Abstract
The AcNPV orf69 gene encodes a protein that contains an S-adenosylmethionine (AdoMet)-dependent methyltransferase signature motif. More significantly, ORF69 shows high conservation at residues diagnostic for (nucleoside 2′-O)-methyltransferase activity. To analyze the function of this protein, which was renamed MTase1, it was overexpressed in Escherichia coli and purified to homogeneity. Photo cross-linking experiments showed that MTase1 bound AdoMet, and functional assays demonstrated cap 0-dependent methyltransferase activity. In vivo expression assays in insect cells showed that MTase1 was synthesized during the late phase of infection and that its expression was dependent on viral DNA replication. Primer extension analysis identified a late promoter motif, ATAAG, at the transcription start site. A mutant virus was constructed by inserting the lacZ gene into the coding region of mtase1. Immunoblot analysis confirmed that MTase1 was not synthesized in these cells, and single-step growth curves revealed that the rate of virus replication in tissue culture was not affected by the absence of MTase1.
Baculoviruses are unique among eukaryotic DNA viruses in their use of both cellular and viral transcription machinery (22, 29). Early genes are transcribed by host RNA polymerase II, and the structure of early promoters therefore mimics that of promoters normally responsive to RNA polymerase II. Late and very late genes are transcribed by a virus-encoded RNA polymerase, which is composed of four protein subunits, i.e., late expression factor 4 (LEF-4), LEF-8, LEF-9, and P47. Transcription of late genes is dependent on a TAAG element that is found at the start of transcription and serves as both a promoter and a transcription start site (14, 21, 42, 44).
Nineteen genes from Autographa californica nuclear polyhedrovirus (AcNPV) are required for expression from late and very late baculovirus promoters in a transient expression system. These genes are referred to as LEFs (48). The LEFs include immediate early transcription factors, DNA replication proteins, the four RNA polymerase subunits, and additional transcription factors (18, 20, 22, 39). Levels of transient gene expression can be further stimulated by addition of other viral genes, such as the AcNPV orf69 gene. ORF69 function is not essential for transient expression but increases levels of viral reporter constructs threefold (38).
Late and very late AcNPV transcripts have a typical methyl-7-guanosine cap at their 5′ ends (47). The cellular capping machinery recognizes its mRNA substrates through interactions with the phosphorylated C-terminal domain of the RNA polymerase II (11, 26, 41). Therefore, even though baculoviruses replicate in the nucleus of infected cells, they had to evolve a novel mechanism for capping late RNAs because the viral polymerase lacks a C-terminal-domain-like motif. The LEF-4 subunit of viral RNA polymerase has guanyltransferase (GTase) and RNA triphosphatase (RTPase) activities (17, 19, 31). The two enzyme activities map to independent domains, which are arranged on the same peptide in a manner similar to that of vaccinia virus and metazoan capping machinery. Thus, baculoviruses target capping enzymes to viral RNAs by incorporating two of the enzymatic activities required for cap formation into the viral RNA polymerase itself.
Whether baculoviruses encode their own version of the third enzyme required for cap formation is as yet unknown. (Guanine-7)-methyltransferase (N7MTase) methylates the N7 on the GMP cap. The presence of an m7GpppN cap increases mRNA stability by protecting it from 5′-to-3′ exonuclease degradation (4, 30). In addition, caps increase the accuracy and efficiency of mRNA splicing, facilitate mRNA transport, and enhance mRNA translation (23, 37, 55, 59). N7MTases have been isolated from a variety of sources, including viruses, fungi, and metazoa (57). All N7MTases belong to the AdoMet-dependent MTase superfamily. One feature that distinguishes viral N7MTases from cellular N7MTases is the presence of RNA polymerase or other capping activities along with the MTase domain on the same polypeptide (49).
Most of the baculovirus late mRNAs have a cap 1 structure (47), and this requires one additional methylation event on the ribose of the first transcribed nucleotide. The enzyme that catalyzes this methylation event is a (nucleoside-2′-O)-MTase (2′OMTase) that is cap dependent, called RNA cap 2′OMTase or cap 1 MTase. The function of ribose methylation is not clear, but in at least some cases, it may stimulate translation by increasing ribosome binding (34, 43).
The protein encoded by AcNPV orf69 is closely related to Escherichia coli FtsJ (38). FtsJ is an rRNA uridine 2′OMTase (6), suggesting that the baculovirus protein might also function in methylation of RNA, possibly as a cap 1 MTase. To address this possibility, the viral protein (renamed MTase1) was expressed in bacteria for biochemical analysis. Immunoblot analyses were also undertaken to determine whether the expression profile of mtase1 was consistent with a role in cap 1 methylation of late viral transcripts.
MATERIALS AND METHODS
Construction of ORF69-His tag clone.
The genomic clone of mtase1 was amplified directly from AcNPV E2 strain DNA by using Pfu DNA polymerase. The 5′ primer (CTGCAGCATATGTTGCAGCAAAAATTAAAT) inserted an NdeI site (underlined) at the ATG codon of MTase1. The 3′ primer (GAATTCGGATCCCTGATTTGAAGCCGCTGG) generated a BamHI site (underlined) downstream of the stop codon. The 0.8-kb PCR product was cloned into the pET15b vector (Novagen) and screened by restriction enzyme digestion. The resulting plasmid was named pET15b-mtase1 and directs the synthesis of MTase1 fused with six consecutive histidine residues at the N terminus. The expression of mtase1 in this vector is regulated by the T7 RNA polymerase promoter.
Purification of MTase1 from E. coli.
MTase1 was expressed in E. coli BL21-slyD1 cells. This is a mutant strain of E. coli that lacks SlyD, a naturally histidine-rich protein that binds to metal affinity columns (51). Two liters of cells containing pET15b-mtase1 was grown at 37°C until the A600 reached approximately 0.6. The expression of MTase1 was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside. After induction, cells were incubated at 30°C overnight. All subsequent procedures were performed at 4°C with all buffers and rotors prechilled to 4°C. Cells were harvested by centrifugation at 4,000 × g for 10 min, resuspended in buffer I (50 mM Tris [pH 7.9], 500 mM NaCl, 0.1% Triton X-100), and lysed by sonication. The lysate was clarified by centrifugation at 100,000 × g for 25 min. The supernatant was loaded onto a column packed with 2 ml of Talon resin (Clontech) previously equilibrated with 40 ml of buffer I. The column was washed with 40 ml of buffer I followed by 40 ml of buffer I plus 10 mM imidazole (pH 7.9). Bound MTase1 was eluted with 10 ml of buffer I plus 250 mM imidazole (pH 7.9). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by staining with Coomassie blue showed a major protein in the 250 mM imidazole fraction that migrated at the expected position for recombinant MTase1. MTase1 was further purified by cation-exchange chromatography. Ten milliliters of the 250 mM imidazole fraction was diluted to 20 ml to give a final buffer concentration of 50 mM Tris (pH 7.9), 250 mM NaCl, 125 mM imidazole, 0.05% Triton X-100, 2% taurine, and 5% glycerol and was loaded onto a Mono S column previously equilibrated with loading buffer (50 mM Tris [pH 7.9], 250 mM KCl, 1 mM dithiothreitol [DTT], 5% glycerol, and 2% taurine). The column was washed with loading buffer and eluted with a 20-ml linear gradient from 250 to 500 mM KCl. MTase1 was eluted in a single peak, which was demonstrated by SDS-PAGE. The protein concentration was determined by Coomassie blue G250 binding assay as recommended by the manufacturer (Pierce).
Photochemical labeling of ORF69 with [methyl-3H]AdoMet.
Photo cross-linking was performed as previously described (1). Reaction mixtures (20 μl) contained 25 mM 3-(N-morpholino)propanesulfonate [pH 7.0], 100 mM NaCl, 2 mM DTT, 30 mM EDTA, and 1.6 μM [methyl-3H]AdoMet. After incubation at 37°C for 60 min, reactions were treated with UV irradiation of 360,000 μJ/cm2 and were then resolved on 12% polyacrylamide gels. Gels were fixed and soaked in 1 M sodium salicylic acid (pH 6.0) for 30 min before drying and exposure to X-ray film at −80°C.
MTase assays.
A plasmid encoding a His-tagged version of the vaccinia virus D1/D12 capping enzyme was obtained from S. Shuman (56). D1/D12 capping enzyme was expressed in bacteria under the control of a T7 RNA polymerase promoter and purified on the Talon (Clontech) affinity matrix. Template RNA was synthesized by in vitro transcription of pBS-SK (Stratagene) previously digested with XbaI. The linearized template was transcribed with T7 RNA polymerase by using standard conditions as recommended by the manufacturer (Promega). The resulting 93-nucleotide RNAs were capped by using the vaccinia capping enzyme D1/D12 (56). Capping reactions contained 50 mM Tris (pH 7.9), 5 mM DTT, 1.25 mM MgCl2, 100 μM GTP, 12 μM RNA ends, and 0.6 μM D1/D12. To monitor the extent of capping, a 10-μl aliquot was removed from each 100-μl reaction and 10 μCi of [α-32P]GTP was added. The percentage of RNAs with caps was calculated by measuring the incorporation of radiolabel into trichloroacetic acid-insoluble material. Under these conditions, we routinely obtain 25 to 40% efficiency of capping. The capping reaction cannot go to completion in the absence of a methyl donor because the GTase reaction is freely reversible (56). RNA was subsequently purified by two rounds of precipitation with 2.5 M LiCl to remove enzyme and free label. A portion of the capped RNA was subsequently methylated with D1/D12 in the presence of 50 μM AdoMet. The percentage of methylated caps was determined by digestion of RNAs with nuclease P1 and separation of caps by thin-layer chromatography on polyethyleneimine (PEI) cellulose developed in 0.45 M ammonium sulfate. Plates were exposed to a phosphorimager screen and quantitated with ImageQuant software. Under these conditions the capped RNA is quantitatively converted to methylated, capped RNA.
Cap 1 MTase activity was measured with a filter binding assay as previously described (54). Assays contained 25 mM HEPES (pH 7.2), 1 mM DTT, 1 μM [methyl-3H]AdoMet (10 Ci/mmol), 5 μg of RNA, and 5 μg of purified MTase1 (Mono S peak fraction) in a final volume of 100 μl. At the indicated times, 10-μl aliquots were removed and spotted onto DE81 filters, washed with 20 mM sodium phosphate (pH 7), and quantitated by liquid scintillation counting.
To confirm that the methyl group was transferred to the ribose of the first transcribed nucleotide and not to the guanine cap, cap species were separated by thin- layer chromatography. In this case the RNA substrate was radiolabeled with [α-32P]GTP as described above. Then the labeled RNA was incubated with 25 mM HEPES (pH 7.2), 1 mM DTT, and 50 μM AdoMet in the presence or absence of D1/D12 and purified MTase1. After 2 h at 37°C, the RNA was precipitated and digested with 5 μg of nuclease P1 prior to analysis by thin-layer chromatography on PEI cellulose plates developed in 0.45 M ammonium sulfate (56).
Preparation of MTase1 antiserum and Western blotting.
Purified MTase1 from Mono S was used to generate polyclonal antiserum in rabbits by using a standard immunization protocol (23a). Western blotting was performed with SuperSignal West Pico Chemiluminescent Substrate as recommended by the manufacturer (Pierce). Separated proteins were electroblotted onto nitrocellulose membranes, and then the membranes were blocked with 10% skim milk in Tris-buffered saline-Tween (TBST) (140 mM NaCl, 25 mM Tris [pH 7.4], 0.2% Tween 20) at 4°C overnight. MTase1 polyclonal antibody was diluted 1:2,000 in TBST and incubated with the membrane for 1 h with shaking. Membranes were washed four times with TBST-1% milk for 5 min each time and were then incubated with secondary antibody diluted 1:10,000 with TBST for 1 h with shaking. The membranes were then washed with TBST-1% milk buffer (four to six times, 5 min each). Equal volumes of stable peroxide solution and luminol/enhancer solution were mixed and incubated with blot for 5 min. Membranes were exposed to film for 30 or 60 s and developed immediately.
Primer extension mapping of mtase1.
Total RNA was isolated from AcNPV-infected cells with guanidine isothiocyanate, followed by centrifugation through saturated cesium chloride (52). The reverse transcription reaction was also performed as described in the same manual with minor modifications. A primer specific for mtase1 (GAGCGAGCGCAAACCACCGAC) was radiolabeled at the 5′ end with 32P and hybridized with 25 μg of RNA. Annealed primer was extended with avian myeloblastosis virus reverse transcriptase (Promega). The reaction mixture (30 μl) contained 50 mM Tris [pH 8.3], 50 mM KCl, 5 mM MgCl2, 5 mM DTT, 0.5 mM spermidine, 0.1 mg of bovine serum albumin/ml, 1 mM deoxynucleoside triphosphates, 50 μg of actinomycin D/ml, and 0.2 U of RNasin/μl. After incubation at 42°C for 1 h, the reaction was terminated by the addition of 170 μl of Tris-EDTA and contaminating RNA was removed by the addition of 2 μg of RNase A. The extension products were extracted with phenol and chloroform, precipitated by ethanol, and analyzed on 6% polyacrylamide-8 M urea sequencing gel. Sequencing ladders were generated by using the same labeled primer and a genomic clone of mtase1.
Construction of mutant virus with an interrupted mtase1 gene and single-step growth analysis.
The AcNPV genomic clone pHindIII-B was digested with EcoRI, EcoRV, and PvuII. A 5.1-kb fragment flanked by EcoRI and EcoRV sites contains the complete mtase1 open reading frame, promoter region, and flanking sequences. The fragment was purified by agarose gel electrophoresis and cloned into pUC18, which had previously been digested with EcoRI and SmaI before ligation. The resulting plasmid was named pMTase. It was then digested with ApaI and incubated with T4 DNA polymerase to cut 3′ overhangs to generate blunt ends. Plasmid pDP502 was digested with BamHI and incubated with Klenow enzyme and deoxynucleoside triphosphates to fill in 5′ overhangs to generate blunt ends. A 3-kb fragment containing a complete copy of the E. coli lacZ gene was purified by agarose gel electrophoresis and ligated with pMTase. The resulting plasmid, named pMTase-LacZ, was cotransfected with wild-type viral DNA into the Sf9 cells using Lipofectin (Invitrogen). Recombinant viruses were identified by screening for blue plaques, plaque purified, and amplified by standard protocols (M. D. Summers and G. Smith, Tex. Agric. Exp. Stn. Bull. 1555, College Station, Tex., 1987). The presence of the lacZ gene within mtase1 was verified by PCR analysis using primers that hybridize to the amino and carboxyl termini of mtase1.
For single-step growth analysis, 2 × 106 cells were grown in 4 ml of TNMFH medium. Wild-type and mutant virus was added at a multiplicity of infection (MOI) of 5 separately. A 200-μl aliquot was taken out at different times postinfection and was used for plaque assay. For each virus, three independent samples were prepared for each time point and the final result is the average of three independent titrations.
RESULTS
AcNPV orf69 encodes a putative MTase.
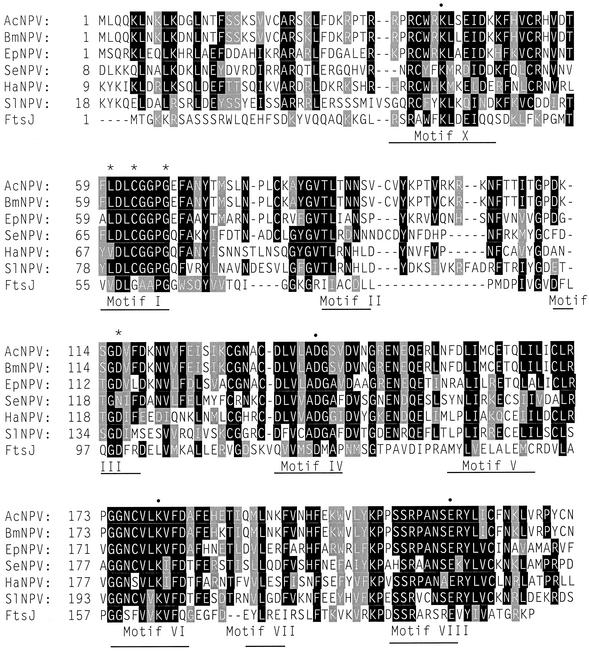
Li et al. (38) previously reported that the AcNPV orf69 gene encodes a protein with homology to FtsJ, which has since been shown to function as a 23S rRNA 2′OMTase (5, 6). Homologs of orf69 have been identified in five other baculoviruses. At least six other baculovirus genomes, which have been completely sequenced, lack orf69 homologs (2, 9, 24, 25, 45). Figure 1 shows the six baculovirus proteins aligned with FtsJ. A number of strongly conserved regions are scattered along the protein sequence. Most of these correspond to nine conserved AdoMet-dependent MTase motifs (40). Structural studies have shown that motif I forms a G loop that binds the methionine moiety of AdoMet (5, 10, 16, 35). Most of the residues within this loop contact AdoMet, and there is strong conservation of sequence between FtsJ and the baculovirus proteins in this region. Three residues within motif I are highly conserved, though not identical, in all members of the AdoMet-dependent MTases. Only one of the three conserved positions lacks identity in the baculovirus proteins. Motif III is also involved in AdoMet binding and contains a single highly conserved aspartic acid residue, which is present in five of the six baculovirus proteins (15, 53).
FIG. 1.
Sequence alignment of AcNPV ORF69 with its homologs. The amino acid sequences of ORF69 from AcNPV were aligned with homologous proteins from five other baculoviruses and FtsJ from E. coli with the MULTALIN multiple alignment program (11a). The baculovirus sequences are A. californica nucleopolyhedrovirus (AcNPV), Bombyx mori nucleopolyhedrovirus (BmNPV), Epiphyas postvittana nucleopolyhedrovirus (EpNPV), Spodoptera exigua nucleopolyhedrovirus (SeNPV), Heliocoverpa armigera nucleopolyhedrovirus (HaNPV), and Spodoptera litura nucleopolyhedrovirus (SlNPV). Gaps in the sequence alignment are denoted by hyphens; residues that are identical in at least four of the sequences are boxed in boldface; sequences that are conserved in at least four sequences are shaded. The indicated motifs represent conserved regions of DNA MTases (40). Residues that are important for binding of AdoMet are represented by asterisks, while those indicated with a bullet (•) are conserved in all 2′OMTases (12). Only the first 233 residues of the alignment are shown.
FtsJ-like proteins are found in a wide variety of species ranging from eubacteria to archaea to eukarya. The crystal structures of FtsJ and three viral proteins that function as RNA cap 2′OMTases have been solved (5, 12, 28, 50). Comparison of the structures and the placement of residues within the active site have allowed the identification of four residues that are diagnostic for the AdoMet-dependent transferases that methylate 2′-O-ribose. The four residues, which are lysine in motif X, aspartate in motif IV, lysine in motif VI, and glutamate in motif VIII, are also conserved in all of the baculovirus proteins. These four residues are not conserved in mRNA cap MTases, tRNA MTases, or DNA MTases (12). Thus, these observations strongly suggest that ORF69 is a 2′OMTase. Based on the sequence homology, we changed the name of orf69 to mtase1, and so the encoded protein should be called MTase1.
MTase activity of AcNPV MTase1.
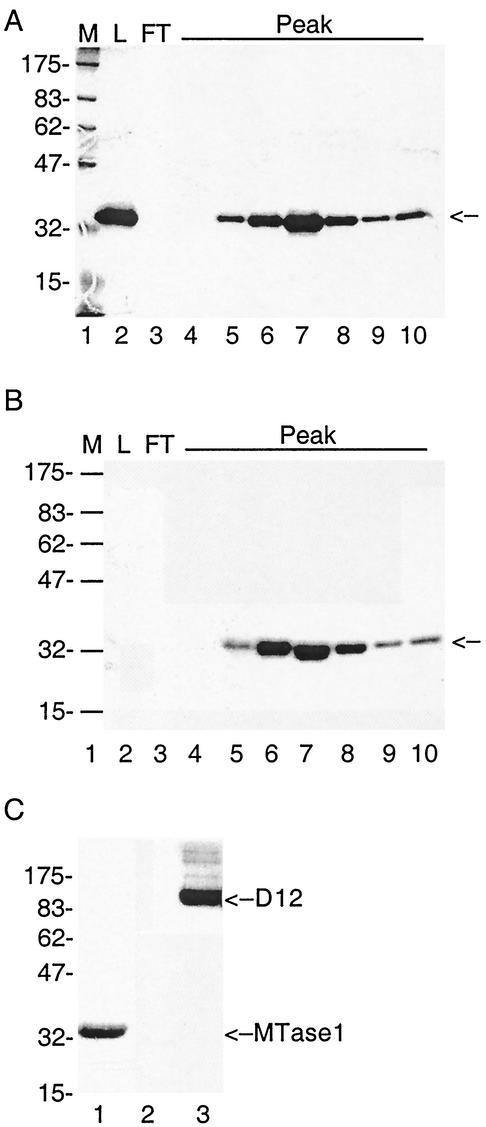
To test biochemical activity of MTase1, we cloned the mtase1 gene into pET15b (Novagen). This clone was designed to enable the expression of N-terminal His6-tagged recombinant MTase1 in E. coli under the control of the T7 RNA polymerase promoter. The protein was expressed in bacteria, and His-tagged MTase1 was purified by immobilized metal affinity chromatography. A protein of the expected size for His-tagged MTase1 (32.443 kDa) bound to Talon and was eluted with imidazole in relatively pure form (Fig. 2A, lane 2). MTase1 was the major protein band that eluted from Talon, but a number of less abundant contaminants were evident. Therefore, the protein was further purified by cation-exchange chromatography. The Talon elution fraction was loaded onto a Mono S column at 250 mM KCl. MTase1 bound to MonoS and eluted in a single sharp peak. Fractions across the peak were analyzed on SDS-12% acrylamide gels, and MTase1 was judged to be homogeneous in the peak fractions (Fig. 2A, lanes 4 to 10).
FIG. 2.
Purification and AdoMet binding activity of MTase1. (A) MTase1 was expressed in E. coli strain BL21-slyD1 and was purified on a Talon metal affinity column. Protein that eluted from Talon with 250 mM imidazole was loaded onto Mono S, and bound proteins were eluted with a salt gradient. Fractions surrounding the major peak of UV absorbance were resolved by SDS-PAGE on 12% acrylamide gels and stained with Coomassie blue. Lane 1, protein molecular markers with sizes in kilodaltons indicated on the left (M); lane 2, MTase1 purified on Talon (L); lane 3, flowthrough from Mono S column (FT); lanes 3 to 10 are aliquots of adjacent fractions from Mono S. The position of MTase1 is indicated by an arrow on the right. (B) UV cross-linking of MTase1 with [methyl-3H]AdoMet. Aliquots of thesame fractions shown in panel A were incubated with 25 mM morpholinepropanesulfonic acid (pH 7.0), 100 mM KCl, 2 mM DTT, 30 mM EDTA, and 1.6 μM [methyl-3H]AdoMet. After incubation for 1 h at 37°C, samples were resolved by SDS—12% PAGE. The gel was fixed, dried, and exposed to film for 4 days at −80°C. An autoradiograph of the gel is shown. Protein molecular weight markers were loaded in lane 1, and the sizes are shown on the left. The position of MTase1 is indicated by an arrow on the right. (C) Specificity of AdoMet binding activity. MTase1 (lane 1), bovine serum albumin (lane 2), and vaccinia virus capping enzyme D1/D12 (lane 3) were incubated with [methyl-3H]AdoMet as described above. The positions of protein molecular markers with sizes in kilodaltons are indicated on the left. The positions of MTase1 and D12 are labeled on the right.
To determine whether MTase1 functioned as an MTase, we first wanted to show that it bound AdoMet. Many MTases can be photochemically labeled in the presence of [methyl-3H]AdoMet and short-wavelength UV (1, 13, 33, 58). MTase1 was incubated with [methyl-3H]AdoMet, and then protein was cross-linked to bound AdoMet by UV irradiation. The Mono S fractions containing MTase1 bound AdoMet, as demonstrated by SDS-PAGE (Fig. 2B, lanes 4 to 10). The density of labeling in each fraction across the Mono S peak corresponded to the amount of MTase1 in each fraction. Protein from the Talon elution fraction (Fig. 2B, lane 2, Mono S load) showed no AdoMet binding activity, although the protein gel indicated that it contained almost the same amount of protein as the peak fraction. A possible explanation is that the high concentration of imidazole in the sample (125 mM imidazole) inhibited AdoMet binding activity, since both molecules are positively charged.
To rule out the possibility of nonspecific binding of protein to AdoMet, positive and negative controls were run in parallel with the peak fraction of MTase1 (Fig. 2C). No signal was detected when bovine serum albumin was used in the assay. An equimolar amount of vaccinia virus D1/D12 bound more AdoMet than MTase1. This could be due to a weaker interaction of MTase1 with AdoMet or could reflect a higher relative occupancy of MTase1 with unlabeled AdoMet from the bacterial cells.
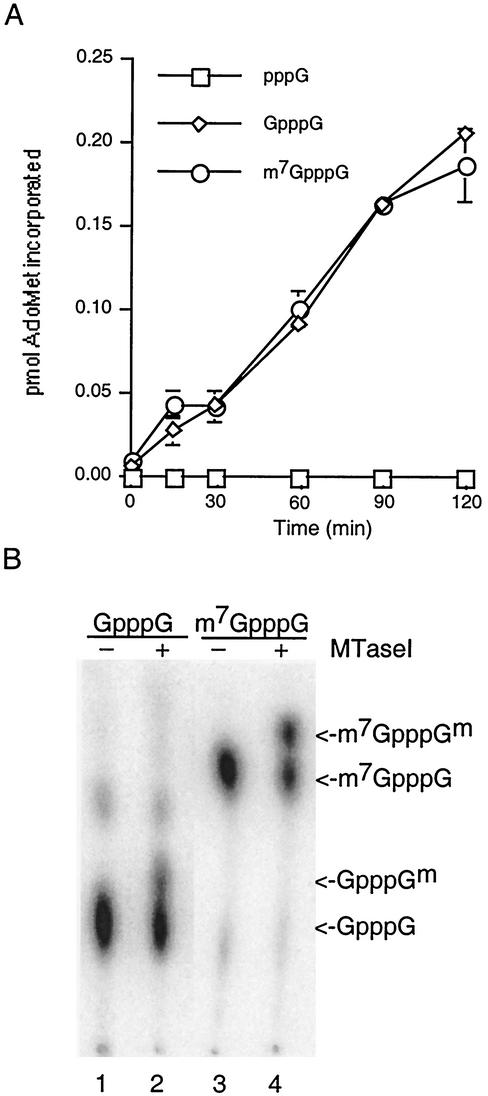
The MTase activity of MTase1 was tested on three different RNA substrates with a filter binding assay (Fig. 3A). The RNAs were derived by in vitro transcription of the vector pBS-SK, and the sequences of the three RNAs were identical except for the structure of their 5′ termini. Triphosphate-terminated RNAs, which are the natural product of in vitro transcription reactions, were incubated with vaccinia virus D1/D12 capping enzyme and GTP to produce the capped RNA substrate GpppGN. Quantitation revealed that less than half the RNAs were capped under these conditions (data not shown); this was expected because the guanylation reaction is freely reversible. A portion of the capped RNAs were subsequently incubated with vaccinia virus D1/D12 capping enzyme in the presence of AdoMet to produce the methylated cap substrate m7GpppGN. This was done under conditions in which the RNAs were quantitatively methylated (see, for example, Fig. 3B, lane 3). An equimolar amount of each RNA was separately incubated with MTase1. The amount of methyl-3H transferred to each RNA as a function of time was determined by spotting aliquots on DEAE filter disks. MTase1 catalyzed transfer of a methyl group from [methyl-3H]AdoMet to a unmethylated capped RNA (GpppG) or methylated capped RNA (m7GpppG) but not to uncapped RNA (pppG). This indicates that the MTase activity is cap specific. The fact that methyl transfer was the same whether or not the N7 position was methylated was consistent with the prediction that MTase1 has ribose 2′-O-MTase activity and not N7MTase activity.
FIG. 3.
MTase activity. (A) Filter binding assays. The extent of methyl transfer from [methyl-3H]AdoMet to RNA substrates is plotted against time. Data points represent the averages of three different experiments with standard errors. (B) Analysis of methylated cap structures. Reactions contained 50 mM HEPES (pH 7.2), 5 mM DTT, 50 μM AdoMet, and 0.25 μM RNA with the cap structure indicated at the top. Omission (lanes 1 and 3) or inclusion (lanes 2 and 4) of 3 μg of MTase1 is indicated above the lanes. Caps were liberated with nuclease P1 and separated by thin-layer chromatography on PEI cellulose in 0.45 M ammonium sulfate (56). The positions of methylated and unmethylated caps are indicated on the right.
To further characterize the methylation reaction, radiolabeled RNA substrates were formed with vaccinia virus D1/D12 capping enzyme in the presence of [α-32P]GTP. Then the GpppG or m7GpppG capped RNAs were incubated in the presence or absence of MTase1 and AdoMet. RNA caps were subsequently liberated with nuclease P1 and separated by thin-layer chromatography on PEI cellulose (Fig. 3B). Development in 0.45 M ammonium sulfate clearly separated the guanine-N7-methylated caps and nucleoside-2′-O-methylated caps. Methylation at N7 introduces a positive charge that partially neutralizes the negative phosphates so that the mobility of m7GpppG is significantly higher than GpppG (compare lanes 1 and 3). Methylation of the ribose, however, alters mass but not charge and so has only a minor effect on mobility (compare lanes 1 and 2 or lanes 3 and 4). Nucleoside methylation was independent of prior methylation of the cap at guanine-N7, consistent with data from the filter binding assays. Together, the results of these experiments demonstrate that baculovirus MTase1 is an RNA cap 2′OMTase.
In vivo expression of MTase1.
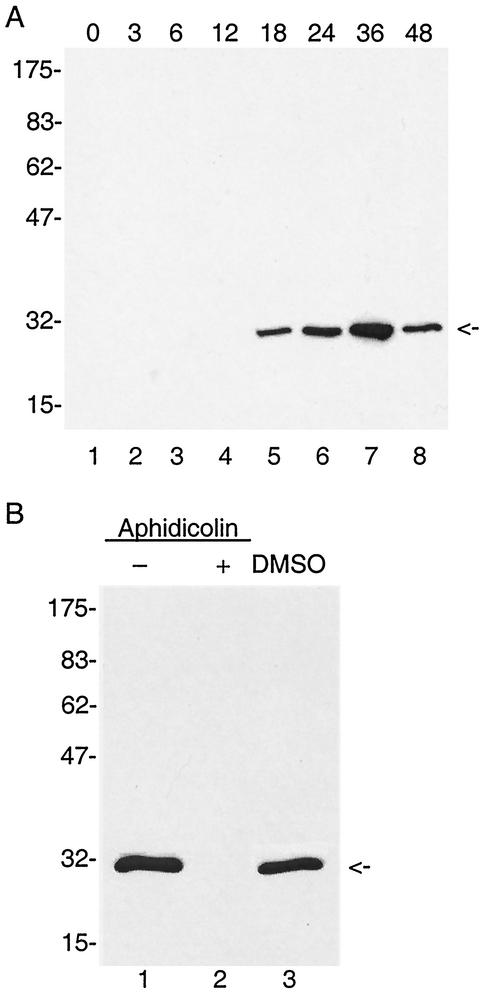
We expected that the baculovirus cap 1 MTase would function in conjunction with the virus-encoded RNA polymerase to methylate late viral RNAs. Therefore, we predicted that mtase1 would be expressed during the early phase of viral infection, as are the other late transcription factors. To analyze the expression pattern of MTase1, polyclonal antibodies were first prepared by immunization of rabbits with MTase1 purified from bacteria. Then total protein from AcNPV-infected Sf9 cells was extracted at different times postinfection. Immunoblot analysis revealed a specific polypeptide migrating with an apparent molecular mass of 30 kDa (Fig. 4A). This is in good agreement with the predicted size of 30.4 kDa for the orf69 gene product (3). The protein was first detected at 18 h postinfection, and maximal expression was reached at 36 h postinfection. At 48 h, MTase1 levels decreased. This pattern of expression suggested that mtase1 is a late gene. To further analyze the timing of expression, infected cells were incubated in the presence of 5 μg of aphidicolin/ml from 1 to 24 h postinfection. Aphidicolin is a DNA polymerase inhibitor and has been shown to inhibit AcNPV viral DNA replication, thus blocking late gene expression (60). No MTase1 signal was detected in cells treated with aphidicolin, although protein was detected in control cells incubated in the absence of drug. Aphidicolin was dissolved in dimethyl sulfoxide (DMSO), and therefore an additional control was performed consisting of infected cells grown in the presence of DMSO. The MTase1 signal in these cells was equivalent to infected cells with no solvent, indicating that DMSO did not interfere with expression of MTase1. This confirmed that the expression of mtase1 was dependent on viral DNA replication.
FIG. 4.
Immunoblot analysis MTase1 expression and its dependence on viral DNA replication. (A) Total protein was extracted from AcNPV-infected Sf9 cells at the indicated times postinfection. Samples from each time point were electrophoresed on a 12% polyacylamide gel, transferred to a nitrocellulose membrane, and probed with MTase1 antiserum. (B) Expression of MTase1 in the presence and absence of aphidicolin. At 1 h postinfection, 5 μg of aphidicolin/ml dissolved in DMSO (lane 2, +) or an equal amount of DMSO only (lane 3, DMSO) was added to infected cells. Lane 1 shows infected cells with no treatment. At 24 h postinfection, cells were harvested, total protein was extracted, and an equal amount of each culture was electrophoresed on a 12% polyacrylamide gel. After transfer to nitrocellulose, blots were probed with MTase1 antiserum. Protein molecular markers are indicated on the left with sizes in kilodaltons.
Transcriptional mapping of the 5′ end of the transcript.
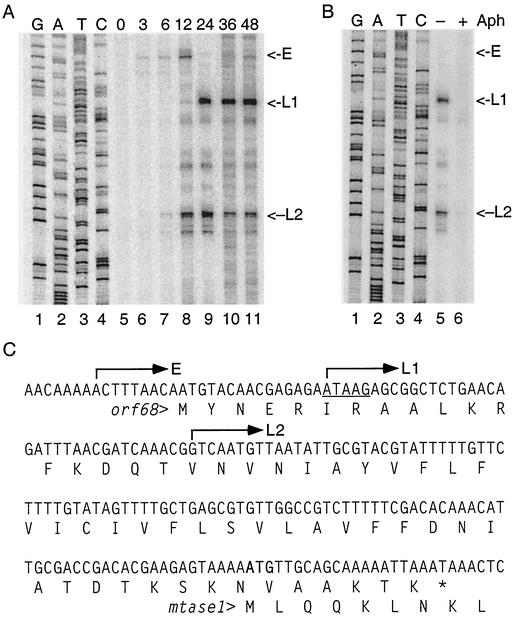
Primer extension mapping was performed to confirm the temporal pattern observed in the protein expression studies and also to map the 5′ ends of mtase1. Total RNA was extracted at different times postinfection from AcNPV-infected Sf9 cells and hybridized with an oligonucleotide complementary to the 5′ end of mtase1. After extension with reverse transcriptase, cDNAs were separated on denaturing polyacrylamide gels along with DNA sequencing reactions performed with the same primer (Fig. 5A). At 3 h postinfection, early transcripts extending through mtase1 were detected at a low level. These RNAs initiate upstream of the orf68 open reading frame and so would be expected to direct the synthesis of ORF68, which is a 22.3-kDa hypothetical protein with no function assigned. ORF68 overlaps the N-terminal 7 amino acids of MTase1 (Fig. 5C). It is unlikely that the early transcripts are used for synthesis of MTase1, because eukaryotic ribosomes are generally monocistronic. This early transcript was also evident at 6 and 12 h postinfection but was not detected at later times.
FIG. 5.
Primer extension mapping of mtase1 mRNA. (A) Total RNA was purified from AcNPV-infected Sf9 cells at the indicated times postinfection. The 5′ ends of mtase1 transcripts were mapped by primer extension analysis by using an oligonucleotide complementary to nucleotides 59331 to 59352 of the AcNPV genome sequence. DNA sequence ladders were generated with the same primer. The early (E) and late (L1 and L2) cDNA products are labeled on the right. (B) Effect of aphidicolin (Aph) on mtase1 transcription. Infected cells were grown in the presence or absence of 5 μg of aphidicolin/ml from 1 to 24 h postinfection. Transcripts are labeled on the right. (C) Position of 5′ ends on the AcNPV genome. The DNA sequence corresponding to the AcNPV genome from nt 59091 to 59270, which encodes orf68 and the mtase1 upstream region, is shown with the relevant translations below. Arrows above sequence denote the transcriptional start sites of each signal. The sequence is antisense to the sequence ladders shown in panels A and B. The late transcription motif ATAAG is underlined. The ATG start codon for MTase1 is in boldface.
At 12 h postinfection, two additional transcripts were observed that initiated within orf68 and so would be expected to direct synthesis of MTase1. The distal transcript (L1) initiated within a conserved ATAAG late promoter motif (Fig. 5C). The proximal transcript (L2), however, did not map to a late promoter. To confirm that these were late transcripts, additional primer extension reactions were performed with RNAs purified from infected cells treated with or without aphidicolin from 1 to 24 h postinfection (Fig. 5B). Both of the late transcripts were sensitive to the addition of aphidicolin, indicating that transcription was dependent on DNA replication. The early transcript was not detected in the presence of aphidicolin because the cells were harvested at 24 h postinfection, and the transcript was not observed at 24 h in the time course. Thus, the primer extension data are consistent with protein expression experiments and indicate that expression of mtase1 is dependent on viral DNA replication, so it should be classified as a late gene.
Mutant virus construction and single-step growth analysis.
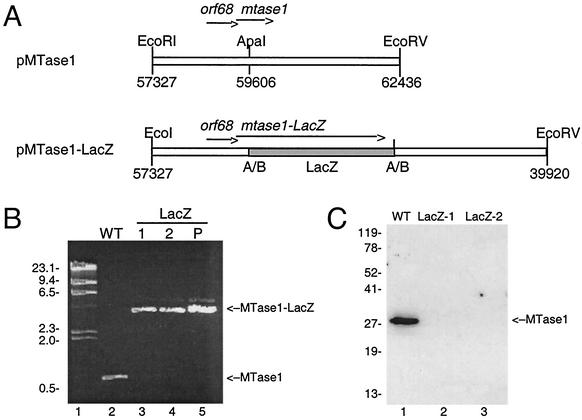
To gain a better understanding of the in vivo function of MTase1, we decided to determine whether it is essential for replication of AcNPV in tissue culture. To do this, a viral mutant containing an interrupted mtase1 gene was constructed (Fig. 6A). A genomic copy of mtase1 with flanking regions was cloned into pUC18. A 3-kb DNA fragment containing the complete lacZ gene was then inserted into the middle of mtase1. The resulting plasmid, pMTase1-LacZ, is expected to direct the synthesis of fusion between the N-terminal 109 amino acids of MTase1 and LacZ. This interrupts MTase1 between motifs II and III, which makes it extremely unlikely that the fusion protein could maintain MTase activity. Wild-type viral DNA was cotransfected with pMTase1-LacZ in insect cells. Two independent blue plaques were purified, and their DNA was amplified by PCR with primers that hybridize to the amino and carboxyl termini of mtase1 (Fig. 6B). A PCR product of approximately 0.8 kb was amplified from wild-type viral DNA, while amplification of the two mutant viral DNAs yielded a band of approximately 3.8 kb (Fig. 6B, lanes 3 and 4). These are the expected sizes for mtase1 alone (786 bp) and mtase1 fused to the 3-kb LacZ insert, respectively. This confirmed that the blue plaques were generated by a homologous double-crossover event replacing mtase1 with the fusion construct. To confirm that MTase1 was not expressed in cells infected with mutant virus, immunoblot analysis was performed. No MTase1 was detected in cells infected with either of the mutant viruses, although cells infected in parallel with wild-type virus showed strong expression of MTase1 (Fig. 6C). The fact that we were able to obtain viable mutants indicates that mtase1 is not essential for viral replication.
FIG. 6.
Construction of β-galactosidase-interrupted mtase1 mutant virus. (A) Schematic diagram of pMTase1-LacZ. A 5.1-kb fragment containing mtase1 and flanking regions was cloned into pUC18 to construct pMTase1. The relative positions and orientations of orf68 and mtase1 are indicated by arrows. Then a 3-kb DNA fragment containing the lacZ gene (shaded box) was inserted into the unique ApaI site in mtase1. The numbers below the restriction sites refer to the AcNPV genomic map positions. (B) PCR analysis. Primers specific for mtase1 were used to amplify wild-type (WT) viral DNA (lane 2), two independent isolates of mutant virus (lanes 3 and 4), or pMTase1-LacZ (lane 5). Samples were analyzed on a 1% agarose gel and stained with ethidium bromide. Lane 1, lambda-HindIII DNA with sizes in kilobases indicated on the left. The positions of relevant products are labeled on the right. (C) Immunoblot analysis. Sf9 cells were infected with wild-type (WT) or mutant virus at an MOI of 5 and were harvested at 24 h postinfection. The positions of molecular weight markers are indicated on the left in kilodaltons, and the position of MTase1 is indicated on the right.
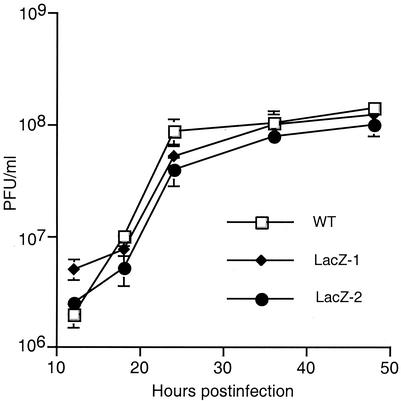
To further examine the effect of mtase1 deletion on virus production, single-step growth curves were performed. Sf9 cells were infected with either wild-type or mutant virus at an MOI of 5. At different times postinfection, viral yields were determined by standard plaque assay (Fig. 7). Both mutant viruses exhibited growth curves similar to those of wild-type virus, indicating that mtase1 has little effect on the rate of budded virus production in tissue culture.
FIG. 7.
Single-step growth curve for mutant and wild-type virus. S. frugiperda cells were infected with wild-type or one of two mutant viruses (LacZ-1 or LacZ-2) at an MOI of 5. At the indicated times postinfection, an aliquot was removed and virus titers were determined by plaque assay. Values represent means of triplicate determinations with error bars.
DISCUSSION
AdoMet-dependent MTases are a large group of proteins that use AdoMet as cofactor to catalyze methylation. Despite the low level of sequence conservation among different members of the superfamily, all of the enzymes adopt a similar α/β core structure to bind the cofactor AdoMet (8). Two conserved sequence motifs (motif I and motif III) that comprise the AdoMet binding site can be identified in most MTases and are diagnostic of AdoMet-dependent MTases (32). The protein encoded by the AcNPV orf69 gene contains two highly conservative regions that correspond to the AdoMet-dependent MTase signature motifs, suggesting that the encoded protein functions as an MTase, and so it was renamed MTase1.
Protein BLAST analysis grouped MTase1 in the FtsJ-like family of proteins. FtsJ is an E. coli heat shock protein that was recently shown to be a 2′OMTase (6). FtsJ specifically methylates a uridine residue in 23S rRNA. Three viral proteins belonging to the FtsJ family are also 2′OMTases, but their substrates are cap 0 mRNAs, and they specifically methylate the ribose of the first transcribed base in mRNA caps (12, 28, 50). Comparison of the structure of these four proteins has allowed the identification of four residues that are diagnostic for 2′OMTases. All four of these residues are conserved in the baculovirus sequences, strongly indicating that MTase1 also methylates RNA.
The three viral cap 1 MTase are vaccinia virus VP39, domain I of Reovirus protein λ2, and the N-terminal portion of the flavivirus NS5 protein (12, 28, 50). All three of these viruses replicate in the cytosol of infected cells and encode a complete set of mRNA capping enzymes. Baculoviruses replicate in the nucleus but still encode at least two of the enzymes required for the formation of mRNA caps. The LEF-4 subunit of baculovirus RNA polymerase contains GTase and RTPase activities. Although a baculovirus-encoded N7MTase has not yet been identified, the presence of the 2′OMTase diagnostic residues suggested to us that MTase1 might also function in methylation of cap 0 mRNAs. It has been shown that most AcNPV transcripts possess a cap 1 structure at the 5′ end of mRNA (47), so either a host or viral protein must catalyze this reaction. We successfully demonstrated transfer of [methyl-3H]AdoMet to cap 0 RNAs, indicating that MTase1 has this function. This activity was dependent on the presence of a guanine cap on the RNA substrate but was independent of methylation at N7 of the guanine cap. Thus, the 2′OMTase activity of baculovirus MTase1 is similar to that of flavivirus NS5, which also does not discriminate between methylated and unmethylated caps (12). Vaccinia virus VP39, on the other hand, is only active on a methylated cap substrate (27).
The presence of virus-encoded 2′OMTase along with RTPase and GTase in the AcNPV genome is somewhat surprising, given that the virus apparently does not encode a good candidate for N7MTase. MTase1 is the only baculovirus protein so far identified that has a MTase domain. It is possible that baculoviruses contain another MTase that has relatively low sequence conservation and so has not been identified by standard database searches. Preliminary results from our lab suggest that levels of N7MTase significantly increase in infected cells, indicating that the virus either encodes this activity or upregulates levels of the host N7MTase.
Also of interest is the fact that only half of the baculoviruses sequenced to date contain MTase1 homologs. This suggests that these viruses are able to use the host enzyme for methylation of viral mRNAs. This is consistent with our finding that MTase1 is not essential for replication of AcNPV. Both of these observations concur with our finding that MTase1 is a late gene, because a late protein cannot itself be required for late gene expression. It is possible that MTase1 is required for optimal expression of late and very late genes. We limited our analysis of the mtase1 mutants to the production of infectious budded virus, which indirectly measures late gene expression because the structural proteins are late genes.
Primer extension assays mapped mtase1 transcripts to an ATAAG motif, which is common to baculovirus late gene promoters. A late transcription start site was also mapped to a noncanonical site. The presence of this endpoint in reverse transcription could be artifactual due to pausing of reverse transcription or could represent authentic transcription initiation. Further experiments will be needed to confirm this. Evidence for early transcripts initiating upstream of orf68 was also observed. These could potentially direct the synthesis of MTase1 during the early phase of infection, as they contain at least the N-terminal part of the open reading frame. This is somewhat unlikely because eukaryotic ribosomes are generally monocistronic, and furthermore immunoblot synthesis did not reveal evidence of MTase1 expression in the presence of aphidicolin. However, given the presence of these early transcripts, we cannot rule out the possibility that low levels of protein are synthesized during the early phase of infection. But, overall, the protein expression and RNA mapping data support the idea that MTase1 is not synthesized in the absence of viral DNA replication.
Mutant virus lacking MTase1 was not only viable, but it replicated at wild-type levels. Yet the results of Li et al. (38) suggest that orf69/mtase1 stimulated late gene expression threefold. Similar results have been found for several of the LEFs, which are essential for transient late expression but are not required for viral replication (7, 20, 46, 48). It is possible that these differences between transient expression and viral infection could reflect the fact that proteins like 2′OMTase offer a limited advantage to expression and that this is more significant in transient expression, which is less efficient than viral infection. The MTase activity of the vaccinia virus RNA cap 2′OMTase VP39 is also not required for replication of vaccinia in tissue culture (36). But the retention of 2′OMTases in multiple members of the Baculoviridae and Poxviridae suggest that virus-encoded MTase activity offers a growth advantage to replication in a natural infection. While this is not evident in experimental tissue culture systems, it should be possible to address this issue in experimental animals.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB 0110925).
We thank Wen Dong for excellent technical assistance.
REFERENCES
- 1.Adams, G. M., and R. M. Blumenthal. 1997. The PvuII DNA (cytosine-N4)-methyltransferase comprises two trypsin-defined domains, each of which binds a molecule of S-adenosyl-L-methionine. Biochemistry 36:8284-8292. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of the Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 4.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 5.Bugl, H., E. B. Fauman, B. L. Staker, F. Zheng, S. R. Kushner, M. A. Saper, J. C. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 6.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. J., and S. M. Thiem. 1997. Differential infectivity of two Autographa californica nucleopolyhedrovirus mutants on three permissive cell lines is the result of lef-7 deletion. Virology 227:88-95. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, C. H., and R. M. Blumenthal. 1999. S-Adenosylmethionine-dependent methyltransferase: structures and functions. World Scientific, River Edge, N.J.
- 9.Cheng, C.-H., S.-M. Liu, T.-Y. Chow, Y.-Y. Hsiao, D.-P. Wang, J.-J. Huang, and H.-H. Chen. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, X., S. Kumar, J. Posfai, J. W. Pflugrath, and R. J. Roberts. 1993. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell 74:299-307. [DOI] [PubMed] [Google Scholar]
- 11.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finta, C., U. Sulima, P. Venetianer, and A. Kiss. 1995. Purification of the KpnI DNA methyltransferase and photolabeling of the enzyme with S-adenosyl-L-methionine. Gene 164:65-69. [DOI] [PubMed] [Google Scholar]
- 14.Garrity, D. B., M. J. Chang, and G. W. Blissard. 1997. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology 231:167-181. [DOI] [PubMed] [Google Scholar]
- 15.Gomi, T., K. Tanihara, T. Date, and M. Fujioka. 1992. Rat guanidinoacetate methyltransferase: mutation of amino acids within a common sequence motif of mammalian methyltransferase does not affect catalytic activity but alters proteolytic susceptibility. Int. J. Biochem. 24:1639-1649. [DOI] [PubMed] [Google Scholar]
- 16.Grimmig, B., and U. Matern. 1997. Structure of the parsley caffeoyl-CoA O-methyltransferase gene, harbouring a novel elicitor responsive cis-acting element. Plant Mol. Biol. 33:323-341. [DOI] [PubMed] [Google Scholar]
- 17.Gross, C. H., and S. Shuman. 1998. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J. Virol. 72:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarino, L. A., W. Dong, and J. Jin. 2002. In vitro activity of the baculovirus late expression factor LEF-5. J. Virol. 76:12663-12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarino, L. A., J. Jin, and W. Dong. 1998. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J. Virol. 72:10003-10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarino, L. A., T.-A. Mistretta, and W. Dong. 2002. Baculovirus lef-12 is not required for viral replication. J. Virol. 76:12032-12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarino, L. A., and M. Smith. 1992. Regulation of delayed-early gene transcription by dual TATA boxes. J. Virol. 66:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamm, J., and I. W. Mattaj. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63:109-118. [DOI] [PubMed] [Google Scholar]
- 23a.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Hayakawa, T., G. F. Rohrmann, and Y. Hashimoto. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, C. K., V. Sriskanda, S. McCracken, D. Bentley, B. Schwer, and S. Shuman. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273:9577-9585. [DOI] [PubMed] [Google Scholar]
- 27.Hodel, A. E., P. D. Gershon, and F. A. Quiocho. 1998. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell 1:443-447. [DOI] [PubMed] [Google Scholar]
- 28.Hodel, A. E., P. D. Gershon, X. Shi, and F. A. Quiocho. 1996. The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 85:247-256. [DOI] [PubMed] [Google Scholar]
- 29.Hoopes, R. R., Jr., and G. F. Rohrmann. 1991. In vitro transcription of baculovirus immediate early genes: accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc. Natl. Acad. Sci. USA 88:4513-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin, J., W. Dong, and L. A. Guarino. 1998. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J. Virol. 72:10011-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koonin, E. V., and K. E. Rudd. 1993. SpoU protein of Escherichia coli belongs to a new family of putative rRNA methylases. Nucleic Acids Res. 21:5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamurthy, V., and D. N. Rao. 1994. Interaction of EcoP1 modification methylase with S-adenosyl-L-methionine: a UV-crosslinking study. Biochem. Mol. Biol. Int. 32:623-632. [PubMed] [Google Scholar]
- 34.Kuge, H., G. G. Brownlee, P. D. Gershon, and J. D. Richter. 1998. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 26:3208-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labahn, J., J. Granzin, G. Schluckebier, D. P. Robinson, W. E. Jack, I. Schildkraut, and W. Saenger. 1994. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc. Natl. Acad. Sci. USA 91:10957-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latner, D. R., J. M. Thompson, P. D. Gershon, C. Storrs, and R. C. Condit. 2002. The positive transcription elongation factor activity of the vaccinia virus J3 protein is independent from its (nucleoside-2′-O-) methyltransferase and poly(A) polymerase stimulatory functions. Virology 301:64-80. [DOI] [PubMed] [Google Scholar]
- 37.Lewis, J. D., and E. Izaurralde. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247:461-469. [DOI] [PubMed] [Google Scholar]
- 38.Li, L., S. H. Harwood, and G. F. Rohrmann. 1999. Identification of additional genes that influence baculovirus late gene expression. Virology 255:9-19. [DOI] [PubMed] [Google Scholar]
- 39.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 41.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris, T. D., and L. K. Miller. 1994. Mutational analysis of a baculovirus major late promoter. Gene 140:147-153. [DOI] [PubMed] [Google Scholar]
- 43.Muthukrishnan, S., B. Moss, J. A. Cooper, and E. S. Maxwell. 1978. Influence of 5′-terminal cap structure on the initiation of translation of vaccinia virus mRNA. J. Biol. Chem. 253:1710-1715. [PubMed] [Google Scholar]
- 44.Ooi, B. G., C. Rankin, and L. K. Miller. 1989. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J. Mol. Biol. 210:721-736. [DOI] [PubMed] [Google Scholar]
- 45.Pang, Y., J. Yu, L. Wang, X. Hu, W. Bao, G. Li, C. Chen, H. Han, S. Hu, and H. Yang. 2001. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 287:391-404. [DOI] [PubMed] [Google Scholar]
- 46.Prikhod'ko, E. A., A. Lu, J. A. Wilson, and L. K. Miller. 1999. In vivo and in vitro analysis of baculovirus ie-2 mutants. J. Virol. 73:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin, J.-C., and R. F. Weaver. 1982. Capping of viral RNA in cultured Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. J. Virol. 43:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy, R., R. Singh, and S. Shimba. 1992. Methylated cap structures in eukaryotic RNAs: structure, synthesis and functions. Pharmacol. Ther. 54:249-267. [DOI] [PubMed] [Google Scholar]
- 50.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 A resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 51.Roof, W. D., S. M. Horne, K. D. Young, and R. Young. 1994. slyD, a host gene required for phi X174 lysis, is related to the FK506-binding protein family of peptidyl-prolyl cis-trans-isomerases. J. Biol. Chem. 269:2902-2910. [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schluckebier, G., M. O'Gara, W. Saenger, and X. Cheng. 1995. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol. 247:16-20. [DOI] [PubMed] [Google Scholar]
- 54.Schnierle, B. S., P. D. Gershon, and B. Moss. 1992. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc. Natl. Acad. Sci. USA 89:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwer, B., and S. Shuman. 1996. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA 2:574-583. [PMC free article] [PubMed] [Google Scholar]
- 56.Shuman, S. 1990. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J. Biol. Chem. 265:11960-11966. [PubMed] [Google Scholar]
- 57.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66:1-40. [DOI] [PubMed] [Google Scholar]
- 58.Som, S., and S. Friedman. 1990. Direct photolabeling of the EcoRII methyltransferase with S-adenosyl-L-methionine. J. Biol. Chem. 265:4278-4283. [PubMed] [Google Scholar]
- 59.Sonenberg, N., H. Trachsel, S. Hecht, and A. J. Shatkin. 1980. Differential stimulation of capped mRNA translation in vitro by cap binding protein. Nature 285:331-333. [DOI] [PubMed] [Google Scholar]
- 60.Tomalski, M. D., J. Wu, and L. K. Miller. 1988. The location, sequence, transcription and regulation of a baculovirus DNA polymerase gene. Virology 167:591-600. [PubMed] [Google Scholar]