence of the DNA replication inhibitor AraC and analyzed by SDS-PAGE, followed by Western blotting with the G7L protein antiserum. The 42-kDa protein was first detected at 6 h after infection, and its band increased in intensity during the remainder of the 24-h time course (Fig. 3A). An additional 15-kDa band was detected by 12 h after infection (Fig. 3A) and most likely corresponds to the C-terminal end of the G7L protein produced by cleavage of the G7L protein at position 238. The antiserum, to a C-terminal peptide of the G7L protein, was not capable of detecting the N-terminal fragment of the G7L protein, which also was not detected in the previous N-terminal sequencing study, presumably because it was blocked. A band migrating slightly faster than the 42-kDa band was consistently seen and might represent initiation at an internal methionine codon. The failure to detect expression of either the 42- or 15-kDa proteins in the presence of AraC (Fig. 3A) confirmed the classification of the G7L protein as a postreplicative protein, as was previously found for the A30L protein.
FIG. 3.
Synthesis and virion localization of the G7L protein. (A) Temporal synthesis of the G7L protein. BS-C-1 cells were mock infected for 8 h (lane U) or infected with VV at a multiplicity of infection of 10 in the absence or presence of AraC and harvested between 0 and 24 h postinfection (hpi). Proteins from total cell extracts were resolved by electrophoresis on an SDS-4 to 20% gradient polyacrylamide gel and analyzed by Western blotting using rabbit G7L peptide antiserum. Proteins were detected by chemiluminescence. The bands corresponding to the full-length and cleaved forms of the G7L protein are indicated on the right. The positions of migrations and molecular masses of marker proteins are indicated on the left. (B) Association of the G7L protein with purified virions. Sucrose gradient-purified VV was incubated in buffer containing 1% NP-40 with or without 50 mM dithiothreitol. After centrifugation, the soluble (S lanes) and insoluble (P lanes) fractions were analyzed by Western blotting using G7L or A14L peptide antiserum. Bands corresponding to the A14L protein or to the full-length and cleaved forms of the G7L protein are indicated on the right. The numbers on the left correspond to the molecular masses of the marker proteins. DTT, dithiothreitol.
Localization of the G7L protein in virus particles.
Biochemical fractionation was used to localize the G7L protein in VV particles. Sucrose gradient-purified virions were treated with NP-40 with or without reducing agent and centrifuged to separate the detergent-soluble membrane proteins from the insoluble core fractions. The samples were analyzed by SDS-PAGE followed by immunoblotting with the G7L protein antiserum. Unlike the A30L protein, which was partially released from the virus particles treated with either NP-40 alone or NP-40 plus dithiothreitol (30), both the full-length and cleaved forms of the G7L protein remained exclusively associated with the insoluble core fraction (Fig. 3B). As a control, the Western blots were probed with antibody to the A14L membrane protein, which was completely released into the soluble fraction with NP-40 and dithiothreitol (Fig. 3B).
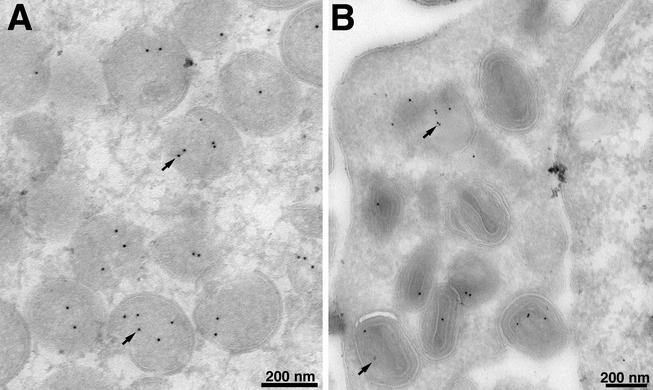
Immunoelectron microscopy was also used to localize the G7L protein. VV-infected cells were stained with a polyclonal antibody to amino acids 26 to 38 of the G7L protein, followed by protein A conjugated to gold spheres. The gold label was found within IV (Fig. 4A) and MV (Fig. 4B).
FIG. 4.
Localization of the G7L protein by immunoelectron microscopy. BS-C-1 cells were infected with VV WR at a multiplicity of infection of 2 PFU per cell. At 22 h, the cells were fixed in paraformaldehyde, cryosectioned, and incubated with an antibody to the peptide corresponding to amino acids 26 to 38 of the G7L protein and then with 10-nm-diameter gold particles conjugated to protein A. Electron micrographs are shown, with the scale indicated by the bars. (A) Field containing large numbers of IV; (B) field containing mature virions. Arrows point to representative gold particles.
Generation of a recombinant VV expressing an inducible copy of the G7L ORF.
To determine the role of the G7L protein during the virus replicative cycle, we constructed a recombinant VV that tightly regulates G7L expression. The starting virus, vT7LacOI, contains an IPTG-inducible copy of the bacteriophage T7 RNA polymerase and constitutively expresses the Escherichia coli lac repressor. The first step in the construction of the vG7Li recombinant virus was the insertion of the G7L ORF regulated by a T7 promoter, the E. coli lac operator, and the untranslated leader sequence of the encephalomyocarditis virus RNA into the HA locus of the vT7LacOI recombinant virus. The resulting intermediate virus, vG7L/G7Li, contained the original G7L ORF as well as the new inducible copy. In the second step, the original G7L ORF was deleted from the vG7L/G7Li virus by homologous recombination with a plasmid that contained the gus ORF under the control of the G7L promoter and the flanking sequences of the G7L gene. The final recombinant virus, vG7Li, had a single inducible copy of the G7L gene (Fig. 5A) and was isolated in the presence of 50 μM IPTG. The stringency of this system depends on regulation of both the T7 RNA polymerase and the G7L gene by the lac repressor.
FIG. 5.
Construction of a conditional lethal inducible G7L VV. (A) Schematic diagram representing the genome of vG7Li. The J2R (thymidine kinase [TK]), G7L, and A56R (HA) loci are shown. Insertions into these loci are displayed below the line. Additional abbreviations: P11, a VV late promoter, P7.5, a VV early late promoter; lacO, E. coli lac operator; lacI, E. coli lac repressor gene; T7 pol, bacteriophage T7 RNA polymerase gene; PT7, bacteriophage T7 promoter; EMC, encephalomyocarditis virus mRNA cap-independent translation enhancer element; gus, E. coli β-glucuronidase gene; gpt, E. coli guanine phosphoribosyltransferase gene. (B) Effect of IPTG on virus plaque formation. BS-C-1 cell monolayers were infected with vT7lacOI, vG7L/G7Li, or vG7Li in the presence (+) or absence (−) of 50 μM IPTG in the methylcellulose overlay. Cells were stained with crystal violet 48 h after infection.
Effect of G7L expression on virus replication.
To determine the effect of IPTG on plaque formation, monolayers of BS-C-1 cells were infected with recombinant viruses in the presence or absence of IPTG. The vT7lacOI and vG7L/G7Li viruses, each containing the original G7L gene, formed plaques both in the presence and absence of IPTG. In contrast, vG7Li, which contains only the inducible copy of the G7L gene, formed plaques in the presence but not in the absence of IPTG (Fig. 5B).
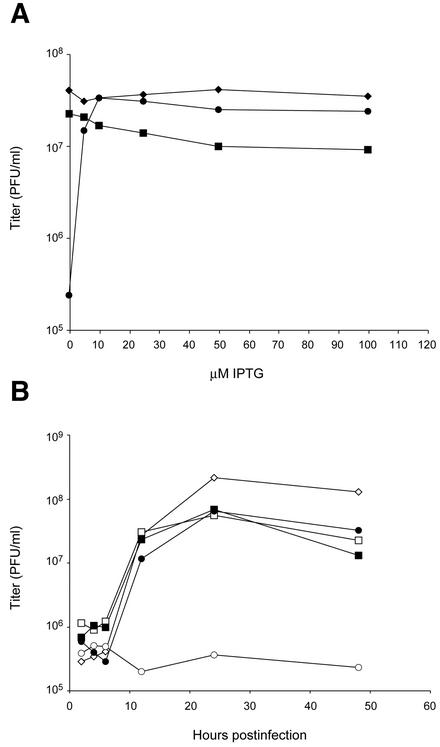
The inhibition of plaque formation may result from effects on virus replication or spread. Further experiments were carried out to determine the requirement for G7L expression on virus replication under one-step growth conditions. Addition of IPTG increased the yield of vG7Li by more than two logs (Fig. 6). A significant increase in virus yield occurred with 5 μM IPTG and reached a maximal yield at 10 μM. Concentrations higher than 10 μM resulted in only a slight decrease in the replication of the two viruses with inducible G7L genes, vG7Li and vG7L/G7Li, suggesting that excess G7L protein is not very toxic (Fig. 6A). Western blotting showed that G7L expression was undetectable in the absence of IPTG and increased progressively from 5 to 100 μM IPTG (to be shown later). A time course experiment indicated that in the presence of 50 μM IPTG, the kinetics of vG7Li replication were similar to those of the other viruses (Fig. 6B).
FIG. 6.
Effect of IPTG on yields of infectious vG7Li. (A) BS-C-1 cells were infected with VV WR (♦), vG7L/G7Li (▪), or vG7Li (•) at a multiplicity of infection of 5 and incubated in the presence of 0 to 100 μM IPTG for 24 h. Virus titers were determined by plaque assay in the presence of 50 μM IPTG. (B) BS-C-1 cells were infected with VV WR (⋄), vG7L/G7Li (□, ▪), or vG7Li (○, •) in the absence (open symbols) or presence (filled symbols) of 50 μM IPTG. Cells were harvested at the indicated times after infection, and virus titers were determined as described for panel A.
Requirement of G7L expression for processing of viral late proteins.
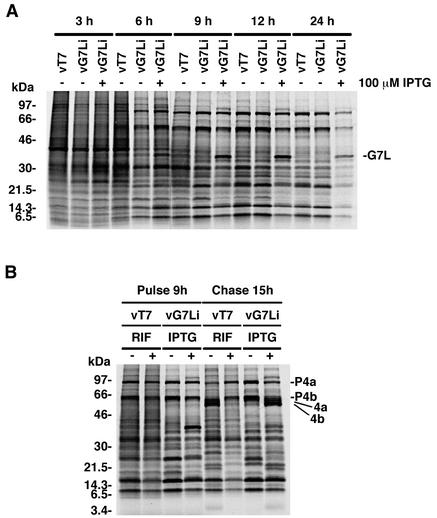
During VV replication, host protein synthesis is severely inhibited, allowing the detection of viral proteins by SDS-PAGE of extracts of metabolically labeled cells followed by autoradiography. Cells infected with vG7Li in the presence or absence of IPTG showed the typical shift from host to early viral to late viral protein synthesis (Fig. 7A). The pattern of protein synthesis of cells infected with vG7Li in the absence of IPTG appeared identical to that of cells infected either with vT7LacOI or with vG7Li in the presence of IPTG, except for the presence of an additional band corresponding to the overexpressed G7L protein in the latter. These results indicated that the inhibition of G7L expression does not have a general effect on viral gene expression.
FIG. 7.
Synthesis and processing of viral proteins. (A) Pulse-labeling of viral proteins. BS-C-1 cells were infected with vT7lacOI (vT7) or vG7Li at a multiplicity of infection of 10 in the presence (+) or absence (−) of 100 μM IPTG and labeled with a mixture of [35S]methionine and [35S]cysteine for 30-min periods starting at 3, 6, 9, 12, or 24 h after infection. Immediately after being labeled, the cells were washed and lysed and the labeled proteins were analyzed by electrophoresis on an SDS-4 to 20% gradient polyacrylamide gel. The gel was dried, and the proteins were visualized by autoradiography. The numbers on the left correspond to the molecular masses of the marker proteins. (B) Proteolytic processing of viral late proteins. BS-C-1 cells were infected either with vT7lacOI in the presence (+) or absence (−) of 100 μg of rifampin (RIF) per ml or with vG7Li in the presence (+) or absence (−) of 100 μM IPTG. At 9 h after infection, the cells were pulse-labeled with a mixture of [35S]methionine and [35S]cysteine for 30 min. Cells were either harvested immediately (pulse) or incubated with excess unlabeled methionine for an additional 15 h (chase). The proteins were denatured with SDS and mercaptoethanol, analyzed by electrophoresis on an SDS-4 to 20% gradient polyacrylamide gel, and visualized by autoradiography. The positions of migration of proteolytically processed structural proteins (4a and 4b) and their precursors (P4a and P4b) are shown on the right.
Several of the major proteins of the VV core, including 4a, 4b, and 25K, are derived from proteolytic processing of higher-molecular-weight precursors during virion morphogenesis. Cleavage of these proteins is inhibited by the drug rifampin, which blocks virion assembly at an early stage, and during infection with conditional lethal viruses that exhibit defects in morphogenesis. To determine whether inhibition of G7L protein expression had an effect on the proteolytic processing of viral proteins, pulse-chase experiments were carried out. Cells were infected with either vT7LacOI control virus in the presence or absence of rifampin or the vG7Li virus in the presence or absence of IPTG. Pulse-chase experiments indicated that processing of the precursors to form the 4a, 4b, and 25K proteins was inhibited in cells infected with the vG7Li virus in the absence of IPTG, similar to the effect observed in cells infected with vT7LacOI in the presence of rifampin (Fig. 7B). These results suggested that infection with vG7Li in the absence of IPTG resulted in an early block in virion morphogenesis at a stage prior to the formation of IMV.
Morphogenesis of vG7Li under nonpermissive conditions.
Electron microcopy was performed to determine the stage at which virion morphogenesis was blocked in the absence of G7L protein expression. Cells infected with vG7Li in the presence of IPTG contained structures corresponding to all stages of virion morphogenesis, including crescent-shaped membranes, IV and IMV (Fig. 8A). In contrast, neither typical IV nor mature virions were found in cells infected with vG7Li in the absence of IPTG. Instead, there was an accumulation of large circular masses of electron-dense viroplasm that were clearly separated from numerous crescent-shaped and circular viral membranes (Fig. 8B). In most cases, the circular membranes did not enclose granular material, resulting in structures that resembled “empty” IV. Some of the crescents and circular membranes appeared as multilayered “onion skin” structures (Fig. 8C), similar to those seen in cells infected with vA30Li in the absence of IPTG. The viral membranes showed a characteristic curvature and rigid appearance, suggesting that the primary defect involved the association of the viral membranes with the granular viroplasm rather than with viral membrane formation per se.
FIG. 8.
Electron microscopy of cells infected with vG7Li in the presence or absence of IPTG. BS-C-1 cells were infected with vG7Li in the presence (A) or absence (B to D) of 50 μM IPTG. After 24 h, the cells were fixed and prepared for transmission electron microscopy. The arrow in panel B points to electron-dense viroplasm, which is separated from IV membranes. Arrows in panel D point to cross-sections of tubules in which the membrane bilayer is clearly discerned. Electron micrographs are shown, with the scale indicated by the bars. Abbreviations: C, crescents; nu, nucleoid within an IV.
Although the IV formed in the absence of IPTG were largely devoid of granular material, they contained tubular elements. The appearance of the tubules depended on the plane of the section; on cross-section they appeared as rings composed of a membrane bilayer (Fig. 8D). Risco et al. (24) previously described tubular membranes associated with assembling IV.
Interaction of the G7L and A30L proteins is direct and does not depend on virus morphogenesis or other viral proteins.
We were curious as to whether the interactions of the G7L and A30L proteins occurred during morphogenesis, perhaps providing the trigger for structural alterations leading to IV formation. Two alternative methods were used to block morphogenesis: addition of the drug rifampin, which prevents viral crescent membrane formation (20), and repression of A17L protein synthesis, which causes an even earlier block in viral membrane formation (25, 39). Cells were infected with vA30Li or vA17Li in the presence or absence of IPTG or with vT7lacOI in the presence or absence of rifampin and metabolically labeled. Infected cell extracts were immunoprecipitated with A30L protein antiserum and analyzed by SDS-PAGE and autoradiography. The A30L and G7L proteins coimmunoprecipitated in cells infected with vA17Li and vT7lacOI under permissive or nonpermissive conditions, indicating that morphogenesis is not required for this protein-protein interaction (Fig. 9A).
FIG. 9.
The interaction of the G7L and A30L proteins is direct. (A) The interaction of the G7L and A30L proteins does not depend on VV morphogenesis. BS-C-1 cells were infected with vA30Li (vA30i) or vA17Li (vA17i) in the presence (+) or absence (−) of IPTG or with the vT7lacOI (vT7) virus in the presence (+) or absence (−) of rifampin (RIF). At 6 h after infection, cells were labeled with a mixture of [35S]methionine and [35S]cysteine. Labeled cells were harvested at 24 h after infection, and cell extracts were incubated with the A30L peptide antiserum and then protein A-Sepharose beads. The bound proteins were resolved in an SDS-4 to 20% polyacrylamide gel and visualized by autoradiography. The G7L protein and A30L protein (lower band of a doublet) are indicted on the right. (B) The interaction of the A30L and G7L proteins is not dependent on other VV late proteins. BS-C-1 cells were infected with vTF7-3 in the presence (+) or absence (−) of AraC and transfected (+) or not transfected (−) with plasmids containing the A30L or the G7L ORF regulated by the bacteriophage T7 promoter. After 6 h, the cells were labeled with a mixture of [35S]methionine and [35S]cysteine for 18 h. Cell extracts were immunoprecipitated with A30L peptide antiserum as described for panel A, and the bound proteins were resolved on an SDS-10 to 20% polyacrylamide gel in Tricine buffer and visualized by autoradiography. (C) The A30L and G7L proteins interact in the absence of other viral proteins. Templates containing (+) or not containing (−) the A30L or G7L ORF under the control of the bacteriophage T7 promoter were transcribed and translated in a reticulocyte lysate in the presence of [35S]methionine. The in vitro-translated proteins were immunoprecipitated with A30L peptide antiserum and analyzed by SDS-PAGE as described for panel A. In all panels, the migration positions and masses of marker proteins are indicated on the left and the positions of the G7L and A30L proteins are indicated on the right.
Additional experiments were carried out to determine whether any other viral late proteins were required for the interaction of the A30L and G7L proteins. Cells were treated with AraC to prevent viral DNA replication and late protein synthesis, infected with the recombinant virus vTF7-3 to express the bacteriophage T7 RNA polymerase, transfected with plasmids containing the G7L and the A30L gene under the control of the T7 promoter, and metabolically labeled. Cell extracts were incubated with A30L protein antiserum, and the complexes of A30L and G7L proteins were detected by SDS-PAGE and autoradiography (Fig. 9B, far-right lane). To prove that AraC had prevented synthesis of viral late proteins, we carried out parallel experiments in which either the G7L or A30L gene-bearing plasmid was not transfected. The G7L protein was not detected under either of these conditions (Fig. 9B, middle two lanes), indicating that AraC had prevented synthesis of the G7L and A30L proteins from their natural late promoters. Therefore, we can assume that AraC also inhibited the synthesis of other viral late proteins. When AraC was not added, coimmunoprecipitation of the A30L and G7L proteins occurred without transfection (Fig. 9B, far-left lane).
The above-described experiments strongly suggested that the interaction between the A30L and G7L proteins was direct, since other viral late proteins were not required. A reticulocyte-based coupled in vitro transcription and translation system was used to provide further support for a direct interaction. When the two proteins were expressed together, the G7L protein coimmunoprecipitated with the A30L protein (Fig. 9C, far-right lane). The 42-kDa band was not detected in the immune complex when the template for either the A30L or G7L protein was omitted (Fig. 9C) or when the G7L and A30L proteins were synthesized in separate reactions and then mixed (data not shown). The latter result suggested that the interaction of the G7L and A30L proteins occurs either during or immediately after translation.
Interaction of the G7L and A30L proteins is required for their stability in infected cells.
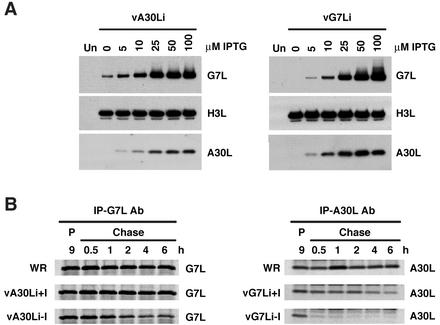
The above data indicating that the interaction of the A30L and G7L proteins occurs during or immediately after their synthesis led us to investigate the stability of each in the absence of the other. Cells were infected with vA30Li or vG7Li in the presence of increasing concentrations of IPTG, and then Western blots were made and probed with G7L or A30L protein antiserum. As expected, synthesis of A30L and G7L was dependent on the presence of IPTG in cells infected with vA30Li and vG7Li, respectively (Fig. 10A). Remarkably, however, the levels of the G7L protein detected in cells infected with vA30Li were directly proportional to the concentration of IPTG and very little was detected in the absence of IPTG (Fig. 10A). Similarly, the levels of the A30L protein were proportional to IPTG concentration in cells infected with vG7Li (Fig. 10A). To ensure that these effects were specific, the nitrocellulose membranes were stripped and reprobed with antiserum to the H3L protein, a late viral membrane protein. In contrast to the A30L and G7L proteins, the levels of the H3L protein were unaffected by IPTG. Together, these results indicated that expression of the A30L protein was required for the synthesis or stability of the G7L protein and that, conversely, expression of the G7L protein was required for the synthesis or stability of the A30L protein.
FIG. 10.
Stability of the A30L and G7L proteins. (A) BS-C-1 cells were either mock infected (lanes Un) or infected with vA30Li (left panel) or vG7Li (right panel) in the absence or presence of increasing concentrations of IPTG. After 24 h, cells were harvested and total cell lysates were analyzed by electrophoresis on an SDS-10 to 20% gradient polyacrylamide gel in Tricine buffer followed by Western blotting using antisera to the A30L, G7L, or H3L protein as indicated. (B) BS-C-1 cells were infected with VV WR, vA30Li, or vG7Li in the presence or absence of 50 μM IPTG. After 9 h, the cells were pulse-labeled with a mixture of [35S]methionine and [35S]cysteine for 15 min. Cells were either harvested immediately (lanes P) or incubated with excess unlabeled methionine for 0.5, 1, 2, 4, and 6 h (Chase lanes). Extracts of cells infected with vA30Li or vG7Li were immunoprecipitated with the G7L (IP-G7L Ab; left panel) or A30L (IP-A30C Ab; right panel) protein antiserum, respectively. The extracts from WR-infected cells were immunoprecipitated with G7L or A30L protein antiserum as indicated. The immunoprecipitated products were analyzed by SDS-PAGE and visualized by autoradiography.
Pulse-chase experiments were performed to determine whether the mutual regulation of the G7L and A30L proteins occurred at the level of synthesis or stability. Cells infected with vA30Li or vG7Li in the absence or presence of IPTG were pulse-labeled and either harvested immediately or incubated further in the presence of unlabeled amino acids prior to being harvested. Extracts from cells infected with the vA30Li virus were incubated with G7L protein antibody, extracts from vG7Li-infected cells were incubated with A30L protein antibody, and control cells infected with VV WR were incubated with both antibodies. The bound proteins were analyzed by SDS-PAGE and visualized by phosphorimaging. The G7L protein was detected after pulse-labeling of cells infected with WR or vA30Li in the presence or absence of IPTG. However, whereas the levels of the G7L protein remained nearly constant in cells infected with WR or with vA30Li in the presence of the inducer, a progressive decrease was observed in cells infected with vA30Li in the absence of IPTG (Fig. 10B). Half of the signal initially detected had disappeared after a 2-h chase. This result was not due to an increase in the level of cleavage of the G7L protein at the AGX site by the virus protease as there was no increase in the levels of the cleaved product of the G7L protein in the absence of IPTG (data not shown). A similar but even more dramatic effect was observed in cells infected with vG7Li in the absence of IPTG. The A30L protein was detected after pulse-labeling, but little remained after a 30-min chase (Fig. 10B). The levels of labeled A30L protein also decreased in cells infected with vG7Li in the presence of IPTG but with slower kinetics than in the cells infected in the absence of the inducer. We concluded that the interaction of the A30L and G7L proteins soon after synthesis is required for their stability.
DISCUSSION
The first distinct newly formed viral structure seen by electron microscopy of thin sections of VV-infected cells appears as dense granular material, destined to become the core, apposed to the concave surface of a crescent viral membrane. The mechanism by which these two different materials come together is unknown, but it seems likely that it is based on protein-protein interactions. Recently, we discovered that repression of the VV A30L protein resulted in the dislocation of the membranes and granular material. The A30L protein is believed to be located in the matrix between the core and membrane based on partial extraction with nonionic detergent and a reducing agent. Our presumption, therefore, was that the A30L protein is a key element in the linkage between the viral core and the membrane. To follow up this idea, we used an unbiased approach to search for proteins that interact with the A30L protein. We lysed cells with a nonionic detergent in order to solubilize the membrane as well as other components and used a rabbit monospecific antiserum directed to a short peptide at the C terminus of the A30L protein to isolate protein complexes. Complexes were also isolated from extracts of cells infected with a viable recombinant VV with HA-tagged A30L protein using a mouse MAb to the HA epitope. By using two different antibodies and also employing an inducible A30L recombinant VV, we could be certain that any candidate protein was specifically associated with the A30L protein. The major protein that interacted with the A30L protein had a mass of 42 kDa. When the masses of tryptic peptides of the 42-kDa protein were screened against the entire protein database, there were seven matches with the VV G7L protein, which has a predicted mass of 41.9 kDa. Confirmation that we had the correct protein was obtained by producing an antibody to a predicted peptide sequence of the G7L protein, which reacted with the 42-kDa protein associated with the A30L protein and also coimmunoprecipitated the A30L protein. Efforts to identify additional proteins associated with the A30L protein, as well as to use the G7L protein as bait to identify additional proteins, are in progress. A protein with an apparent mass of 16 to 17 kDa which coimmunoprecipitated with the A30L protein was identified as the product of the J1R protein ORF by mass spectrometry of tryptic peptides, and this was confirmed by Western blotting with J1R protein antiserum provided by Wen Chang (P. Szajner, unpublished data). Interestingly, repression of the J1R protein led to a phenotype that was similar to that occurring with repression of A30L or G7L expression (7).
The only previous reference to the G7L protein was found in a study of core protein N-terminal sequences (31). A sequence corresponding to a C-terminal fragment of the G7L protein was found, but neither the full-length nor the N-terminal fragment was identified, evidently because the amino acid at the N terminus was blocked. The site of cleavage corresponded to an AGL sequence at position 238 of the G7L protein, which is conserved among all members of the chordopoxvirus subfamily analyzed thus far. Using G7L protein antibody, however, we detected the full-length and cleaved forms in extracts of cells and VV cores, indicating that cleavage is incomplete during morphogenesis. It may be interesting to mutate the AGL sequence to determine whether cleavage is essential for morphogenesis. Inspection of the G7L protein ORF indicated that the initiation codon was overlapped by the late transcription initiator element TAAATG, strongly suggesting late expression, a prediction that was confirmed by a time course experiment.
To investigate the role of the G7L protein during VV replication, we constructed the recombinant virus vG7Li, from which the endogenous G7L ORF was deleted and replaced by an inducible copy under the stringent control of the E. coli lac repressor. Synthesis of the G7L protein, as well as virus replication, was dependent on IPTG. Metabolic labeling experiments indicated that the inhibition of G7L expression had no effect on the synthesis of viral proteins but that the proteolytic processing of certain structural proteins was severely inhibited. Previous studies have shown that inhibition of proteolytic processing is commonly associated with a block of VV morphogenesis at a stage prior to the formation of IMV. Electron microscopy of cells infected with vG7Li virus in the absence of IPTG confirmed a dramatic defect in VV morphogenesis. Cells infected with vG7Li in the absence of the inducer contained large masses of granular material separated from crescents and circular viral IV membranes, which lacked electron-dense viroplasm. Some of these viral membranes were present as concentric multilayered structures with an onion skin appearance. Within the IV, we visualized tubular elements that appeared on cross-section as rings comprised of a bilayer membrane. Risco et al. (24) described similar tubules associated with assembling IV. However, the absence of granular material in IV formed in cells infected with vG7Li under nonpermissive conditions provides particularly favorable conditions for study of the tubules.
The viral structures present in the cytoplasm of cells infected with vG7Li under nonpermissive conditions were virtually identical to those found in the cells infected with the vA30Li virus in the absence of IPTG. The explanation for the identical phenotypes of the vA30Li and vG7Li viruses under nonpermissive conditions became apparent when we discovered that the A30L and G7L proteins were very unstable unless they were coexpressed during infection. The association between these two proteins is independent of viral morphogenesis and occurred in the absence of other viral late proteins or when assembly was inhibited. In vitro transcription-translation studies further suggested that the association might be cotranslational, as the complex formed only when the two proteins were coexpressed.
In conclusion, the present and previous data indicate that protein-protein interactions are needed for the association of the dense viroplasm with IV membranes. Of the two interacting proteins identified thus far, the G7L protein appears to be a component of the core whereas the A30L protein may be a component of the matrix between the core and membrane. Efforts to identify additional interacting proteins are in progress.
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 66:2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 3.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L protein kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulter, E. A., and G. Appleyard. 1973. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog. Med. Virol. 16:86-108. [PubMed] [Google Scholar]
- 7.Chiu, W. L., and W. Chang. 2002. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J. Virol. 76:9575-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Fonseca, F. G., A. Weisberg, E. J. Wolffe, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and post-translational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Effects of deletion or stringent repression of the H3L envelope gene on vaccinia virus replication. J. Virol. 74:7518-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 13.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe, H., Veeranna, K. T. Shetty, and H. C. Pant. 1998. Characterization of the phosphorylation sites of human high molecular weight neurofilament protein by electrospray ionization tandem mass spectrometry and database searching. Biochemistry 37:3931-3940. [DOI] [PubMed] [Google Scholar]
- 17.Morgan, C., S. Ellison, H. Rose, and D. Moore. 1954. Structure and development of viruses observed in the electron microscope. II. Vaccinia and fowl pox viruses. J. Exp. Med. 100:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moritz, R., J. Eddes, J. Hong, G. Reid, and R. Simpson. 1995. Rapid separation of proteins and peptides using conventional silica-based supports: identification of 20 gel proteins following in-gel proteolysis, p. 311-319. In J. W. Crabb (ed.), Techniques in protein chemistry, vol. 6. Academic Press, San Diego, Calif. [Google Scholar]
- 19.Moss, B., and E. N. Rosenblum. 1973. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J. Mol. Biol. 81:267-269. [DOI] [PubMed] [Google Scholar]
- 20.Moss, B., E. N. Rosenblum, E. Katz, and P. M. Grimley. 1969. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature 224:1280-1284. [DOI] [PubMed] [Google Scholar]
- 21.Nagayama, A., B. G. T. Pogo, and S. Dales. 1970. Biogenesis of vaccinia: separation of early stages from maturation by means of rifampicin. Virology 40:1039-1051. [DOI] [PubMed] [Google Scholar]
- 22.Payne, L. G. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 23.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmström, F. Frischknecht, M. Zettl, T. Zimmerman, and M. Way. 2001. Kinesin dependent movement on microtubules precedes actin based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 24.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szajner, P., A. S. Weisberg, and B. Moss. 2001. Unique temperature-sensitive defect in vaccinia virus morphogenesis maps to a single nucleotide substitution in the A30L gene. J. Virol. 75:11222-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szajner, P., A. S. Weisberg, E. J. Wolffe, and B. Moss. 2001. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 75:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi, T., M. Oie, and Y. Ichihashi. 1994. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology 202:844-852. [DOI] [PubMed] [Google Scholar]
- 32.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 33.Traktman, P., A. Caligiuri, S. A. Jesty, and U. Sankar. 1995. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., and S. Shuman. 1995. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J. Virol. 69:6376-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, G. A., C. K. Stover, B. Moss, and T. R. Fuerst. 1995. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc. Natl. Acad. Sci. USA 92:6773-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., and B. Moss. 1992. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 187:643-653. [DOI] [PubMed] [Google Scholar]