Abstract
mid1 is required for the proper placement of the contractile actin ring for cytokinesis at a medial site overlying the nucleus. Here we find that mid1 protein (mid1p) shuttles between the nucleus and a cortical medial broad band during interphase and early mitosis. The position of this broad band, which overlies the nucleus, is linked to nuclear position even in cells with displaced or multiple nuclei. We identified and created mutations in an NLS and in two crm1-dependent NES sequences in mid1p. NES mutations caused mid1p accumulation in the nucleus and loss of function. An NLS mutations greatly reduced nuclear localization but did not perturb cytoplasmic localization or function. mid1p localization to the medial broad band was also not dependent on mid1p PH domain or microtubule and actin cytoskeletons. Overexpression of mid1p produced ectopic cell growth at this band during interphase and abnormal karmellae-like nuclear membrane structures. In plo1–1, mid1p formed a medial broad band but did not incorporate into a tight ring, suggesting that polo kinase plo1p is required for activation of mid1p function. Thus, the mid1p broad band defines a compartment at the medial cell surface, whose localization is linked to the position of the nucleus, and whose function may be to position the plane of cell division.
INTRODUCTION
In most eukaryotic cells (with the notable exception of plant cells), cytokinesis occurs through the action of an actin-based contractile ring (See Fishkind and Wang, 1995, Field et al., 1999, and Hales et al., 1999). The proper placement of the contractile ring is critical for proper cell division. The positioning of the division plane regulates the shape, size, and orientation of cells and the segregation of determinants during the development. Coordination of cytokinesis with nuclear division ensures the correct distribution of genomic material between sister cells.
Fission yeast Schizosaccharomyces pombe is an excellent model system for cytokinesis. S. pombe are simple, rod-shaped cells that divide by medial fission using a medial actin-myosin based ring. This contractile ring forms in early mitosis before nuclear division at the position of the predivisional nucleus. In late mitosis, the septum forms at the position of the ring as the ring closes. Genetic analysis has identified many components of the contractile ring (Nurse et al., 1976; Gould and Simanis, 1997), including profilin (cdc3p; Balasubramanian et al., 1992), tropomyosin (cdc8p; Balasubramanian et al., 1994), type II myosin heavy chains (myo2p and myp2p/myo3p; Kitayama et al., 1997; Bezanilla et al., 1997; May et al., 1997; Motegi et al., 1997), a myosin light chain (cdc4p; McCollum et al., 1995; Naqvi et al., 1999), a formin (cdc12p; Chang et al., 1997), an IQGAP (rng2p; Eng et al., 1998), cdc15p and imp2p (Fankhauser et al., 1995; Demeter and Sazer, 1998), and polo-like kinase (plo1p; Ohkura et al., 1995; Bahler et al., 1998a). Because many of these components are conserved through evolution, molecular mechanisms controlling ring assembly in S. pombe are relevant to cytokinesis in larger eukaryotes.
The molecular mechanisms that control the position of the division plane are poorly understood. In animal cells, position of the contractile ring is determined by the position of the mitotic spindle poles or midzone, which are thought to produce positioning signals (Rappaport, 1986; see also Fishkind and Wang, 1995 and Oegema and Mitchison, 1997). The mechanism of division plane placement in S. pombe may be somewhat different, as the ring assembly begins in early mitosis before establishment of a mitotic spindle, and proper positioning of the ring occurs even in the absence of a mitotic spindle (Chang and Nurse, 1996). Indeed, assembly of a medial contractile ring can even be induced in interphase cells through overexpression of cdc15 or plo1 (Fankhauser et al., 1995; Ohkura et al., 1995), suggesting that the appropriate spatial cues are present even in interphase cells. An important clue for understanding the mechanism of ring placement comes from the observation that the position of the ring is often coupled to the position of the interphase nucleus (see Chang and Nurse, 1996). For instance, mutants defective in microtubule organization sometimes display a displacement of both the nucleus and the division plane (Toda et al., 1983; Radcliffe et al., 1998). Thus, the nucleus may somehow instruct the cell surface where to assemble the ring. Alternatively, the ring and nucleus may be positioned independently through a microtubule-dependent mechanism.
The identification of genes required for proper placement of the contractile ring provides an opportunity to study the molecular basis of division site placement. mid1 is representative of mutants, including plo1 (Bahler et al., 1998a) and pos 1, 2, 3 (Edamatsu and Toyoshima, 1996), that form rings located at random locations and at random angles in the cell (Chang et al., 1996; Sohrmann et al., 1996). In the mid1 mutant, the nucleus is still properly positioned at the middle of the cell, and the organization of microtubules is grossly normal (Chang et al., 1996). A mid1 null allele has a phenotype similar to the temperature-sensitive alleles and is viable (Sohrmann et al., 1996). Therefore, mid1p has a specific function in spatial regulation of ring assembly and may be required for the proper coupling between the nucleus and the ring positioning.
mid1 encodes a 120–130 kDa protein possessing putative nuclear localization signals (NLS), two PEST regions, and a plekstrin homology domain (PH domain; Sohrmann et al., 1996). No obvious homologues in other organisms have been found yet, although mid1p has some weak similarities with the Drosophila contractile ring protein, anillin (Bahler et al., 1998a). Previous immunofluorescence studies show that mid1p is in the nucleus during interphase, in a broad medial cortical band around the nucleus in early mitosis, and in a tight cortical medial ring later in mitosis (Sohrmann et al., 1996; Bahler et al., 1998a).
Because mid1p is essential for the correct placement of the ring and is present both in the nucleus and the ring, an attractive model is that mid1p may function as a molecular link that positions the ring near the nucleus. Here, we find that mid1p shuttles between the nucleus and a broad central band on the cell surface linked to the position of the nucleus. mid1p is established in this cortical band through most of interphase and thus precedes other cytokinesis factors at the medial cell surface. Mid1p overexpression induced abnormal cell surface growth in the vicinity of the nucleus, suggesting that mid1p has the ability to recruit actin and other proteins involved in cell growth to this medial region during cytokinesis. Thus, mid1p defines a novel medial cortical compartment that responds to the position of the nucleus through most of the cell cycle.
MATERIALS AND METHODS
Yeast Strains and Genetic Methods
Standard S. pombe genetic techniques were performed as described in Moreno et al., 1991. All S. pombe strains were isogenic to 972. S. pombe strains used are listed in Table 1. SP1601 strain (leu1–32 ade6-M216 ura4-D18 dmf1::ura4+ h−) is a generous gift from V. Simanis (Sohrmann et al., 1996). YDM109 strain (ura4D18 leu1–32 plo1–1 h−) is a generous gift of J. Bähler and D. McCollum. Yeast transformations were performed by electroporation (Kelly et al., 1993) or by the lithium acetate-DMSO method for integration of linear DNA (Bahler et al., 1998b).
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| FC 418 | ade6-M210 ura4-D18 leu1-32 h− | F. Chang lab |
| AP 103 | same as FC 418 + pAP35 (pnmt*-mid1 LEU2) | This work |
| AP 104 | same as FC 418 + pAP36 (pnmt*-mid1-HA LEU2) | This work |
| AP 105 | same as FC 418 + pAP37 (pnmt*-HA-mid1 LEU2) | This work |
| AP 106 | same as FC 418 + pAP38 (pnmt-mid1 LEU2) | This work |
| AP 107 | same as FC 418 + pAP39 (pnmt-mid1-HA LEU2) | This work |
| AP 108 | same as FC 418 + pAP40 (pnmt-HA-mid1 LEU2) | This work |
| AP 131 | same as FC 418 + pREP41X (pnmt* LEU2) | This work |
| AP 122 | same as FC 418 + pAP67 (pnmt*-ΔNES2mid1 LEU2) | This work |
| AP 126 | same as FC 418 + pAP73 (pnmt-GFP-mid1 LEU2) | This work |
| FC 769 | cdc11-119 leu 1-32 h− + pAP73 (pnmt-GFP-mid1 LEU2) | This work |
| AP 46 | ade6-M210 ura4-D18 leu1-32 mid1-GFP∷kan MX h− | This work |
| SP 1601 | ade6-M216 ura4-D18 leu1-32 dmf1∷ura4+h− | Sohrmann, 1996 |
| AP 76 | same as SP1601 + pAP94 integrated (pmid1-ΔPHmid1 leu1+) | This work |
| AP 80 | same as SP1601 + pAP95 integrated (pmid1-NLS*mid1 leu1+) | This work |
| AP 82 | same as SP1601 + pAP96 integrated (pmid1-ΔNES2mid1 leu1+) | This work |
| AP 85 | same as SP1601 + pAP97 integrated (pmid1-mid1 1-506 leu1+) | This work |
| AP250 | same as SP1601 + pAP115 integrated (pmid1-ΔNES1mid1 leu1+) | This work |
| AP251 | same as SP1601 + pAP116 integrated (pmid1-ΔNES1+2mid1 leu1+) | This work |
| YDM 109 | ura4D18 leu1-32 plo1-1 h− | Bähler, 1998a |
| AP 282 | same as YDM 109 + pRep41x | This work |
| AP 284 | same as YDM 109 + pAP95 integrated (pmid1-NLS*mid1 leu1+) | This work |
| AP 370 | ade6-M216 ura4-D18 leu1-32 plo1-1 dmf1∷ura4+ + pAP95 integrated (pmid1-NLS*mid1 leu1+) h− | This work |
mid1 Overexpression
A 2.9 kb XhoI-BamHI fragment, including 56 bp 5′ and 78 bp 3′ of mid1/dmf1 open reading frame (ORF), was amplified by PCR from pDmf1 (generous gift from V. Simanis) using the high-fidelity Pfu DNA polymerase and was inserted into pREP41X. To remove potential point mutations generated by PCR, a XhoI-SacI fragment from this plasmid was subcloned into pBSIIKS+, and the BglII-XbaI fragment inside mid1 gene (2.5 kb) was replaced by the BglII-XbaI fragment from pDmf1 plasmid to obtain pAP32. The 5′ and 3′ regions of mid1 were confirmed by nucleotide sequencing. Finally, pnmt-mid1 (full strength nmt1 promoter; pAP35) and pnmt*-mid1 (medium strength nmt1 promoter; pAP38) were constructed by inserting the XhoI-SacI fragment from pAP32 into pREP3X or pREP41X.
In a similar way, to add a C-terminus or N-terminus HA tag to mid1p, a XhoI-NotI and a NotI-BamHI fragment containing mid1/dmf1 ORF were amplified by PCR from pDmf1 and ligated into pSLF272 and pSLF273 (generous gift from S. Forsburg). A XhoI-SacI fragment was subcloned into pBSIIKS+ and the BglII-XbaI sequence of mid1 replaced by the same fragment from pDmf1 plasmid to create pAP 33 and pAP 34. 5′ and 3′ regions of mid1 were checked by sequencing. XhoI-SacI fragments from pAP33 and pAP 34 were inserted into pREP3X and pREP41X to generate pnmt*mid1-HA (pAP36 and pAP39) and pnmt*HA-mid1 (pAP37 and pAP40). Finally, the NotI-BamHI fragment from pAP34 containing mid1 sequence was inserted into pSGP573 (generous gift from S. Forsburg) to create a pnmtGFP-mid1 construct (pAP70), and a XhoI-SacI fragment from pAP70 was inserted into pREP3X to generate pAP73.
Construction of mid1-gfp Strain
Strain AP46 (mid1-gfp) was obtained by PCR-based targeted recombination of GFP(S65T)kanMX (Bahler et al., 1998b) at mid1/dmf1 locus. Briefly, linear double strand DNA was amplified by PCR from pFA6a-GFP(S65T)-kanMX6 (generous gift of J. Bähler) and was transformed into a ura4-D18/ura4-D18 leu1–32/leu1–32 ade6-M210/ade6 M216 h+/h− diploid strain. Stable G418 resistant diploids were selected and sporulated to generate the haploid AP46.
mid1 Mutagenesis
Site directed mutagenesis was performed by PCR using high fidelity Pfu DNA polymerase (Stratagene), except when fragments were amplified from genomic DNA, in which case Taq polymerase (Promega, Madison, WI) was used instead.
NLS*-mid1
Production of NLS*-mid1, in which RKKRK (aa 691 to 695) was replaced by QNSQS, was achieved by creating a HindIII-BamHI fragment containing the mutated sequence using the forward oligonucleotide 5′tgacaagcttttcaaccgacagaacagccaatcactcaacaaggctgc that was inserted in place of the wild-type fragment in pAP32.
ΔNES1-mid1
NES1 sequence (LNVATDLLESLDL; aa 69 to 81) was deleted by creating a XhoI-NsiI fragment carrying the deletion using the reverse oligonucleotide 5′-gccatgcatgcttctttccgaaaggtttccaaggg. This fragment was inserted in place of the wild-type fragment in pAP32.
ΔNES2-mid1
NES2 sequence (LGNLTLTCLYI; aa 763 to 773) was deleted using two rounds of PCR. First a 5′ fragment of 325 bp containing the deletion using the reverse oligo 5′ctggaaccgaaagctcaggtgtacgtttccctatagagtt and a 3′ fragment of 525 bp overlapping the 5′ fragment on 20 bp right downstream of the deleted region was amplified. These two fragments were used in the second round of PCR to create a 830 bp fragment, which was inserted at HindIII-BamHI sites in pAP32. The XhoI-SacI fragment from the resulting plasmid was subcloned in pREP41X to create pAP67 (nmt*ΔNES2-mid1).
ΔNES1 + 2-mid1
NES1 and NES2 deletions were combined by subcloning a XcmI-XbaI fragment containing the deletion of NES2 into the pBS plasmid containing the deletion of NES1.
ΔPH-mid1
Deletion of the C-terminal region of mid1p containing the PH domain (aa 798 to 920) was created by constructing a fragment of 410 pb with a stop codon and a XbaI site after the codon of aa 797 using the reverse oligo 5′-gctctagaaacgtgtcgcaagtccatcg. This fragment was inserted at HindIII-XbaI sites in pAP32.
mid1 1–506 XhoI-SalI fragment corresponding to mid1p aa 1 to 506 was amplified by PCR and inserted at XhoI site in pREP41X to create pAP60. To create pBS mid1 1–506, a XhoI-SacI fragment was then subcloned into pBSIIKS+.
In all cases, the presence of the correct mutation/deletion was confirmed by DNA sequencing. To place these mutated sequences under the control of mid1 endogenous promoter in an integrating vector, a fragment of genomic DNA with a XhoI site in 5′ containing 1kb upstream of mid1 ORF, as well as the first 280pb of mid1 ORF, was amplified by PCR using oligonucleotides forward 5′-cacactcgagtcgtaggatcagagtcaaagc and reverse 5′-gaatcgaggtgcccatagcc. This fragment was inserted at XhoI-BglII sites in the pBS plasmids carrying the various mutated/deleted constructs of mid1, except when NES1 was deleted. Instead, NES1 deletion was recreated during the amplification of mid1 promoter sequence using the mutagenic oligonucleotide described earlier and the resulting fragment inserted at XhoI-NsiI sites. In all cases a KpnI-SacI fragment of the new plasmids was subcloned in the integrative vector pJK148 to create pAP 94 (pmid-ΔPH-mid1), 95 (pmid-NLS*-mid1), 96 (pmid-ΔNES2-mid1), 97 (pmid-mid1–1-506), 115 (pmid-ΔNES1-mid1) and 116 (pmid-ΔNES1 + 2-mid1). All plasmids were targeted by a NdeI digest in the leu1 gene of pJK148 before transformation into Δmid1 strain SP1601 to create strains AP76, 80, 82, 85, 250, and 251. pAP95 (pmid-NLS*-mid1) was also integrated into YDM 109 strain (strain AP284). Strain AP284 was then crossed to an ade6-M216 ura4-D18 leu1–32 dmf1::ura4+ h+ strain to obtain strain AP370.
Immunofluorescence and Microscopy
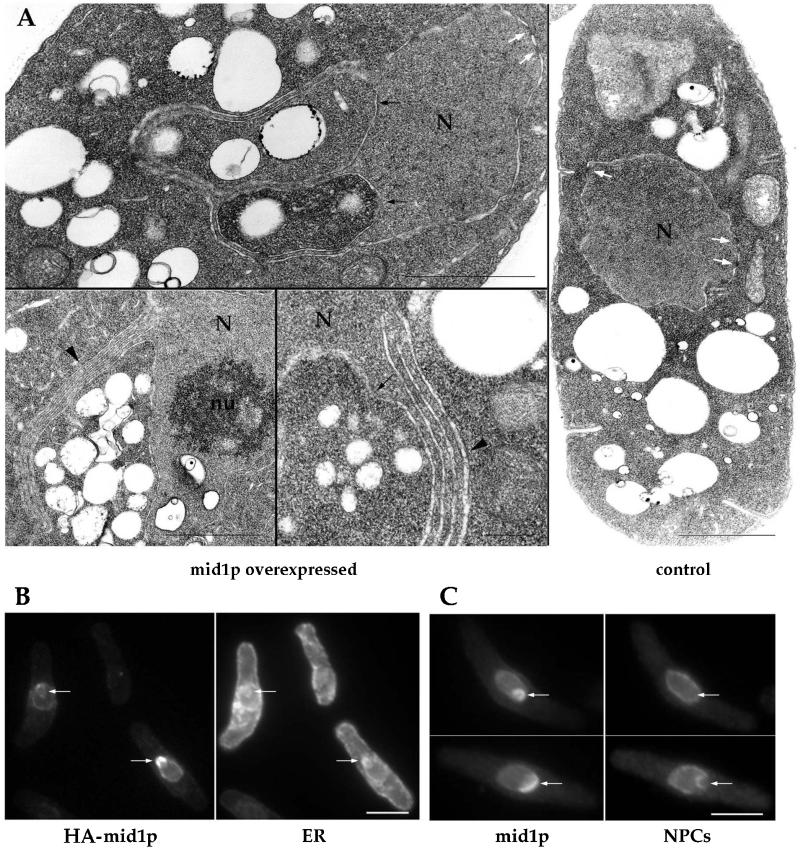
For immunofluorescence, cells were fixed with 4% formaldehyde and 0.25% glutaraldehyde for 30 min by addition of a 2:1 mix of 16% EM-grade formaldehyde (Polyscience, Warrington, PA) and PEM buffer (0.1 M NaPipes pH 6.8, 1 mM EGTA, 1 mM MgCl2) and of 8% EM-grade glutaraldehyde (Polyscience) to exponentially growing cultures. Cells expressing endogenous levels of wild-type or mutated mid1 were fixed with 4% formaldehyde only. Cells were further processed as described (Snell and Nurse, 1993). Anti-mid1p serum (Sohrmann et al., 1996; generous gift of V. Simanis) was used at 1:50 dilution on cells overexpressing mid1. In these conditions, cortical and nuclear mid1p could only be detected upon overexpression. On cells expressing endogenous levels of the protein, affinity purified Igs were used. Igs were affinity-purified from the serum by retro-elution on an his6-tagged mid1 300–506 fragment, produced from a QE9 plasmid (QIAGEN, Valencia, CA) in XL1 Blue bacteria. Briefly, inclusion bodies were purified, solubilized in Laemmli buffer, submitted to SDS-PAGE on 8% gels, and transferred to nitrocellulose. A band containing the his6-tagged mid1 fragment was isolated, saturated in TBS (10 mM Tris pH 7.4 150 mM NaCl) with 3% BSA and 1% Tween, and incubated for 1 h at room temperature with 1 ml of diluted serum (1:10 in TBS). The nitrocellulose was then washed three times in TBS-BSA-Tween, and Igs eluted in 400 μl of 100 mM glycine pH 1.9 for 1 min. pH was then immediately neutralized by addition of 100 μl of 1 M Tris pH 8. Igs were then kept at 4°C in presence of sodium azide and used in immunofluorescence at a 1:5 dilution with a Cy3-conjugated anti-rabbit secondary antibody at 1:200 (Sigma, St. Louis, MO). Tubulin was stained using mAb TAT1 (1:10; generous gift of K. Gull) and a fluorescein-coupled anti-mouse Ab. Endoplasmic reticulum was stained using an anti-BiP serum (1:100; generous gift of A. Pidoux) and a Cy3-conjugated anti-rabbit antibody. HA-tagged mid1p was detected using mAb 3F10 (1:1000; Boehringer Mannheim, Indianapolis, IN) and an Oregon green-conjugated anti-rat antibody (1/200; Molecular Probes, Eugene, OR). Nuclear pore complexes were stained with mAb414 (1:50; BabCo) and Cy3-conjugated anti-mouse antibody (1:200; Sigma). For this antibody, cells were fixed with formaldehyde only.
For midGFP imaging, AP46 cells were grown in 2 ml of YE5S at 20°C without shaking. Cells were concentrated 20-fold by brief centrifugation, and 2 μl cell slurry was mounted on slides in medium. GFP fluorescence of randomly chosen cells was then captured (3 s. exposure) after focusing on the DIC image.
Rhodamine phalloidin staining of actin was achieved as described by Sawin and Nurse, 1998 except that fixation time was reduced to 5 min. Calcofluor staining was performed on live cells concentrated by brief centrifugation by addition of 50 μg/ml Calcofluor (Sigma). Leptomycin B (LMB) stock solution (100 μg/ml in ethanol; generous gift of M. Yoshida) was diluted 4000-fold in culture medium to obtain a 25 ng/ml final concentration. Methyl-benzidazole-carbamate (MBC; Aldrich, Milwaukee, WI), thiabendazole (TBZ, Sigma) and latrunculin-A (LatA, Molecular Probes) studies were performed as described in Chang, 1999, except that the cells were first grown on agar plates, then resuspended into liquid media for the drug treatment. MBC (methyl 2-benzimidazolecarbamate; Aldrich) was used at 25 μg/ml in 1% DMSO.
Microscopic images were acquired with an E800 Nikon microscope with an 100x PlanApo 1.4NA 100X objective and a Sensicam (Cooke, Auburn Hills, MI) or an Orca100 (Hamamatsu, Hamamatsu, Japan) cooled CCD cameras. Figure 4C was obtained using a CSU-100 real time confocal (Yokogawa, Perkin Elmer-Cetus, Norwalk, CT). Image acquisition and analysis was performed using Phase 3 or Open Lab (Improvision) imaging software.
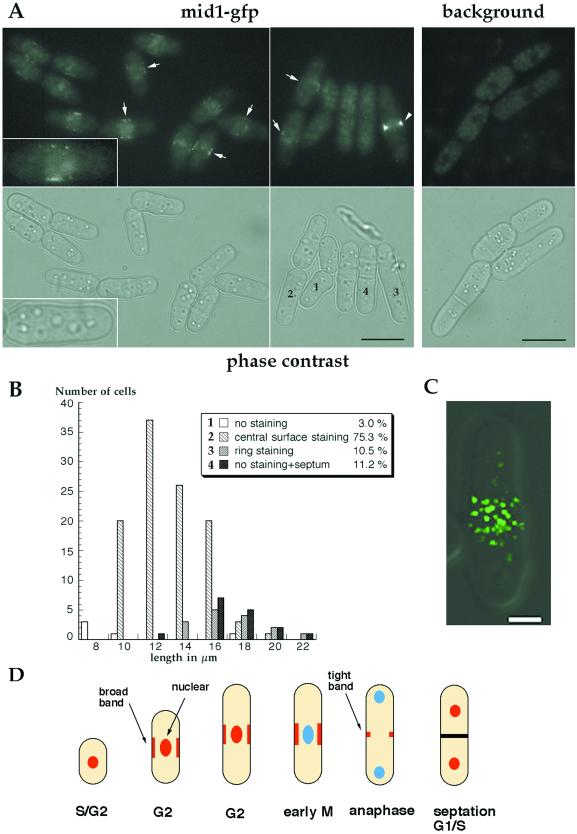
Figure 4.
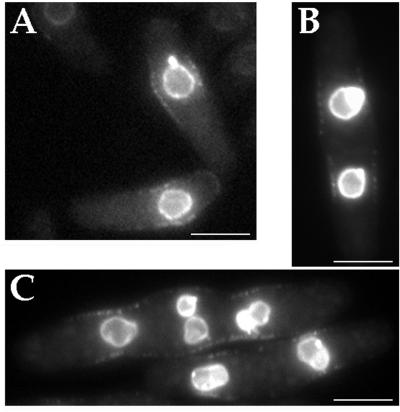
Localization of mid1p in a central broad band in wild-type cells. (A) Localization of mid1-GFP fusion protein. Fluorescent and phase images of cells expressing a functional mid1-GFP fusion (AP46), grown in YE5S at 20°C without shaking. Inset: 2-fold magnification. Most cells show a dim central broad band staining (arrows) and a faint staining in the region of the nucleus (like cell #2). Bright, tight rings are seen in longer cells without septum (arrow head, cell #3). Remaining cells have no obvious staining and have a septum (cell #4) or no septum (cell #1). Background panel correspond to wild-type cells grown and observed in similar conditions that exhibit autofluorescence. Bars: 10 μm. (B) Correlation between cell length and staining. Length of cells was correlated to the pattern of mid1-GFP fluorescence (a cell representative of each of the four categories labeled 1 to 4, is shown in A). Cells presenting a central broad band staining represent 75% of the population and their length range from 10 to 16 μm. n = 150. (C) Confocal image of mid1-GFP broad band (green) in a single focal plane at the cell surface overlaid over a DIC image (gray). Cells were grown as described in A, B. Bar: 2 μm. (D) Schematic representation of mid1p localization patterns during the cell cycle. mid1p is represented in red, and nucleus without mid1p in blue.
Western Blots
Crude boiled extracts were prepared as described by Correa-Bordes and Nurse (1995). Protein concentration was assayed by coomassie blue staining. Equal amounts of extract were loaded on 8% SDS-PAGE and blotted on nitrocellulose according to Towbin et al., 1979. mid1p was revealed using anti-mid1p affinity purified Igs (1:50), peroxidase-coupled anti-rabbit Igs (1:5000; Jackson ImmunoResearch, West Grove, PA), and a chemoluminescence kit (Pierce, Rockford, IL).
Electron Microscopy
Electron microscopy was performed according to Doye et al., 1994: exponentially growing cells were washed in K-phosphate buffer pH 6.5 with 0.5 mM MgCl2. Cells were resuspended in the same buffer supplemented with 2% formaldehyde and 2% glutaradehyde, incubated for 90 min at 4°C, and washed 3 times in 100 mM phosphate-citrate buffer pH 5.8. Cell walls were digested by incubation with 3 μg/ml Zymolyase 20T (NEN) in phosphate-citrate buffer for 2 h at 37°C. Cells were then washed 3 times in 0.1 M Na-Acetate pH 6.1 at 4°C and postfixed for 15 min at 4°C with 2% Osmium Tetroxyde in 1:2 Na-Acetate buffer. Cells were then rinsed 3 times in H2O, stained with 1% Uranyl-Acetate for 1 h at room temperature in the dark, washed again 3 times in H2O, and dehydrated in 70% and 90% ethanol for 15 min. After pelleting in a microtube, the cell pellet was incubated 2 times, for 10 min each time, in 100% ethanol and 2 times, for 10 min each time, in propylenoxyde, then incubated overnight in a 1-to-1 mix of Epon and propylenoxide. The next day, the pellet was incubated in pure epon for 1 h. Then epon was exchanged for fresh epon, and the tube incubated at 60°C for three days to induce epon polymerization. Thin sections of embedded cells were produced with an ultramicrotome (Leica, Deerfield, IL), stained with lead and uranyl for 30 min, and observed on a Philips electron microscope .
RESULTS
mid1 Overexpression Induces Ectopic Growth at the Middle of the Cell
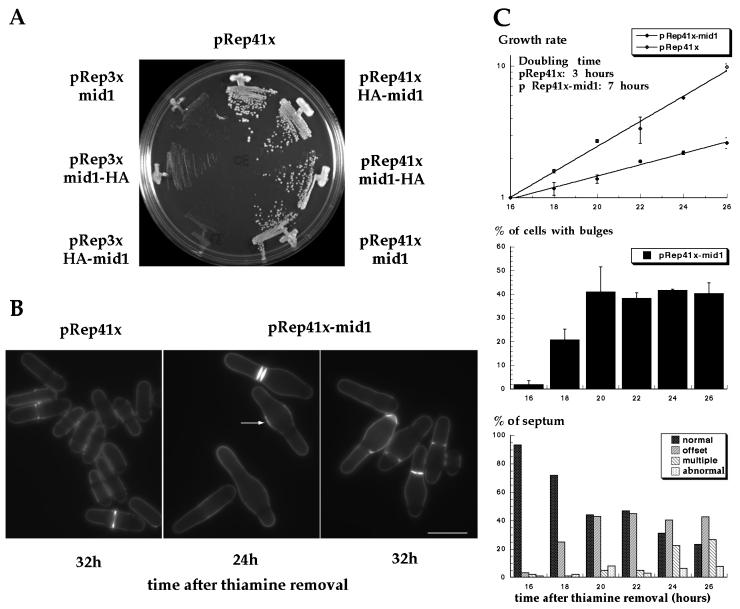
To investigate the function of mid1p, we examined the cellular effects of mid1 overexpression. mid1 was placed under the control of full strength or medium strength thiamine-repressible nmt1 promoter on the multicopy plasmids pREP3X or pREP41X (see MATERIALS AND METHODS). Although mid1 overexpression from the strong nmt1 promoter was lethal, mid1 expression from the medium strength nmt1 promoter did not affect colony formation (Figure 1A). Cells carrying pREP41X-mid1 exhibited a striking phenotype: they formed bulges near the cell center (Figure 1B). Medial bulges were exhibited in 40% of the cells 20 h after removal of thiamine (Figure 1C middle). Cells were longer than normal, suggestive of a cell cycle delay in interphase (Figure 1B), and the generation time of the population was increased approximately two-fold (Figure 1C top). Cells also exhibited abnormal cell division patterns: septa were often offset at the edge of the medial bulge, and occasionally multiple or even more abnormal septa formed (Figure 1C bottom). In general, septa were perpendicular to the long axis of the cell and were different from the tilted septa in cells carrying loss-of-function mid1 alleles. No defects in nuclear positioning were apparent, as nuclei were positioned properly at the middle of the cell or in the bulge region (see Figure 2).
Figure 1.
Effects of mid1 overexpression. (A) Effect on cell growth. Wild-type cells overexpressing mid1p or HA-tagged mid1p were streaked on EMM plates without thiamine and grown at 30°C for 4 days. (B) Effects on cell morphology and septum formation. Cells overexpressing mid1p (pREP41X-mid1) were grown at 30°C for 24 h and 32 h after thiamine removal from the medium and were stained with calcofluor. Note formation of bulges at the cell center, increased calcofluor staining in the bulge region (arrow) and abnormal placement of septa. Bar: 10 μm. (C) Quantitative analysis. Effects of mid1 overexpression from pREP41X on growth rate (top), bulge formation (middle), and on septum position (bottom) were assayed from 16 h to 26 h after thiamine removal of the medium. Growth rate was determined by cell count with an hemacytometer. For growth rate and bulge formation, mean (± SEM) from two independent experiments are presented. For septum position, 100 cells or more were scored at each time point.
Figure 2.
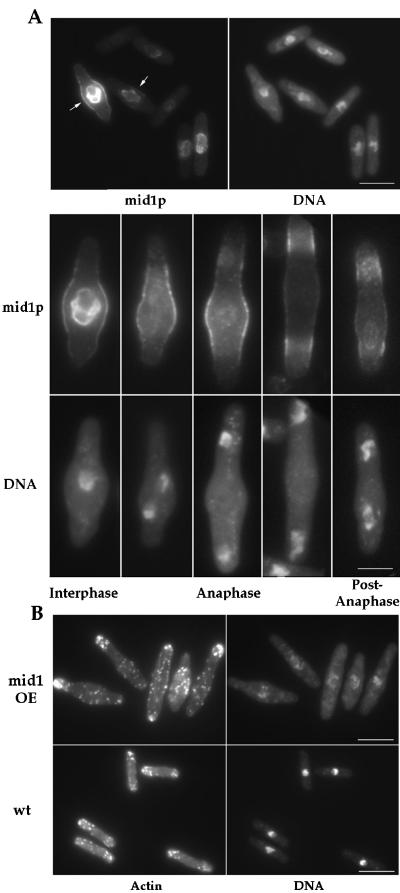
Localization of mid1p and F-actin in mid1 overexpressing cells. (A) mid1p localization. Cells overexpressing mid1 grown at 30°C were fixed 22 h after thiamine removal from the medium and double stained with anti-mid1p antibody and DAPI. Interphase cells have nuclear and central cortical staining (arrows). In mitotic cells, nuclear staining disappears, and the cortical staining splits in two bands. Bars: top, 10 μm; bottom 5 μ m. (B) F-actin localization. Cells overexpressing mid1 induced for 22 h, and wild-type cells grown at 30°C were fixed and stained for F-actin with rhodamine phalloidin. Note increased concentration of actin patches at the cell center and the abnormal DNA staining in the mid1 overexpressing cells. Bars: 10 μm.
Mid1p localization in these overexpressing cells was analyzed by immunofluorescence (Figure 2A). In interphase cells, mid1p was concentrated at rim of the nucleus and in a broad band at the central cell surface overlying the nucleus in the region of the bulge. This cell surface staining was finely punctuate or reticular. Timecourse studies showed that the central accumulation of mid1p preceded the development of bulges (our unpublished results). During mitosis, nuclear staining disappeared but the broad band persisted. In contrast to wild-type cells, tight ring staining was not observed. At the end of anaphase, two bands, each overlying a nucleus, were observed. Nuclear staining reappeared in telophase when nuclei had migrated back toward the center of the new sister cells. Similar patterns were seen in cells overexpressing a GFP-mid1 fusion protein (our unpublished results; Figure 10).
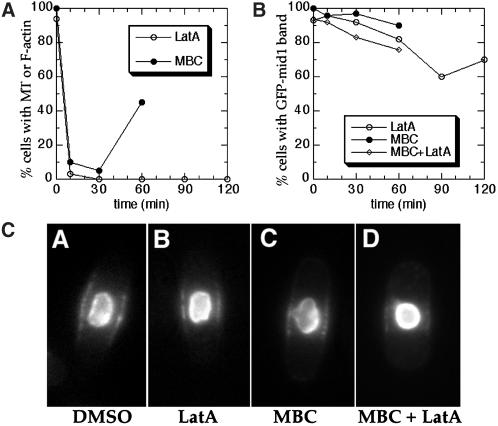
Figure 10.
Maintenance of mid1p broad band localization in the absence of F-actin and microtubule cytoskeletons. Wild-type cells carrying a nmt-GFP-mid1 construct (AP126 strain) were grown on minimal plates containing thiamine, inoculated into liquid cultures with thiamine, then treated with 200 μm LatA in 1% DMSO, 25 μg/ml MBC in 1%DMSO, both LatA and MBC in 2% DMSO, or 1% DMSO only. Samples were harvested at various time points after addition of drug. (A) Efficacy of the drug treatment. Open circles ○ represent percentage of LatA-treated cells with detectable F-actin structures as determined by rhodamine phalloidin. Filled circles ● represent percentage of MBC-treated cells with a microtubule (MT) over 2 μm in length. (n = 100 for each point). (B) Living cells with bright GFP-mid1 in the nucleus were assayed for a visible GFP-mid1 band by GFP fluorescence. All of the cells (100%) retained bright GFP-mid1 band staining after 1% DMSO treatment after 150 min. GFP-mid1-bands in cells treated with LatA were dimmer after 60 min. (C) GFP fluorescence images of representative AP126 cells 30 min after drug treatments.
Overexpression of mid1p affected cell polarity. In contrast to wild-type cells, which only grow at cell tips, cells overexpressing mid1p appeared to grow in the area around the nucleus, forming bulges in some cells. Other cells grew in more delocalized manner at the sides of the cells. F-actin staining showed delocalization of actin patches, with concentration of actin patches in the bulge regions in some cells (Figure 2B). Some bulges exhibited increased calcofluor staining, which is characteristic of sites of active cell wall growth (Mitchison and Nurse, 1985). Bulge formation was not a consequence of nuclear abnormalities, as overexpression of NLS*-mid1, a mid1 mutant that does not accumulate in the nucleus (see below) also induced similar cell shape changes (our unpublished results). Thus, mid1p may have the ability when overexpressed to promote active cell growth in the region around the nucleus.
mid1 Overexpression Induces “Karmellae” Formation
Cells overexpressing mid1 also exhibited abnormal nuclear morphology. mid1p staining patterns at the nuclear rim revealed enlarged, mis-shaped nuclei. DAPI staining was less intense and less regular (Figure 2B). Ultrastructural analysis by electron microscopy revealed striking nuclear membrane abnormalities that were never observed in control cells. Nuclear membranes were distorted and formed outpouches that appeared to contain cytoplasm (Figure 3A). In addition, nuclear membranes were folded into stacks of two to seven sets of membrane pairs (Figure 3A). These abnormal membrane folds were similar in appearance to karmellae structures induced by overproduction of transmembrane NE/ER proteins such as HMG-CoA reductase (Wright et al., 1988).
Figure 3.
Association of mid1p with NE/ER compartments and formation of karmellae-like structures. (A) Ultrastructural analysis of mid1 overexpressing cells. Cells overexpressing mid1 (24 h after thiamine removal) and control cells (grown in presence of thiamine) were fixed and processed for electron microscopy. Note invaginations of cytoplasm in the nucleus (black arrows) and karmellae-like multiple layers of nuclear envelope (black arrowheads). White arrows: nuclear pores. N: nucleus. nu: nucleolus. Bars: 1 μm, except bottom middle panel: 200 nm. (B-C) Colocalization of mid1p with the ER and nuclear pores. Cells overexpressing HA-tagged mid1p or mid1p were grown at 30°C, shifted to thiamine-free media for 22 h, and stained with mAb 3F10 (anti-HA) and anti-BiP antibody (ER marker) (B) or with the anti-mid1p antibody and mAb 414 (nuclear pore marker) (C). Note increased concentration of mid1p in regions that contain BiP but not nuclear pores (arrows). Bars: 5 μm.
To determine where mid1p may be located in these abnormal nuclear membranes, we stained cells overexpressing mild levels of mid1p or HA-tagged mid1p for mid1p and either the nuclear envelope (NE) or the endoplasmic reticulum (ER), which is in part perinuclear (Pidoux and Armstrong, 1993). Although some mid1p colocalized with the NE marker (nuclear pore protein) around the nucleus, mid1p staining was concentrated in a subset of NE regions that did not contain nuclear pores but did contain the ER marker (BiP; Figure 3 B,C arrows). Since the folded membrane stacks in the electron micrographs also did not contain nuclear pores, these data suggest that mid1p may be concentrated in these stacks (see Figure 3A,B). Thus, overexpression of mid1 causes its accumulation at nuclear membranes and perturbs their morphology, suggesting that mid1p has an affinity for these membrane compartments.
Mid1p Is Located in a Central Cortical Compartment During Interphase
Next, we asked whether mid1p is present in a central band when expressed at endogenous levels. Immunofluorescence in fixed samples reveals central broad band staining only in early mitotic cells (Bahler et al., 1998a). Since fixation can alter localization patterns, we reinvestigated mid1p localization by examining a mid1-GFP fusion in living cells. A GFP-kanMX cassette was inserted into the genomic mid1 locus by homologous recombination to produce a C-terminal mid1-GFP fusion (Bahler et al., 1998b; see MATERIALS AND METHODS). This construct was expressed from the mid1 promoter and was judged fully functional by the correct placement of septa in cells expressing this fusion as the only mid1 protein. Most interphase cells exhibited punctuate mid1-GFP fluorescence in a medial broad band at the cell surface (Figure 4A, arrows) as well as some nuclear fluorescence. Other cells exhibited a bright central ring (Figure 4A, arrowhead) or no cell surface staining at all. Confocal microscopy (Figure 4C) revealed that the mid1-GFP broad band consisted of multiple, discrete dots on the medial cell surface. An estimation of the number of discrete dots was 60–120 per cell in longer interphase cells.
Mid1-GFP localization through the cell cycle was evaluated by correlating staining pattern with cell length and septation state. The mid1-GFP signal was not amenable to other methods of cell cycle analysis such as time-lapse microscopy because of sensitivity to photobleaching, or to double staining in immunofluorescence studies because of sensitivity to fixation. As fission yeast grow throughout interphase and septate in a reproducible manner, cell length and septum formation can be used to approximate cell cycle stage (Mitchison and Nurse, 1985). The large majority of cells (75.3%) in the asynchronous population had both nuclear and central broad band staining. These cells were of variable length, generally ranging from 10 to 16 μm. This length distribution and the fact that most cells (>70%) in an asynchronous population are in interphase indicate that cells have central broad band localization through most of interphase. Cells showing a ring staining (10.5%) or no staining but also a septum (11.2%) were longer, ranging from 14 to 20 μm. Finally, cells without obvious staining and without a septum (3%) were very short, from 8 to 10 μm. This analysis of the mid1-GFP fusion coupled with other studies using fixed samples showed that mid1p is present in: 1) a central broad band through much of G2 phase and during very early mitosis; 2) a tight ring later in mitosis; 3) no specific cell surface localization in G1 and S phases, between septum formation and cell separation. The appearance of the broad band during interphase is significant because it suggests that mid1p is established at the future site of medial ring assembly well before other ring proteins, which only arrive during mitosis.
mid1p Localization at the Cell Surface Is Dependent on Nuclear Positioning
Since the broad band of mid1p at the cell surface overlies the nucleus and has roughly the same width as the nucleus, we tested whether the position of this band is coupled to nuclear position. To displace the nucleus away from the middle of the cell, we treated cells with a microtubule inhibitor, methyl 2-benzimidazolecarbamate (MBC), as nuclear positioning is a microtubule-dependent process (Toda et al., 1983). In these studies, cells overexpressing a functional GFP-mid1 fusion were used, since liquid culture conditions led to loss of the mid1-GFP signal when it was not overexpressed even in untreated cells. In cells where the nucleus was displaced from the middle, the GFP-mid1p broad band was displaced with the nucleus (Figure 5A). This linkage between nucleus and broad band positions was further tested by examining cells with multiple nuclei. Cells with multiple, separate nuclei without a septum were generated by treating a septation mutant cdc11 with MBC, which inhibits nuclear clustering (see Hagan and Yanagida , 1997). In these cells, there were multiple GFP-mid1 broad bands, each one in the vicinity to a nucleus. Thus, localization of mid1p on the cell surface is dependent on the distance from the nucleus. These results are consistent with the previous findings that cells with displaced or multiple nuclei ultimately divide at position of the predivisional nucleus (Toda et al., 1983; Marks et al., 1987; Chang and Nurse, 1996).
Figure 5.
Mid1p band on cell surface is linked to nuclear position in cells with displaced or multiple nuclei. (A) Wild-type cells carrying pAP73 (pnmt-GFP-mid1) were grown on minimal plates with thiamine at room temperature, inoculated in liquid media, incubated for one hour at 30°C and then treated with 25 μg/ml MBC for 2 h at 30°C, and imaged for GFP fluorescence. Bar: 5 μm. (B-C) cdc11 cells carrying pAP73 (nmt-GFP-mid1) were grown on minimal plates with thiamine at room temperature, inoculated into culture and incubated for 4 h at 36°C, treated with MBC at 36°C for 90 min, and imaged for GFP fluorescence. Some cells have multiple, separate nuclei but no cell division septum. Bars: 5 μm.
Identification of mid1 NLS and NES Sites Involved in Nuclear Shuttling
An attractive model for how mid1p broad band position may be coupled to nuclear position is through nuclear shuttling. It has been suggested that mid1p may shuttle in and out of the nucleus (Sohrmann et al., 1996). In principle, this shuttling could concentrate mid1p near the nucleus, so that it would preferentially bind to the cell surface near the nucleus when mid1p exits the nucleus. mid1p would then recruit other components of the medial ring to the cell middle during mitosis. Nuclear shuttling may also be intrinsic to mid1p function, for instance, to take another factor in or out of the nucleus. This model is especially attractive because it offers a potential mechanism for determining how the position of the site of cell division is coupled to nucleus position.
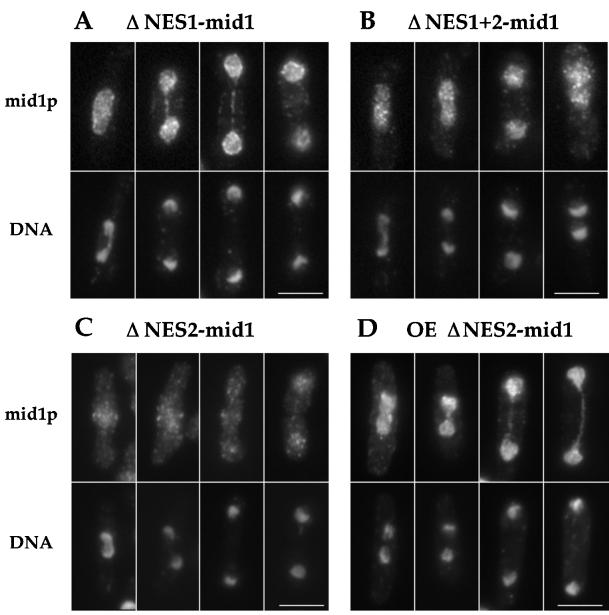
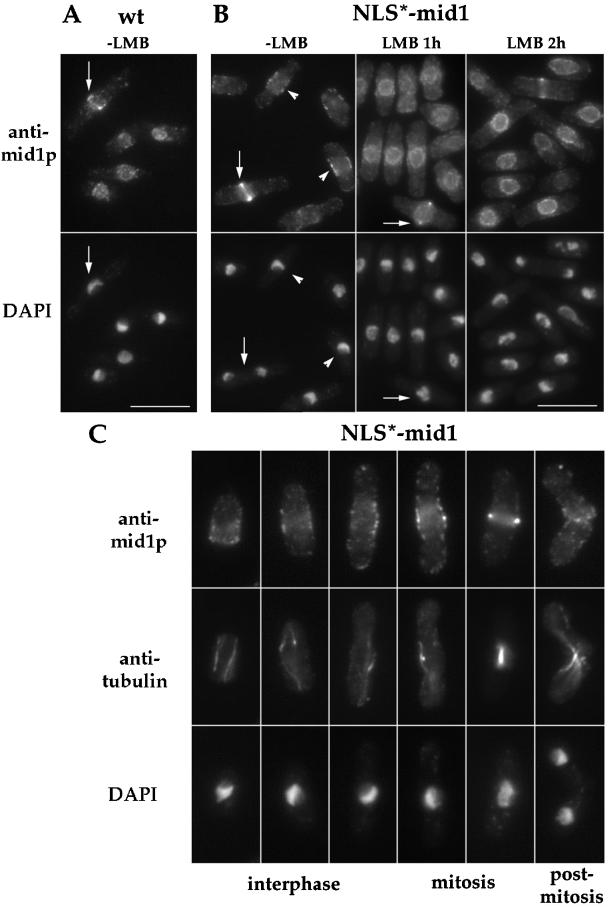
To test this hypothesis, we identified the nuclear import and export signals in mid1p. First, two leucine-rich nuclear export sequences (NES) were identified on the basis of similarity of rev-like NES sequences (Fischer et al., 1995; Wen et al., 1995; see MATERIALS AND METHODS). Small deletions in these sequences were generated by site-directed mutagenesis, and the mutant genes expressed under the control of the mid1 promoter were integrated into a mid1 deletion strain. ΔNES1-mid1p had a clear defect in nuclear export– in contrast to wild-type mid1p, which exits the nucleus during mitosis, it remained in the nucleus throughout the cell cycle, although some faint rings were occasionally seen (Figure 6, A-B). ΔNES2-mid1p had a weaker, but demonstrable nuclear export defect: it retained some weak cortical staining and weak rings in addition to nuclear staining (Figure 6C), but was clearly concentrated in the nucleus during mitosis upon overexpression (Figure 6D). ΔNES1+ 2-mid1p was localized like ΔNES1-mid1p. The function of these mutant proteins was assayed by their ability to position the septum. ΔNES1-mid1p was partially functional (20.5% abnormal septa); Figure 7 and Table 2) at 30°C, and ΔNES2-mid1p was fully functional. ΔNES1+2-mid1p was not functional (74.6% abnormal septum). Nuclear export of mid1p was sensitive to leptomycin B (LMB), a drug that blocks the nuclear export factor crm1p (see Figure 8; Nishi et al., 1994; Kudo et al.; 1999), showing that the nuclear export of mid1p is crm1 dependent. Although there is a possibility that these deletions may affect other functions beside nuclear export, these data suggest that mid1p has two NES sequences required for efficient crm1-dependent nuclear export and that mid1p needs to be in the cytoplasm to carry out its function.
Figure 6.
Localization of ΔNES1-mid1p, ΔNES2-mid1p, and ΔNES1+2-mid1p. (A-C) Strains AP250 (ΔNES1-mid1, A), AP251 (ΔNES1+2-mid1, B), and AP82 (ΔNES2-mid1, C) were grown at 30°C, fixed and double-stained with affinity purified antimid1p antibody (top) and DAPI (bottom). (D) Wild-type cells overexpressing ΔNES2-mid1 from pREP41X (AP122) were grown at 30°C, fixed 24 h after thiamine removal from the medium, and stained with antimid1p antibody (top) and DAPI (bottom). Note that ΔNES1-mid1p and ΔNES1 + 2-mid1p clearly remain located in the nucleus during mitosis. Very faint central rings are sometimes detected. Nuclear export defect is obvious upon overexpression of ΔNES2-mid1. Multinucleated interphase cells (like the fourth cell) are common in ΔNES1 + 2-mid1 strain, as this construct fails to complement mid1 deletion. Bars: 5 μm.
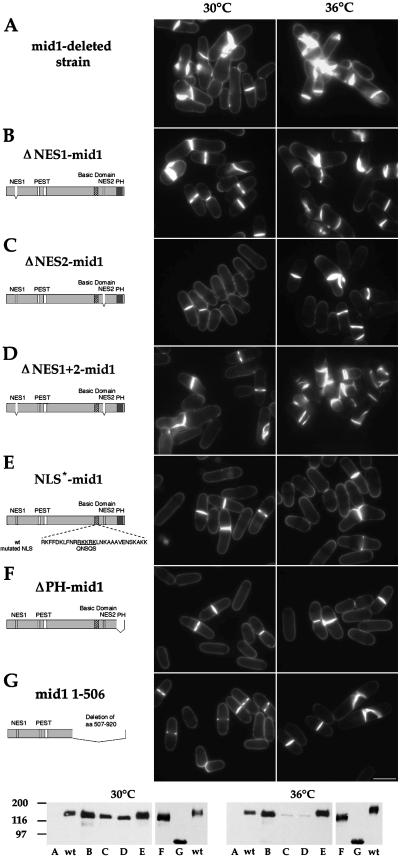
Figure 7.
Complementation of mid1 deletion by mutated mid1 constructs. Strains SP1601 (A), AP250 (B), AP82 (C), AP251 (D), AP80 (E), AP76 (F), and AP85 (G) were grown at 30°C (left) or 36°C (right) and stained for cell wall and septum with calcofluor (see Table 2 for statistics). Bar: 10 μm. Diagrams of mid1 mutant constructs are shown on the right. Bottom: Western blot analysis of expression levels of mid1 mutant constructs at 30 and 36°C. wt: wild-type strain FC 418. A-G: see above.
Table 2.
Ability of mid1 mutant constructs to complement mid1 and plo1 mutants
| Strains | Complementation (% abnormal
septa)
|
Localization | |
|---|---|---|---|
| 30°C | 36°C | ||
| wt | NA (0.5) | NA (1.5) | Nucleus/broad band tight ring |
| mid1-Δ strain | NA (98.5) | NA (99.8) | NA |
| ΔNES1-mid1 | + (20.5) | − (75.8) | Nucleus + occasional faint rings |
| ΔNES2-mid1 | ++ (1.0) | − (79.0)a | Nucleus + faint rings |
| ΔNES1+2-mid1 | − (74.6) | − (98.9)a | Nucleus + occasional faint rings |
| NLS*-mid1 | ++ (0.5) | ++ (1.0) | Broad band tight ring |
| ΔPH-mid1 | ++ (nd) | ++ (4.0) | Nucleus/broad band tight ring |
| mid1 1-506 | ++ (nd) | ± (53.6) | Diffuse punctuate staining |
| 25°C | 36°C | ||
| plo1-1 + pRep 41x | NA (1.8) | − (94.8) | |
| plo1-1 + NLS*-mid1 | NA (2.5) | − (95.5) | |
Wild type strain, mid1Δ strain and mid1Δ strain carrying integrated constructs expressing mid1 mutants were grown at 30°C or 36°C, stained with calcofluor, and assayed for septation defects by microscopic analysis. Plo1-1 strain with vector pREP41x only or an integrated copy of NLS*-mid1 construct were grown in minimal medium at 25°C or shifted from 25°C to 36°C for 4 h. Mean percentages of cells with abnormal septa were calculated from two separate experiments using two independent clones (n > 200 cells with a septum). NA, not applicable; nd, not determined.
Functionality in these mutants was difficult to assess because Western blot analysis showed that these constructs were expressed at a lower level than wild-type mid1p at 36°C (see Figure 7).
Figure 8.
Localization of NLS*-mid1p and effect of LMB treatment. (A-B) Wild-type strain (A) and strain AP80 (NLS*-mid1, B) were grown at 30°C, treated or not with 25 ng/ml LMB for 1 and 2 h, and stained with anti-mid1p Igs and DAPI. NLS*-mid1p is not detected in the nucleus, but is present at one cell tip in very short cells, at the central broad band in longer interphase cells (arrowheads) or in very early mitosis, and in a tight ring during anaphase (arrows). LMB treatment induces nuclear accumulation of NLS*-mid1p. Bars: 10 μm. (C) Strain AP80 (NLS*-mid1) was triple stained for mid1p, tubulin, and DNA. Different cell cycle phases are depicted. Note in the fourth cell with a short mitotic spindle, that mid1p cortical staining is brighter at the very center, suggesting that the cortical staining is starting to form into a tight ring. 1.5-fold enlargement compared with top panels.
Next, a basic nuclear localization sequence (NLS) was identified and mutated (see MATERIALS AND METHODS). Conservative changes in the NLS (aa 691 to 695; denoted NLS*) caused a significant defect in nuclear import: no NLS*-mid1p was detectable in the nucleus when expressed under the control of mid1 promoter (Figure 8), or when overexpressed. The NLS*-mid1 protein was detected by immunofluorescence in a central broad band during both interphase and mitosis, consistent with an increased cytoplasmic pool of NLS*-mid1p during interphase compared with wild-type mid1p. The NLS*-mid1 mutant had no detectable phenotype: cells divided symmetrically (Figure 7; Table 2). Thus, this NLS is not required for mid1p function or cytoplasmic localization. However, treatment of the NLS*-mid1 mutant with the nuclear export drug LMB still led to nuclear accumulation, suggesting that this mutant protein is still able to enter the nucleus at a low rate (Figure 8). In search of a second NLS sequence, we also deleted a second basic region located in N-terminal of the protein (PNRKRM, aa 102 to 108), but this did not cause any demonstrable defect in nuclear import either by itself or in combination with the first NLS mutation (our unpublished results).
Thus, it is likely that mid1p has at least two sequences required for nuclear import, a basic NLS sequence, which accounts for most of nuclear import activity, and a minor second sequence that has yet to be identified. Because there is still some residual nuclear shuttling in the NLS*-mid1 mutant, the function of nuclear shuttling still remains an open question.
The C-terminus of mid1p including the PH Domain Is Not Required for mid1p Function
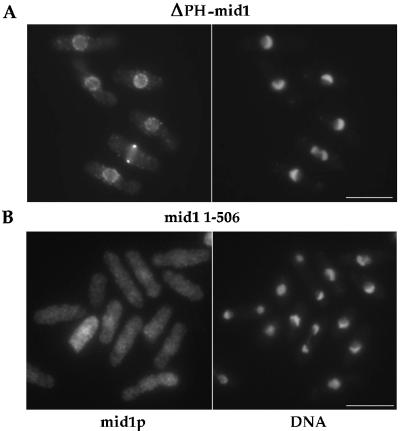
The C-terminus of mid1p possesses similarities to PH domains, which have been shown to target some proteins to the cell surface, possibly through direct interactions with phospholipids (Lemmon et al., 1996). We tested whether mid1p may be targeted by its PH domain. Surprisingly, ΔPH-mid1p expressed under the control of mid1 promoter was localized in an identical manner to wild-type mid1p (Figure 9A) and was fully functional (Figure 7 and Table 2). Overexpression of ΔPH-mid1p produced changes in cell shape and nuclear morphology identical to the wild-type protein (our unpublished results). Thus, mid1p PH domain is not essential either for its function or for targeting mid1p to the cell cortex and the nuclear envelope.
Figure 9.
Localization of ΔPH-mid1p and mid1 1–506p. Strain AP76 (ΔPH-mid1) and AP85 (mid1 1–506) were grown at 30°C and stained with anti-mid1p Igs and DAPI. Bars: 10 μm.
We further constructed an even larger C-terminal deletion of mid1p (mid1 1–506) that removes NES2, NLS, and the PH domain. mid1 1–506 expressed as the only mid1 gene was fully functional at 30°C and partially functional at 36°C (53% abnormal septa; Table 2 and Figure 7). mid1 1–506 protein showed a punctuate staining that was fairly evenly distributed over the entire cell (Figure 9B). No tight ring or broad band concentration was seen even in mitosis, although it is possible that the diffuse distribution of mid1p could mask its localization to these specific sites. Thus, the N-terminal half of mid1p is sufficient for function, and mid1p function does not require it to be concentrated at the broad band or tight ring. A model that provides an explanation for this unexpected result is presented in the DISCUSSION section.
Maintenance of mid1p Localization in the Broad Band Is Independent of F-actin and Microtubules
We next tested whether the microtubule or actin cytoskeleton may be required for mid1p localization to the broad central band. Cells overexpressing a functional GFP-mid1 construct were treated with the anti-actin drug latrunculin A (Lata) and/or the antimicrotubule drug MBC. Treatment with 200 μM LatA caused complete loss of detectable F-actin structures (Figure 10a) in 10 min or less. These cells retained GFP-mid1 bands (Figure 10, B-C) as judged by GFP fluorescence. After 60 min LatA treatment, bands were generally dimmer and some started to disappear, suggesting that some F-actin dependent process may be required for long term maintenance of the band. Treatment with MBC caused interphase microtubules to shrink to multiple dots on the nucleus (Figure 10A) by 10 min. The effect of MBC was transient, as microtubules began to grow back by the 60-min timepoint (Figure 10A). In MBC-treated cells, GFP-mid1 bands persisted (Figure 10, B-C). Even after treatment with both LatA and MBC, most cells retained GFP-mid1 bands (Figure 10, B-C). Drug treatment did not affect viability, as 96% of cells treated with LatA and MBC were viable after 90 min treatment. Thus, F-actin and microtubules are not required for the short-term maintenance of mid1p localization to the central band, and this localization of mid1p is not directly dependent on association with cytoskeletal structures. Consistent with this result is that there is no obvious accumulation of any F-actin or microtubules in this region during interphase. However, these studies do not address whether the actin or microtubule cytoskeletons may be involved in the initial establishment of the band or in transport of mid1p to this site.
Polo Kinase plo1p Is Required for mid1p Cytoplasmic Function and Incorporation into Tight Ring
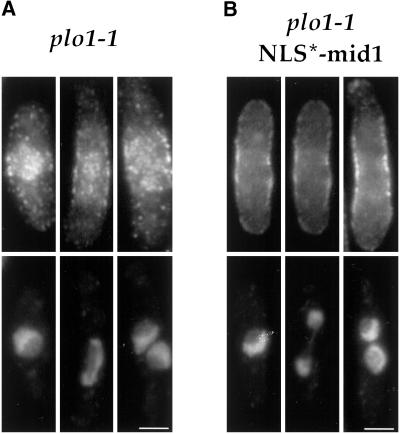
Bahler et al. (1998a) reported that the polo kinase plo1p has a role in placement of the ring, possibly through interaction and phosphorylation of mid1p. Although a deletion of plo1 produces numerous defects in mitosis and cytokinesis (Ohkura et al., 1995), some temperature sensitive alleles of plo1, such as plo1–1, have fairly specific mid1-like phenotypes (Bahler et al., 1998a). In the plo1–1 mutant, mid1p was found to be predominantly nuclear even in mitosis (Bahler et al., 1998a), suggesting that plo1p has a primary role in the nuclear export of mid1p. We tested this model using the NLS*-mid1 mutant allele. If the primary defect of mid1p in plo1–1 mutants is in nuclear export, then expression of NLS*-mid1 may be able to rescue the plo1–1 phenotype. When NLS*-mid1 construct was expressed in addition to wild-type mid1p in plo1–1 strain, cells still exhibited division defects and thus plo1–1 was not rescued by the NLS*-mid1p (Table 2). In addition, when expressed in plo1–1 mutant as the only mid1 protein, NLS*-mid1 still localized in a broad band at the central cell surface during most of the cell cycle including mitosis (Figure 11B). However, no tight rings were seen during anaphase, indicating a strong defect in tight ring formation. Thus, a defect in nuclear export of mid1p is not the sole cause of division site positioning defects in plo1–1 mutants.
Figure 11.
Localization of mid1p and NLS*-mid1p in plo1–1 mutant. Exponential cultures of plo1–1 mutant (A; YDM 109 strain) and plo1–1 mutant expressing NLS*-mid1 as the only mid1 protein (B; AP370 strain) were grown at 25°C, shifted for 2 h at 36°C, and fixed and stained with anti-mid1p Igs (top) and DAPI (bottom). mid1p is present in the nucleus and in a central broad band in interphase (left), mitotic (middle), and post mitotic (right) cells. NLS*-mid1p forms a broad cortical band in interphase and mitosis. Bars : 2.5 μm.
We also reexamined mid1p distribution in the plo1–1 mutant (Figure 11A). Although some weak defects in nuclear export were seen, mid1p was clearly localized to the medial broad band in interphase and mitotic cells, but did not form tight rings. Since in the plo1–1 mutant, tight rings of actin and cdc4p are present at random positions (Bahler et al., 1998a), the mid1p medial broad band was present but dissociated from the position of other ring components. Thus, mid1p did not function to position other components of the tight ring at a medial position and did not incorporate into the misplaced tight rings.
These results show that, although plo1p may have some role to increase nuclear export of mid1p at mitosis, it is also required for the cytoplasmic function of mid1p. plo1p may be required for the interaction of mid1p with other components of the ring, which is required both for mid1p function to position these other ring components and for its incorporation into the tight ring.
DISCUSSION
Mid1p Localization to a Novel Subcellular Cortical Compartment Linked to the Position of the Nucleus
In considering the mechanism of how the cell division ring is positioned, the temporal order of localization can indicate which proteins are important for initially marking the division site. Mid1p is a protein required for proper medial positioning of the cell division ring. Our studies show that, in addition to being localized at the nucleus and tight medial band, mid1p is present in a medial broad cortical band through most of the cell cycle, probably beginning in early G2 phase. The mid1p interphase broad band staining is roughly the width of the nucleus and is visualized in a reticular pattern or dots at the cell surface. During ring formation in early mitosis, when we imagine mid1 functions, additional mid1p is placed in this broad band as mid1p is exported from the nucleus. We detected this broad band in living cells using a functional mid1-GFP fusion expressed at endogenous levels. This pattern was not seen previously in wild-type interphase cells by immunofluorescence, possibly because of low protein abundance or poor preservation after fixation. Visualization of the broad band by immunofluorescence in cells expressing NLS*-mid1p from the mid1 promoter, where cytoplasmic levels of mid1p are more elevated, further supports this localization pattern. In addition, upon overexpression, mid1p broad band was easily seen by both immunofluorescence and GFP fusion through the whole cell cycle, including mitosis. Thus, to our knowledge, mid1p is the first cytokinesis factor to arrive at the medial site at the cell surface, as it precedes other known cytokinesis factors by at least an hour. This temporal order is consistent with mid1p function in placement of the cell division site.
A striking aspect of this broad band is that its position at the cell surface is coupled to the position of the nucleus. As shown in cells with multiple or displaced nuclei, mid1p surface localization is dependent on close proximity to the nucleus. How the nucleus and medial cell surface may be linked is not yet clear. We are not aware of any cytological structure linking the two. The actin and microtubule cytoskeletons do not obviously connect the nucleus and cell surface, and drug inhibitor studies show that actin and microtubule cytoskeletons are not required for maintenance of mid1p at the site.
Mid1p Exhibits Nuclear Shuttling
The highly regulated localization patterns of mid1p suggest that the spatial regulation of mid1p may be critical to its function and regulation. In particular, why should a cytokinesis factor be present in the nucleus? Several other cytokinesis factors, such as anillin (Field and Alberts, 1995; Aroian et al., 1997), are also in the nucleus during interphase and at the contractile ring during mitosis. Possibilities include: 1) mid1p has a function inside the nucleus; 2) nuclear localization may serve to sequester the protein during interphase; 3) shuttling may localize mid1p in the vicinity of the nucleus at the cell surface.
Mid1p overexpression phenotype suggests that sequestering mid1p in the nucleus may ensure that cells do not form bulges inappropriately during interphase. However, under laboratory conditions, cells expressing NLS*-mid1p at wild-type levels exhibited no discernible phenotype.
An attractive hypothesis is that nuclear shuttling may be responsible for mid1p localization at the cell surface. For instance, mid1p may enter the nucleus, exit the nucleus, then bind to a receptor at the cell surface; shuttling would concentrate the mid1 protein in the vicinity of the nucleus. We have demonstrated that mid1p does shuttle in and out of the nucleus, and we have identified signals responsible for shuttling. mid1p has two crm1-dependent nuclear export sequences and at least two sequences responsible for nuclear import. NES mutations caused nuclear accumulation and loss of mid1p function and cytoplasmic localization. These mutants suggest that mid1p must exit the nucleus and perform a function in the cytoplasm. NLS* mutation greatly reduced the amount of mid1p in the nucleus, but did not affect function or cytoplasmic localization. This suggests that a high concentration of mid1p in the nucleus may not be required for mid1p function. However, the NLS* mutation did not entirely block nuclear import, as shown by its nuclear accumulation after LMB treatment. There does not seem to be a second obvious NLS (basic-rich sequence). Rather, this suggests an interesting alternative mechanism for mid1p import into the nucleus. For example, it is possible that mid1p binds to another protein that is imported into the nucleus. The existence of multiple sequences involved in nuclear export and import may reflect the complex regulation of mid1p localization during the cell cycle.
The function of nuclear shuttling is still not clear. Certainly, mid1p must exit the nucleus to function in the cytoplasm. However, our data do not conclude whether mid1p needs to pass through the nucleus for function or localization, as there is residual shuttling in the NLS* mutant. The role of nuclear shuttling in linking the nucleus and cell surface remains an intriguing possibility that awaits further tests.
Mid1p Localization to the Ring and Polo Kinase
Mid1p localization changes in midmitosis from a broad punctuated band to a tight ring, after position of the ring has been determined (Bahler et al., 1998a, and our unpublished observations). The function of mid1p in the tight ring is not yet clear. This tight band colocalizes with other components of the medial ring, and localization of mid1p to this ring is dependent on other medial ring genes such as cdc12, cdc3, and cdc8 (Sohrmann et al., 1996). We favor a model in which mid1p functions in early mitosis in the broad band to position other ring components and then, after the ring position has been determined, incorporates into the ring by virtue of its association with other ring components. Alternatively, mid1p may function in the tight ring; however, the late incorporation of mid1p into the ring after the position of the ring has already been established is not consistent with this latter hypothesis. Overexpression of mid1 led to the persistence of the broad band through mitosis without formation of the tight band, but actin and cdc12p rings (our unpublished results) and the septum were still of normal thickness.
The polo kinase plo1 appears to have roles both in nuclear export and in activation of mid1p. plo1–1 mutants have a similar phenotype as mid1 mutants, and genetic and 2-hybrid data suggest that plo1p and mid1p interact and function together (Bahler et al., 1998a). Bahler et al. (1998a) suggested that plo1p has a role in the nuclear export of mid1p. Our results demonstrate that plo1p has additional effects on mid1p. In a plo1–1 mutant, a portion of mid1p still forms a medial broad band. Even though mid1p is medially placed, the tight actin ring forms at other positions, and mid1p does not incorporate into these misplaced rings. Thus, at mitosis plo1p may phosphorylate mid1p, which activates its ability to associate with other ring components. This association may be required both for mid1p function and for its tight ring formation.
The mid1 1–506 C-terminus truncation further tests the function of mid1p localization patterns. Its distribution was diffusely cytoplasmic throughout the cell cycle, with no broad band or tight ring staining, suggesting that determinants required for the association of mid1p with the cortex are located in C-terminus of mid1p. Surprisingly, this mutant was fully functional at 30°C. Thus, the concentrated distribution of mid1p in the tight band and broad band may not be essential for mid1p function. However, since it is possible that mid1–506p is still present in these locations but its detection is masked by the diffuse cytoplasmic pool, these findings do not rule out the possibility that a subset of mid1–506p is functioning at these cellular locations. For instance, mid1–506p may be acting with other proteins that are still properly localized in this band. These results do imply that it is unlikely that mid1p itself acts as a unique spatial cue for ring positioning.
Interaction of mid1p with Cell Growth Machinery and Membranes
Overexpression of mid1p led to a novel cell shape phenotype: mid1p concentrated in the broad cortical band induced cell growth, leading to bulges around the middle of the cell and redistribution of actin patches. This phenotype suggests that mid1p in the broad band has the ability to interact with and recruit actin and other proteins involved in cell tip growth to the cell middle for the purposes of cytokinesis.
Overexpression of mid1 also induced proliferation and folding of membranes around the nucleus in karmellae-like structures. Karmellae have been found to form as a result of overexpressing the integral ER membrane enzyme HMG-CoA reductase in S. cerevisiae and S. pombe (Wright et al., 1988, Lum and Wright, 1995) or other ER integral membrane proteins. To date, only ER transmembrane proteins have been shown to induce karmellae. However, no obvious transmembrane domains are detectable in the mid1p amino acid sequence, and the mid1p PH-like domain, which has been implicated with membrane association in other proteins, is dispensable for mid1p function, localization, or karmellae formation. This effect on membranes may provide clues suggesting possible association of mid1p with an ER/NE integral membrane protein responsible for mid1p function and aspects of localization, or its possible role in membrane events such as membrane fusion during cytokinesis.
CONCLUSION
In fission yeast, the middle of the cell may be defined by the position of the nucleus. Analysis of mid1 has revealed a novel, unexpected, cortical compartment that overlies the nucleus. Our findings illustrate how the nucleus can delineate a zone on the cell surface and they address general questions of how local regions or compartments may be delineated in the single cell. Similar processes are critical in development. For instance during Drosophila development, the position of an asymmetrically-placed nucleus in the oocyte is thought to determine the future dorsal-ventral axis of the embryo (Ray and Schupbach, 1996).
ACKNOWLEDGMENTS
We are indebted to V. Simanis for providing us with dmf1/mid1 plasmid pDW232, mid1 deletion strain SP1601, and anti-mid1p serum. Electron microscopy was performed at Institut Curie with the valuable help of G. Péau. We thank Annie Rousselet and Claude Celati for their assistance; Ray Lustig for technical support; Scott Randall (Improvision), Peter Franklin (Perkin Elmer-Cetus), and Phong Tran for imaging assistance; S. Forsburg and J. Bähler for epitope tagging vectors; J. Bähler and D. McCollum for plo1–1 strain; K. Gull for TAT1 mAb; A. Pidoux and J. Armstrong for anti-BiP serum; M. Yoshida for leptomycin B; L. Pon, G. Gundersen, and members of the Chang lab for comments on the manuscript; and H. Worman, V. Doye, and M. Bornens for helpful discussion. F.C. was supported by NIH Grant R01-GM5–35540, ACS Research Project Grant, March of Dimes Basil O'Conner Starter Scholar award, ACS Institutional Research Grant #177E from the Herbert Irving Comprehensive Cancer Center, and by a grant from the Howard Hughes Medical Institute to Columbia University for New Investigators. A.P. was supported by postdoctoral fellowships from Association pour la Recherche sur le Cancer, from Ministère des Affaires Etrangères, France (Bourse Lavoisier), and by a grant from the Leukemia and Lymphoma Society.
Abbreviations used:
- GFP
green fluorescent protein
- HMG-CoA reductase
3-hydroxy-3-methylglutaryl coenzyme A reductase
- LatA
latrunculin-A
- LMB
leptomycin B
- MBC
methyl 2-benzimidazolecarbamate
- NE
nuclear envelope
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- ORF
open reading frame
- PH domain
plekstrin homology domain
- TBZ
thiabendazole
REFERENCES
- Aroian RV, Field C, Pruliere G, Kenyon C, Alberts BM. Isolation of actin-associated proteins from Caenorhabditis elegans oocytes and their localization in the early embryo. Embo J. 1997;16:1541–1549. doi: 10.1093/emboj/16.7.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998a;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A r, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998b;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Helfman DM, Hemmingsen SM. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature. 1992;360:84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a Second Myosin-II in Schizosaccharomyces pombe: Mol Biol Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Identification and characterization of fission yeast mutants defective in actin ring assembly and placement. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell divsion ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr Biol. 1999;12:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S. phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Demeter J, Sazer S. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J Cell Biol. 1998;143:415–427. doi: 10.1083/jcb.143.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+RNA transport and nuclear pore distribution. Embo J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamatsu M, Toyoshima YY. Isolation and characterization of pos mutants defective in correct positioning of septum in Schizosaccharomyces pombe. Zoolog Sci. 1996;13:235–239. doi: 10.2108/zsj.13.235. [DOI] [PubMed] [Google Scholar]
- Eng K, Naqvi NI, Wong KC, Balasubramanian MK. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fishkind DJ, Wang YL. New horizons for cytokinesis. Current Opinion in Cell Biology. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1997;110:1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Hales KG, Bi E, Wu JQ, Adam JC, Yu IC, Pringle JR. Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol. 1999;11:717–725. doi: 10.1016/s0955-0674(99)00042-3. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S-phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Lum PY, Wright R. Degradation of HMG-CoA reductase-induced membranes in the fission yeast Schizosaccharomyces pombe. J Cell Biol. 1995;131:81–94. doi: 10.1083/jcb.131.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS. Spatial association of F-actin with growth polarity and septation in the fission yeast Schizosaccharomyces pombe. In: Poole R K, Trinci A P C, editors. Spatial Organization in Eukaryotic Microbes. Oxford: IRL Press; 1987. pp. 119–135. [Google Scholar]
- May KM, Watts FZ, Jones N, Hyams JS. Type II myosin involved in cytokinesis in the fission yeast. Schizosaccharomyces pombe. Cell Motil Cytoskeleton. 1997;38:385–396. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- McCollum D, Balasubramanian M, Gould K. Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Method Enzymol. 1991;194:795–723. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motegi F, Nakano K, Kitayama C, Yamamoto M, Mabuchi I. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 1997;420:161–166. doi: 10.1016/s0014-5793(97)01510-x. [DOI] [PubMed] [Google Scholar]
- Naqvi NI, Eng K, Gould KL, Balasubramanian MK. Evidence for F-actin-dependent, and -independent. mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. Embo J. 1999;18:854–862. doi: 10.1093/emboj/18.4.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Oegema K, Mitchison TJ. Rappaport rules: cleavage furrow induction in animal cells. Proc Natl Acad Sci USA. 1997;94:4817–4820. doi: 10.1073/pnas.94.10.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Armstrong J. The BiP protein and the endoplasmic reticulum of Schizosaccharomyces pombe: fate of the nuclear envelope during cell division. J Cell Sci. 1993;105:1115–1120. doi: 10.1242/jcs.105.4.1115. [DOI] [PubMed] [Google Scholar]
- Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol Biol Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol. 1986;101:245–281. doi: 10.1016/s0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- Ray RP, Schupbach T. Intercellular signaling and the polarization of body axes during Drosophila oogenesis. Genes Dev. 1996;10:1711–1723. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, V., and Nurse, P. (1993). Investigations into the control of cell form and polarity: the use of morphological mutants in fission yeast. Development Supplement 289–299. [PubMed]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Toda T, Umesono K, Hirata A, Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983;168:251–270. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wright R, Basson M, D'Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]