Abstract
Glycoprotein D (gD) interacts with two alternative protein receptors, nectin1 and HveA, to mediate herpes simplex virus (HSV) entry into cells. Fusion of the envelope with the plasma membrane requires, in addition to gD, glycoproteins gB, gH, and gL. Coexpression of the four glycoproteins (gD, gB, gH, and gL) promotes cell-cell fusion. gD delivered in trans is also capable of blocking the apoptosis induced by gD deletion viruses grown either in noncomplementing cells (gD−/−) or in complementing cells (gD−/+). While ectopic expression of cation-independent mannose-6 phosphate receptor blocks apoptosis induced by both stocks, other requirements differ. Thus, apoptosis induced by gD−/− virus is blocked by full-length gD (or two gD fragments reconstituting a full-length molecule), whereas ectopic expression of the gD ectodomain is sufficient to block apoptosis induced by gD−/+ virus. In this report we took advantage of a set of gD insertion-deletion mutants to map the domains of gD required to block apoptosis by gD−/− and gD−/+ viruses and those involved in cell-cell fusion. The mutations that resulted in failure to block apoptosis were the same for gD−/− and gD−/+ viruses and were located in three sites, one within the immunoglobulin-type core region (residues 125, 126, and 151), one in the upstream connector region (residues 34 and 43), and one in the C-terminal portion of the ectodomain (residue 277). A mutant that carried amino acid substitutions at the three glycosylation sites failed to block apoptosis but behaved like wild-type gD in all other assays. The mutations that inhibited polykaryocyte formation were located in the upstream connector region (residues 34 and 43), at the α1 helix (residue 77), in the immunoglobulin core and downstream regions (residue 151 and 187), and at the α3 helix (residues 243 and 246). Binding of soluble nectin1-Fc to cells expressing the mutant gDs was generally affected by the same mutations that affected fusion, with one notable exception (Δ277-310), which affected fusion without hampering nectin1 binding. This deletion likely identifies a region of gD involved in fusion activity at a post-nectin1-binding step. We conclude that whereas mutations that affected all functions (e.g., upstream connector region and residue 151) may be detrimental to overall gD structure, the mutations that affect specific activities identify domains of gD involved in the interactions with entry receptors and fusogenic glycoproteins and with cellular proteins required to block apoptosis. The evidence that glycosylation of gD is required for blocking apoptosis supports the conclusion that the interacting protein is the mannose-6 phosphate receptor.
The entry of herpes simplex virus 1 (HSV-1) into cells consists of at least three sequential steps (6, 43). In the first, the virus attaches to sulfated proteoglycans present on cell surfaces. This interaction is mediated by glycoproteins B and C (gB and gC). In the next step, glycoprotein D (gD) interacts with one of at least two known receptors, a member of the tumor necrosis receptor family designated herpesvirus entry mediator A (HveA) (30) or a member of the nectin family designated nectin1 or HveC (14, 19). gD (or the gD-receptor complex) and three additional viral glycoproteins enable the fusion of the envelope with the plasma membrane. gD also performs two additional functions. Expression of gD blocks superinfection, a function designed to block superinfection of the infected cells with the virus progeny of that cell (5, 20). In addition, gD blocks apoptosis in cells infected with mutants lacking the gene encoding gD (47).
A central question investigated in this report is whether the antiapoptotic functions of gD overlap those of entry into cells, specifically those mediating the fusion of the envelope with the plasma membrane and binding to nectin1. In this report, we took advantage of a set of well-characterized gD insertion-deletion mutants generated by Chiang et al. (10) to map the domains necessary to block apoptosis and those critical for membrane fusion. The following observations are relevant to this report.
(i) We found that the distribution of HveA is mainly associated with cells of lymphoid origin (30). The interactions of gD with HveA have been studied at both the structural and genetic levels. Structural studies indicate that HveA binds at the amino terminus (8, 9, 21, 34). Genetic studies indicate that the interaction between gD and HveA is abolished by mutations at the amino terminus of gD and in the C-terminal portion of the ectodomain (residues 222 to 224) (15, 46).
(ii) Nectins form a family of intercellular adhesion molecules (17, 24, 37, 38, 41, 43). Structurally, they are members of the extended immunoglobulin superfamily. Nectin1 appears to be a major HSV receptor because of its ability to mediate entry as well as cell-to-cell spread of all HSV-1 and HSV-2 strains and its ubiquitous distribution in human tissues targeted by HSV (13, 14, 19). The nectin site that interacts with gD is in the N-terminal V-type domain (11, 12, 22, 25). Mutations at the amino terminus (residues 24 to 25) that abolish binding of gD with HveA do not affect the binding of gD to nectin1 (21). Studies in several laboratories have led to the conclusion that expression of gD on the cell surface sequesters nectin1 and HveA and blocks superinfection of the infected cell. This function, called restriction to infection or interference, stems from the ability of gD to interact in cis rather than in trans with the gD receptor, to sequester it, and to make it unavailable to superinfecting HSV (1, 5, 7, 18, 20).
(iii) Fusion of the envelope with the plasma membrane requires, in addition to gD, the glycoproteins B, H, and L. A cell-cell fusion assay based on coexpression of the four glycoproteins from expression vectors in HSV-susceptible cells was developed by Browne and coworkers (2). Inasmuch as the assay has two requirements in common with virion entry into cells (namely, coexpression of gD with gB, gH/gL, and the presence of one of the gD receptors in the target cell [36]), the cell-cell fusion mediated by the four glycoproteins is thought to mirror the basic fusion mechanism that takes place at virus entry into cells. A major advantage of the cell-cell fusion assay is that it can be readily quantified, whereas virus entry is usually quantified in an indirect manner, based on expression from the viral genome of a reporter gene (e.g., β-galactosidase) placed under the control of an α promoter.
(iv) gD− mutants grown in complementing cells (gD−/+ stocks) contain gD in the envelope but lack the gD gene. gD− stocks produced in noncomplementing cells infected with gD−/+ virus (gD−/− stocks) lack both the gD gene and gD in the envelope. Both gD+/− and gD−/− stocks induce apoptosis that is characterized by translocation of annexin V and nucleosomal degradation of cellular DNA and blocked by gD delivered in trans (47).
The mechanism by which apoptosis is induced by gD−/− and gD−/+ stocks differs (47). gD−/− virus is taken up into endosome-like vesicles and degraded. Cells exposed to large numbers of gD−/− virus particles undergo apoptosis that is blocked by either intact gD or a heterodimer consisting of the ectodomain and transmembrane domain linked by disulfate bonds to the transmembrane domain and cytoplasmic tail of gD. Apoptosis induced by gD−/− virus is also blocked by chloroquine and by the cation-independent mannose-6 phosphate receptor (48). Earlier studies have shown that gD colocalizes with and binds mannose-6 phosphate receptor (3). More recent literature has shown that the mannose-6 phosphate receptor regulates the sorting of lysosomal enzymes and plays a role in apoptosis (50).
The picture that emerges from these studies is that gD interacts with and retains the mannose-6 phosphate receptor in the compartment containing the endocytosed virus particles, thereby preventing the release of lysosomal enzymes. Apoptosis induced by gD−/+ stocks is also blocked by gD and the mannose-6 phosphate receptor but not by chloroquine. Moreover, apoptosis induced by gD+/− stocks is blocked by the ectodomain of gD. Because gD−/+ stocks produce gD−/− particles in noncomplementing cells, one hypothesis that explains the accumulating data is that the secreted form of gD can interact with the mannose-6 phosphate receptor in a late endosomal compartment subverted for viral exocytosis to block the release of lysosomal enzymes (47-49).
Two sets of reagents have helped define the functional topology of gD. The first consists of a panel of monoclonal antibodies characterized for their ability to bind different sites of gD (33-35, 46). The second reagent consists of a panel of linker-scanning and deletion mutants of gD (10). Cells expressing the mutant forms of gD were used to complement a gD deletion virus (F-gD-β) (23). The complemented virus was in turn assayed for the ability to infect Vero cells which express a simian isoform of nectin1 (10). Virus entry was quantified as percent complementation relative to that of virus grown in cells expressing wild-type gD. These studies led to the definition of four functional regions, I to IV, located at residues 27 to 43, 125 to 161, 225 to 246, and 277 to 310, respectively (10).
The crystal structure of a soluble truncated form of gD (gD285t) bound to HveA (see Fig. 8 in reference 9) showed that the gD ectodomain is composed of several structural regions. The N terminus (residues 1 to 32) of the molecule has no specific structure in the unbound state. It constitutes the HveA binding site and, upon binding, folds over itself, forming a hairpin. The core region (residues 56 to 184) adopts a V-like immunoglobulin structure made of two beta sheets composed of nine parallel and antiparallel β strands. It is preceded by a connector sequence (residues 33 to 55) containing two β strands. The immunoglobulin core region is followed by two additional β strands. The downstream portion (residues 185 to 259) carries two α-helical structures and folds back towards the N-terminal hairpin. Its major α helix (α3) supports the hairpin. The structure of the most C-terminal portion of the ectodomain beyond residue 259 remains to be determined. The crystal structure of gD bound to nectin1 has not yet been solved.
The objectives of the studies presented in this report were twofold. The first was to map the domains of gD that are required to block apoptosis. Since the blocking of apoptosis appears to involve interaction with the mannose-6 phosphate receptor, this would be a first step in delineating the sites of interaction of gD with the mannose-6 phosphate receptor. These studies would also help determine whether the sites required for blocking apoptosis by gD−/− virus stocks differ from those required by the gD−/+ stocks. A second objective was to determine whether the domain required for blocking apoptosis overlapped the gD domains required to initiate cell-cell fusion and binding to nectin1. The studies were based on the panel of linker-scanning and deletion mutants described previously (10). We report that the domains of gD required to block apoptosis induced by gD−/− stocks could not be differentiated from those required for blocking the effects of gD−/+ stocks. However, the domains of gD required to block apoptosis were distinct from those involved in membrane fusion. In addition, the ability of gD to block apoptosis depends on its N-linked oligosaccharides, supporting the hypothesis that the antiapoptotic function of gD involves its interaction with the mannose-6 phosphate receptor.
MATERIALS AND METHODS
Cells and viruses.
SK-N-SH cells were obtained from the American Type Culture Collection (Rockville, Md.). The 143 cell line lacking thymidine kinase (TK−) was a gift of Carlo Croce. All cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Insect cell line Sf9 (Spodoptera frugiperda) was obtained from PharMingen (San Diego, Calif.). Unless otherwise indicated, cultures were seeded less than 20 h prior to infection and assayed at 60 to 70% confluence for apoptosis and about 80% confluence for cell fusion. gD−/− and gD−/+ mutant viruses were produced as described in detail elsewhere (23, 47).
Construction of recombinant baculoviruses expressing gD linker-insertion and linker-insertion-deletion mutants.
The 19 plasmids including gD linker-insertion and linker-insertion-deletion mutants were described elsewhere (10). The locations of insertion and the inserted amino acids are shown in Table 1. The EcoRI-PstI fragments encoding the 19 mutant gDs were amplified by PCR with primers CGGAATTCATGGGGGGGGCTGCCGCCAG and AACTGCAGCTAGTAAAACAAGGGCTGGTGCG and inserted into the EcoRI and BglII sites of baculovirus transfer vector pAc-CMV (pMTS-1), a derivative of the pAcSG2 baculovirus transfer vector (PharMingen) in which the cytomegalovirus promoter sequence was inserted as a 756-bp fragment in the XhoI and EcoRI sites. All constructs were sequenced to ensure fidelity.
TABLE 1.
gD linker-insertion and linker-insertion-deletion mutants
| Sequential no. | Plasmid | Mutationa and location of insertion | Amino acids inserted |
|---|---|---|---|
| 3 | D1-H28 | ∇12 | GKIFP |
| 5 | D1-H110 | Δ12-77 | GKIFP |
| 8 | D1-N29 | ∇34 | GKIFL |
| 9 | D1-H27 | ∇43 | EDLP |
| 12 | D1-H78 | ∇77a | GKIFP |
| 13 | D1-N155 | ∇77b | EDLP |
| 16 | D1-N65 | ∇83 | GRSS |
| 17 | D1-H104 | ∇84 | GKIFP |
| 20 | D1-N22 | ∇125 | GRSS |
| 21 | D1-H98 | ∇126 | GKIFP |
| 25 | D1-N37 | ∇151 | WKIFL |
| 31 | D1-N119 | ∇187 | GRSS |
| 37 | D1-H24 | ∇243 | GRSS |
| 39 | D1-H54 | ∇246a | GKIFP |
| 40 | D1-N1 | ∇246b | EDLP |
| 42 | D1-H26 | ∇277 | GKIFP |
| 43 | D1-H75 | Δ277-310 | GKIFP |
| 44 | D1-N2 | ∇287 | EDLP |
| 48 | D1-H15 | ∇310 | GKIFRKIFP |
Δ, deletion; ∇, insertion.
Construction of baculovirus recombinants expressing triple mutant gD(QAA) lacking N-glycosylation sites.
The triple mutant gD(QAA) (42) was amplified by the same primers from plasmid pDS145 and then inserted into baculovirus transfer vector pAc-CMV. The procedures for generation of recombinant baculovirus and infection of mammalian cells were as described elsewhere (47).
Construction of plasmids expressing HSV glycoproteins.
The complete gB coding sequence was PCR amplified with primers 5′-GGCACGAGGCCTCCCCGTAGTCCCGCCATGC (forward) and 5′-GATGCACATGAGATCTAACACCCGTGG (reverse) and cloned as a StuI-BglII fragment in the corresponding sites of pMTS-1. The complete gH coding sequence was PCR amplified with primers 5′-CCCCC ATGGG CGGCC GCTTA TGGTT CGTG (forward) and 5′-ACCAC AGACA CGGTT AACGT TTATT CG (reverse) and cloned as a NotI-HpaI fragment in the NotI and SmaI sites of pMTS-1. The complete gL sequence was PCR amplified with primers 5′-GACTG TAGGC CTCTG TGGGT GCCGA C (forward) and 5′-GTTCGCCCCCGGTACCCGGACGGGTTTC (reverse) and cloned as a StuI-Asp718 fragment in the MTS-1 vector. The pEA99 plasmid carrying the gD coding sequence in pcDNA3.1(−) was described previously (28) and obtained by amplification of the gD coding sequence of HSV-1(F) gD (25 bp upstream of the start codon and 34 bp downstream of the stop codon) with primers 5′-TATCCTTAAGGGATCCTTTGTGTGGTGCG (forward) and 5′GGTCTGCGGGGTAAGCTTGGGACCTTA (reverse).
Baculoviruses expressing HSV-1 glycoproteins.
Recombinant baculoviruses expressing HSV-1 gB, gH, and gL were obtained by cotransfecting the corresponding baculovirus transfer vectors gB-MTS, gL-MTS, and gH-MTS with BaculoGold DNA (Pharmingen) in Sf9 insect cells as specified by the Pharmingen protocol. The baculovirus expressing gD was described previously (47).
Double infections.
Subconfluent cultures of SK-N-SH cells in 25-cm2 flasks were first exposed at 37°C for 2 h to 10 PFU of recombinant baculovirus per cell and then to either 100 PFU equivalent of gD−/− or 10 PFU of gD−/+ mutant HSV-1 viruses per cell. The cultures were then maintained for an additional 24 h at 37°C in medium containing 2.5 mM sodium butyrate.
DNA fragmentation assay.
The assays for fragmentation of cellular DNA were described elsewhere (47).
Fusion assay.
Subconfluent cultures of BHK or 143TK− cells grown on glass coverslips in 24-well plates were transfected with a DNA mixture containing 80 ng each of the HSV1-glycoprotein-expressing vectors and the pcDNA3.1(−) Myc-His/Lac vector (Invitrogen) for constitutive expression of β-galactosidase. The total amount of DNA transfected per well was kept at 400 ng by addition of empty MTS vector DNA. Transfections were performed with Lipofectamine (Life Technologies) according to the manufacturer's instructions. After incubation at 37°C for 20 h, the transfection mixture was replaced with fresh medium; cells were incubated for an additional 30 h and then fixed with 0.2% glutaraldehyde and 0.2% paraformaldehyde in phosphate-buffered saline.
Syncytia were detected by light microscopy observation of β-galactosidase-expressing cells after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) with a Leitz microscope equipped with a Kodak DC120 photocamera and Kodak Digital Science 1D LE 3.0 software. The percentage of cells expressing β-galactosidase was quantified by means of the Photoshop Histogram program as detailed previously (13). In each case, the value obtained for a replicate sample containing only gD or the Myc-His/Lac vector was subtracted. All mutants were assayed in three independent experiments with two different plasmid DNA preparations.
Binding of soluble nectin1-Fc to mutant gD-expressing cells.
Binding was measured by cell enzyme-linked immunosorbent assay. Briefly, BHK cells were transfected in 24-well trays with 300 ng of DNA from each gD mutant. At 24 or 48 h after transfection, cells were blocked for 30 min at room temperature with 10% fetal bovine serum diluted in phosphate-buffered saline, incubated for 60 min at 4°C with nectin1-Fc (0.6 ng/μl), and fixed with 4% paraformaldehyde. Fixed cells were sequentially incubated with biotinylated anti-human immunoglobulin G (Sigma), extravidin-horseradish peroxidase (Sigma), and a substrate solution containing o-phenylenediamine (Sigma). The reaction was stopped with sulfuric acid, and the optical density was read at 490 nm in a Bio-Rad microplate reader. Each sample was run in duplicate.
RESULTS
Experimental design of antiapoptotic studies.
An earlier report from one of our laboratories showed that the gD ectodomain was both required and sufficient to block apoptosis induced by gD−/+ virus, whereas full-length gD (or two gD fragments reconstituting a full-length gD) was required to block apoptosis induced by gD−/− virus (47, 49). In order to fine map the functional region of the gD ectodomain required to block apoptosis, we constructed 19 recombinant baculoviruses encoding a gD gene which carried the linker-insertion or linker-insertion-deletion mutations reported in Table 1. In this system, the gD gene is under the control of the cytomegalovirus promoter and is the only gene expressed by recombinant baculoviruses in mammalian cells. The mutation sites covered the major structural and functional regions of gD. The recombinant baculoviruses were tested for the ability to block apoptosis induced by either gD−/+ and gD−/− virus stocks by infecting SK-N-SH cells with 10 PFU of recombinant baculovirus 2 h prior to infection with HSV.
The region responsible for blocking apoptosis induced by gD−/+ maps to three discontinuous sites.
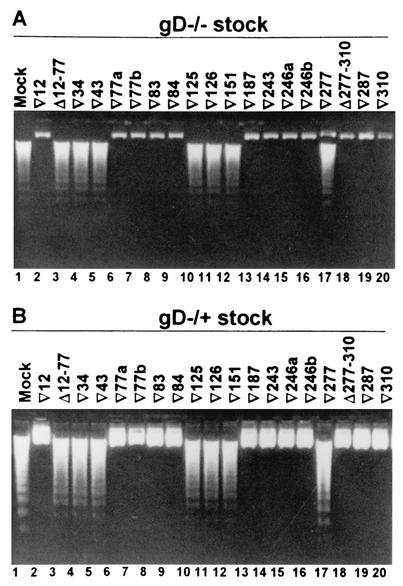
SK-N-SH cells infected with recombinant baculoviruses were infected with 100 PFU equivalents of gD−/− virus per cell, harvested 24 h later, and examined for the presence of fragmented DNA. The results reported in Fig. 1A and summarized in Table 2 show that the gD mutations that enabled apoptosis mapped at three sites, (i) Δ12-77, ▿34, and ▿43, (ii) ▿125, ▿126, and ▿151, and (iii) ▿277. These mutations fall into functional regions I, II, and IV, which correspond to structures outside the immunoglobulin core, within the core, and downstream of the core, respectively. Of the two deletion mutants (Δ12-77 and Δ277-310), only the former failed to block apoptosis.
FIG. 1.
Agarose gel containing electrophoretically separated DNA from cells infected first with recombinant baculoviruses expressing linker-insertion and linker-insertion-deletion mutant gDs and then with the gD−/− and gD−/+ virus stocks. Replicate cultures of subconfluent SK-N-SH cells in 25-cm2 flasks were infected with 10 PFU of the indicated recombinant baculovirus per cell for 2 h, and then the cells were exposed to 100 PFU equivalents of gD−/− (A) or 10 PFU of gD−/+ (B) virus stock per cell. The cells were harvested at 24 h after gD−/+ infection and processed as described in Materials and Methods.
TABLE 2.
Effect of mutations on antiapototic and fusion activity of gD
| Sequen- tial no. | Mutation | Apoptosis inhibition | BHK fusion | 143TK− fusion | Binding to nectin1 | % Comple- mentationa |
|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | 100 | |
| 3 | ∇12 | + | + | + | ± | 96 |
| 5 | Δ12-77 | − | − | − | − | ND |
| 8 | ∇34 | − | − | − | − | 2 |
| 9 | ∇43 | − | − | − | − | 9 |
| 12 | ∇77a | + | + | + | − | 16 |
| 13 | ∇77b | + | − | − | − | 29 |
| 16 | ∇83 | + | + | + | + | 77 |
| 17 | ∇84 | + | + | + | + | 115 |
| 20 | ∇125 | − | + | ± | ± | 21 |
| 21 | ∇126 | − | + | ± | ± | 17 |
| 25 | ∇151 | − | − | − | − | 0 |
| 31 | ∇187 | + | − | − | − | 79 |
| 37 | ∇243 | + | − | − | − | 3 |
| 39 | ∇246a | + | − | − | − | 1 |
| 40 | ∇246b | + | + | + | + | 1 |
| 42 | ∇277 | − | + | − | ± | 44 |
| 43 | Δ277-310 | + | − | − | + | 0 |
| 44 | ∇287 | + | + | + | + | 59 |
| 48 | ∇310 | + | + | + | + | 105 |
Data from reference 10. ND, not determined.
gD domains required to block apoptosis induced by gD−/+ cannot be differentiated from those required to block apoptosis by gD−/− virus.
In this series of experiments, the experimental design was similar to that described above except that cells were exposed to 10 PFU of gD−/+ virus per cell. The results of the DNA fragmentation assay are shown in Fig. 1B. A summary of the results is reported in Table 2 and schematically represented against the crystal structure of gD in Fig. 5. The results indicate that the gD mutants ineffective in blocking the apoptosis induced by the gD−/+ virus were also ineffective in blocking apoptosis induced by the gD−/− virus.
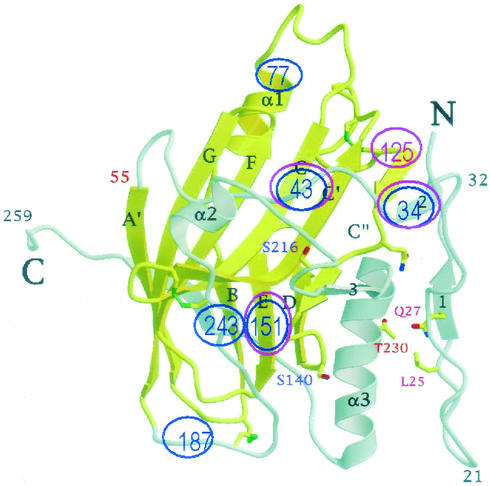
FIG. 5.
Schematic backview of the crystal structure of gD (from reference 9) in which the mutations that block apoptosis and those that affect cell-cell fusion have been highlighted in fuchsia and in blue, respectively.
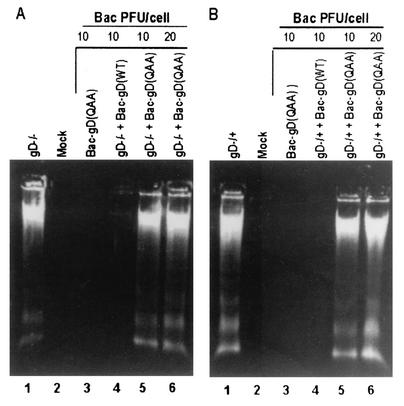
Triple mutant gD(QAA), which lacks N-glycosylation sites, failed to protect SK-N-SH cells from the apoptosis induced by gD−/− and gD−/+ viruses.
An earlier report showed that the cation-independent mannose-6 phosphate receptor was involved in the apoptosis pathway induced by gD deletion virus (47). To test the hypothesis that gD specifically interacts with the cation-independent-mannose-6 phosphate receptor through its phosphorylated mannose residues, we tested the ability of the triple mutant gD(QAA) (42) to block apoptosis induced by either gD−/− or gD−/+ mutant stocks. The triple mutant carries amino acid substitutions that preclude N-linked glycosylation. SK-N-SH cells were exposed to 10 PFU or 20 PFU of gD(QAA) recombinant baculovirus per cell and held for 2 h before infection with 100 PFU of gD−/− (Fig. 2A) virus or 10 PFU of gD−/+ (Fig. 2B) virus per cell. The results of the DNA fragmentation assay showed that the triple mutant gD(QAA) failed to block apoptosis induced by either the gD−/− or gD−/+ virus (Fig. 2A and B, lanes 5 and 6), consistent with the expectation that nonglycosylated, forms of gD lacking mannose residues would not interact with the mannose-6 phosphate receptor.
FIG. 2.
Agarose gel containing electrophoretically separated DNA from cells infected with recombinant baculoviruses expressing mutant gD(QAA) and then with the gD−/− and gD−/+ virus stocks. Subconfluent SK-N-SH cells in 25-cm2 flasks were infected with the indicated amount of recombinant baculovirus per cell for 2 h. Then the cells were exposed either to 100 PFU equivalents of gD−/− virus stock per cell (A) or to 10 PFU of gD−/+ virus stock per cell (B). The cells were harvested at 24 h after gD−/+ infection and processed as described in Materials and Methods.
Experimental design of cell-cell fusion assay.
In this series of experiments, the gD linker-insertion and linker-insertion-deletion mutants were assayed for their ability to induce cell-cell fusion. As indicated in the introduction, cells cotransfected with gD, gB, gH, and gL expression plasmids give rise to polykaryocytes provided that the target cells express one of the gD receptors. For the cell-cell fusion assay, the glycoproteins were expressed from plasmids rather than from the baculoviruses because the latter induced the highest extent of cell fusion (E. Avitabile and G. Campadelli-Fiume, unpublished results). In order to quantify the number of cells recruited into polykaryocytes, a plasmid encoding a lacZ gene under the control of the cytomegalovirus promoter was cotransfected together with the glycoprotein-encoding plasmids. Upon staining with X-Gal, the polykaryocytes acquire a green stain. Micrographs were taken of each sample, and the areas of the monolayers with green staining corresponding to polykaryocytes were quantified by Photoshop Istogram software as detailed in Materials and Methods.
Cell-cell fusion mediated by gD mutants.
The cell-cell fusion assay was performed in three different cell lines, BHK, human 143TK−, and SK-N-SH. BHK cells express a hamster homolog of human nectin1, 143 cells express human nectin1, and SK-N-SH express both human nectin1 and HveA (14, 29, 47). We observed that different cells lines exhibit strong variations in the ability to undergo fusion when cotransfected with wild-type gD and gB, gH, and gL. Thus, BHK cells give rise to massive polykaryocytes, whereas those formed in 143TK− cells contain few nuclei. Because of their small size, the polykaryocytes in 143TK− cells were scored as either absent (−) or present (+). The results with 143TK− cells were therefore scored following microscopic observations by three independent investigators.
SK-N-SH cells proved resistant to glycoprotein expression following transfections. We tried to overcome this obstacle by expressing the four glycoproteins through baculovirus infection and treatment with 2 mM sodium butyrate. In this way, the four glycoproteins were expressed, but we could only observe very tiny syncytia. A comparative study of the gD mutants could not be carried out in these cells.
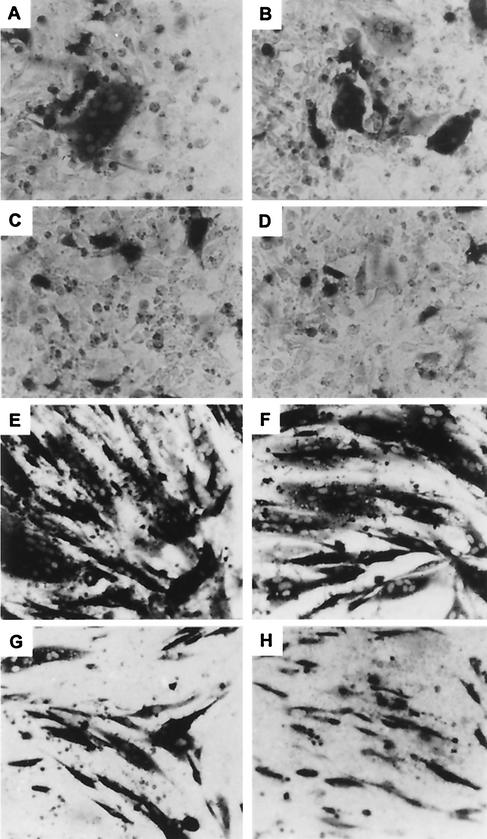
Representative results with BHK and 143TK− cells are shown in Fig. 3A to D for 143TK− cells and Fig. 3E to H for BHK cells. Quantitative results of a typical experiment with BHK cells are shown in Fig. 4A, summarized in Table 2, and schematically represented against the crystal structure of gD in Fig. 5. Notwithstanding the differences in the sizes of polykaryocytes formed in the two cell lines, there was an overall correspondence between the fusion ability of the gD mutants in BHK and 143 cells. Some minor discrepancies may reflect differences in the inherent composition of the plasma membranes that are also reflected in the size of the polykaryocytes and/or the ease of detection of polykaryocytes in BHK cells. The fusion activity was arbitrarily considered positive when it accounted for 25% of that attained with wild-type gD (dashed line in Fig. 4A). The samples whose syncytia constituted between 15 and 25% of the areas of syncytia obtained with wild-type gD were marked as ±. Overall, mutations at four sites disrupted the ability to form polykaryocytes dependent on the interaction of gD with human or hamster nectin1. They mapped at residues 34 and 43, located upstream of the core region; residues 77 and 151, within the immunoglobulin core; and residues 187, 243, and 246 downstream of it (Fig. 5). The former mutations (34 and 43) coincide with those that cause inability to block apoptosis, whereas the latter (77b, 187, 243, and 246d) do not (with the exception of mutant 151). Both deletion mutants (Δ12-77 and Δ277-310) were ineffective in cell-cell fusion.
FIG. 3.
Representative examples of polykaryocytes in 143TK− cells (A to D) or BHK cells (E to H) induced in BHK cells. 143TK− cells were cotransfected with plasmids encoding gB, gH/gL, and a mutant gD or wild-type gD, together with a plasmid encoding lacZ. Transfected cells were stained with X-Gal. (A) Wild-type gD. (B) ▿77a. (C) ▿77b. (D) gD alone. BHK cells were cotransfected with plasmids encoding gB, gH/gL, and a mutant gD or wild-type gD, together with a plasmid encoding lacZ. Transfected cells were stained with X-Gal at 40 h after transfection. (E) Wild-type gD. (F) ▿77a. (G) ▿77b. (H) Background resulting from transfection of wild-type gD in the absence of other glycoproteins.
FIG. 4.
Quantitation of fusion activity induced in BHK cells by gD mutants (A) and quantitation of binding of mutant gDs to nectin1-Fc (B). (A) BHK cells were cotransfected with plasmids encoding gB, gH/gL, and a mutant gD or wild-type gD (gD wt), together with a plasmid encoding lacZ. Transfected cells were stained with X-Gal at 40 h after transfection. Micrographs were taken with a Leitz microscope equipped with a photocamera and Kodak Digital Science 1D LE 3.0 software. Percentage of cells expressing β-galactosidase was quantified by means of the Photoshop Histogram program, as previously detailed (13). In each case, the value obtained for a replicate sample containing only gD or the Myc-His/Lac vector was subtracted. Thin bars denote standard deviation. Samples that gave rise to syncytia whose areas were between 15 and 25% of those attained with wild-type gD were considered ±. Samples whose syncytia had an area >25% of the area obtained with wild-type gD (dashed line) were considered positive and are indicated as such in Table 2. (B) Quantitation of binding of mutant gDs to nectin1-Fc. Duplicate samples of BHK cells were transfected with 300 ng of DNA from each gD mutant. At 24 h posttransfection, cells were reacted with nectin1-Fc, followed by biotinylated anti-human immunoglobulin G and extravidin-horseradish peroxidase. Samples whose binding activity was below 25% of that attained with wild-type gD are within the limits of detection and are marked as negative in Table 2. ±, binding around the 25% cutoff value (dotted line). OD, optical density.
The gD(QAA) mutant displayed a high level of fusion activity, consistent with its ability to complement the infectivity of the ΔgD virus (42).
Binding of mutant gDs to soluble nectin1-Fc.
The experiment testing binding of mutant gDs to soluble nectin1-Fc was designed to identify the mutations that affect binding of soluble nectin1-Fc to mutant gDs and to verify whether some mutations can affect cell-cell fusion without decreasing nectin1 binding. Mutant gDs were expressed in BHK cells by transfection. Transfected cells were then incubated with soluble nectin1-Fc, generated as described before (12). The results are shown in Fig. 4B and summarized in Table 2. As none of the mutants was previously reported to be defective in cell surface expression, lack of binding to nectin1 should reflect alteration in the nectin1 binding site. Overall, the mutations that reduced binding coincided with those that reduced fusion, with two sets of exceptions. The 77a, 125, and 126 mutants strongly reduced binding without significantly affecting fusion. In this case, binding may be below the limit of detection but sufficient to mediate fusion. A remarkable difference was that observed with the Δ277-310 mutant. This deletion abolished fusion but left a significant level of binding.
DISCUSSION
gD interacts with several structurally dissimilar proteins. In addition to the two entry receptors (HveA and nectin1), several lines of evidence indicate that gD interacts with the cation-independent mannose-6 phosphate receptor (4, 48) and at least one of the complex of glycoproteins responsible for the fusion of the envelope with the plasma membrane (2, 45). An additional complexity of the interaction of gD with its ligands is that at least in the case of nectin1, gD is predicted to bind the protein both in cis, i.e., both located on the same membrane, and in trans, i.e., the proteins juxtaposed but on different membranes. The development of a molecular model of gD function requires both the building of a structure-function map of gD and resolution of the crystal structure of gD, alone or in complex with various proteins with which it is predicted to interact. This report focuses on the domains required to block apoptosis and the fusion of the membranes of adjacent cells.
As reported elsewhere, gD−/− and gD−/+ viruses differ in the minimal structure of gD that is delivered in trans and required to block apoptosis. Whereas gD−/− stocks require a protein comprising all of the sequences of gD, albeit not in a single polypeptide, only the ectodomain of gD is required to block the apoptosis induced by gD−/+ mutant stocks (46-49). Notwithstanding these differences, the mutations that resulted in failure to block apoptosis by gD−/− stocks also failed to block apoptosis by gD−/+ stocks. The observations that gD−/− and gD−/+ stocks require identical domains to block apoptosis support the conclusion that gD blocks apoptosis by interacting with the same cellular proteins but, as described elsewhere, in two different compartments of the infected cell (47-49).
It is noteworthy that these mutations were located at three positions, one outside the immunoglobulin core region and located in the connector region between the hairpin and the immunoglobulin core (key residues 34 and 43), the second within the immunoglobulin core region (residues 125, 126, and 151), and the third in the carboxyl terminus of the ectodomain (residue 277). Of these, the insertions at residues 34, 43, and 151 affected all assayed functions of gD and their effects most likely reflect a detrimental effect on the structure of gD and not a site required for a specific interaction of gD with a cellular protein involved in apoptosis. Thus, residue 151 falls into β-strand E of the immunoglobulin core (Fig. 5). This insertion is likely to disrupt the β-strand itself, to perturb its bonds with the adjacent β-strands, and thus to affect the structure of the immunoglobulin core. The antiapoptotic domain therefore appears to involve residues or regions around residues 125 to 126 and 277.
Several lines of evidence reported elsewhere indicate that gD interacts with the mannose-6 phosphate receptor to block apoptosis (4, 48, 50). One requirement for this interaction is that gD must be at least glycosylated. In this report we show that a gD mutant in which all three N-glycosylation sites were mutated failed to block apoptosis while maintaining the competence to mediate cell fusion and nectin1 binding, two properties consistent with its ability to complement the infectivity of ΔgD virus (42). This observation is consistent with the idea that gD and the mannose-6 phosphate receptor colocalize and interact in the endosomal compartment and that this interaction is necessary to block apoptosis resulting from release of lysosomal enzymes (3, 4, 48).
The results of the cell-cell fusion in hamster (BHK) and human (143 and SK-N-SH) cells showed remarkable differences in the extent to which different cell lines were prone to form polykaryocytes, including the number of cells recruited into average-sized polykaryocytes. This in turn may affect the readout of the results, as mutant gDs with reduced ability to form polykaryocytes may appear to be negative in a cell line with an innate aversion to polykaryocytosis. This conclusion is in accord with observations published earlier to the effect that with the exception of a few virus strains, (e.g., the MP strain), the formation of polykaryocytes by syn mutant strains is cell type dependent (40). Irrespective of the innate differences, the effects of gD mutations on cell-cell fusion in BHK and 143TK− cells did not differ substantially (Table 2).
All mutant gDs were previously shown not to be defective in cell surface expression (10). Taking into account that the mutations likely to reflect specific interactions are those that did not simultaneously affect all the measured functions of gD, the mutations affecting fusion were located in the additional α1 helix present within the immunoglobulin core (residue 77), in the core downstream region that folds back towards the N terminus and carries the α3 helix (residues 187, 243, and 246), and in the Δ277-310 mutant. In particular, residues 77 and 187 appear to be located in unstructured, highly exposed regions of gD (Fig. 5) that may represent convenient sites of contact. In turn, the sites involved in fusion may reflect two sets of interactions, those between the receptor (nectin1) and gD and those between gD alone (or the receptor-gD complex) and the set of proteins involved in the cell-cell fusion.
We observed an overall agreement between the mutations that disrupted fusion activity and those that reduced binding to nectin1, with two exceptions. In those cases where binding was low but fusion was unaffected (mutations 77a, 125, 126, and 277), it is likely that binding was low or below the limits of detection but sufficient to mediate fusion. This is similar to what was observed with murine nectin1, a receptor competent to mediate HSV entry into cells, although its binding to gD is, at best, very weak (26-28). The dissociation between fusion and binding was observed with the Δ277-310 mutant, which was capable of binding but incapable of fusion. One hypothesis that could explain this result is that mutagenesis of this site enables attachment but affects a site involved in a postattachment function required for cell-cell fusion. This hypothesis is consistent with several lines of evidence indicating that the C-terminal portion of the ectodomain carries epitopes recognized by neutralizing monoclonal antibodies and is important for the ability of soluble gD to compete with virus entry (10, 31, 32, 46). More specifically, earlier studies have shown that the mutant lacking residues 277 to 310 has a very high affinity for both HveA and nectin1.
It can be seen from Table 2 that the mutations that hampered fusion activity and/or nectin1 binding generally reduced virus entry. Two exceptions were ▿187 and ▿246a, which in some respects was no surprise. They may reflect the fact that fusion and infectivity were assayed in different cellular systems, implying that the receptor interacting with gD as well as the fusion factors provided by the cells differed between the two systems. Furthermore, the cell-cell fusion assay mirrors virus entry but is not necessarily an identical process and is not subject to the control exerted by additional viral proteins. Thus, while all fresh isolates by definition enter cells, few if any form polykaryocytes in cells in culture. Failure to fuse cells may reflect different structural requirements, since syn mutations map to several different glycoproteins. Altogether, these results highlight differences in the structural requirements for fusion activity and for virus entry that may help in dissecting the two phenomena.
In essence, our data highlight three functional domains in gD located at nonoverlapping regions, one antiapoptotic domain, one nectin1-binding domain involved in gD fusion, and one site involved in fusion independent of nectin1-binding activity, likely at a post-nectin1-binding step. The additional independent domain characterized in previous studies is that involved in HveA interaction, located at the N terminus of the molecule (15, 16, 46). Interestingly, most of the mutations that affect functional domains lie predominantly outside the immunoglobulin core structure. The model that emerges from our studies is that the immunoglobulin core provides a scaffold for the molecule and binding sites for the proteins with which gD interacts lie predominantly in additional structural domains of gD.
Acknowledgments
We thank Roselyn Eisenberg and Gary Cohen (University of Pennsylvania) for the generous gift of plasmids encoding the mutant gDs. We thank Elisabetta Romagnoli for invaluable help with cell cultures.
The studies done at the University of Chicago were aided by grants from the National Cancer Institute (CA78766, CA71933, CA83939, CA87661, and CA88860), U.S. Public Health Service. The studies at the University of Bologna were supported by grants from Cofin-MUIR, FIRB, 2001 and 2002, University of Bologna.
REFERENCES
- 1.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 91:5406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 3.Brunetti, C. R., R. L. Burke, B. Hoflack, T. Ludwig, K. S. Dingwell, and D. C. Johnson. 1995. Role of mannose-6-phosphate receptors in herpes simplex virus entry into cells and cell-to-cell transmission. J. Virol. 69:3517-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunetti, C. R., K. S. Dingwell, C. Wale, F. L. Graham, and D. C. Johnson. 1998. Herpes simplex virus gD and virions accumulate in endosomes by mannose 6-phosphate-dependent and -independent mechanisms. J. Virol. 72:3330-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume, G., M. Arsenakis, F. Farabegoli, and B. Roizman. 1988. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J. Virol. 62:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume, G., S. Qi, E. Avitabile, L. Foà-Tomasi, R. Brandimarti, and B. Roizman. 1990. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J. Virol. 64:6070-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carfi, A., H. Gong, H. Lou, S. H. Willis, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. Sect. D Biol. Crystallogr. 58:836-838. [DOI] [PubMed] [Google Scholar]
- 9.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, H.-Y., G. H. Cohen, and R. J. Eisenberg. 1994. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J. Virol. 68:2529-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocchi, F., M. Lopez, P. Dubreuil, G. Campadelli-Fiume, and L. Menotti. 2001. Chimeric nectin1-poliovirus receptor molecules identify a nectin1 region functional in herpes simplex virus entry. J. Virol. 75:7987-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocchi, F., M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume. 1998. The V domain of herpesvirus immunoglobulin-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl. Acad. Sci. USA 95:15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi, F., L. Menotti, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2000. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (PRR1/HveC/HIgR) and nectin2 (PRR2/HveB). J. Virol. 74:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex viruses 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly, S. A., J. J. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC(nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 17.Fabre, S., N. Reymond, F. Cocchi, L. Menotti, P. Dubreuil, G. Campadelli-Fiume, and M. Lopez. 2002. Prominent role of the Ig-like V domain in trans-interactions of nectins: nectin3 and nectin4 bind to the predicted C-C′-C"-D β-strands of the nectin1 V domain. J. Biol. Chem. 277:27006-27013. [DOI] [PubMed] [Google Scholar]
- 18.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 19.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. M., and P. G. Spear. 1989. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J. Virol. 63:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krummenacher, C., A. H. Rux, J. C. Whitbeck, M. Ponce-de-Leon, H. Lou, I. Baribaud, W. Hou, C. Zou, R. J. Geraghty, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, M., M. Aoubala, F. Jordier, D. Isnardon, S. Gomez, and P. Dubreuil. 1998. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood 92:4602-4611. [PubMed] [Google Scholar]
- 25.Martinez, W. M., and P. G. Spear. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex virus types 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J. Virol. 76:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menotti, L., E. Avitabile, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2001. Comparison of murine and human nectin1 binding to herpes simplex virus glycoprotein D (gD) reveals a weak interaction of murine nectin1 to gD and a gD-dependent pathway of entry. Virology 282:256-266. [DOI] [PubMed] [Google Scholar]
- 27.Menotti, L., R. Casadio, C. Bertucci, M. Lopez, and G. Campadelli-Fiume. 2002. Substitution in the murine nectin1 receptor of a single conserved amino acid at a position distal from herpes simplex virus gD binding site confers high affinity binding to gD. J. Virol. 76:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menotti, L., M. Lopez, E. Avitabile, A. Stefan, F. Cocchi, J. Adelaide, E. Lecocq, P. Dubreuil, and G. Campadelli-Fiume. 2000. The murine homolog of human Nectin1δ serves as a species nonspecific mediator for entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA 97:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milne, R. S., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315-328. [DOI] [PubMed] [Google Scholar]
- 30.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 31.Muggeridge, M. I., V. J. Isola, R. A. Byrn, T. J. Tucker, A. C. Minson, J. C. Glorioso, G. H. Cohen, and R. J. Eisenberg. 1988. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, with deletion mutants and monoclonal antibody-resistant mutants. J. Virol. 62:3274-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muggeridge, M. I., T. T. Wu, D. C. Johnson, J. C. Glorioso, R. J. Eisenberg, and G. H. Cohen. 1990. Antigenic and functional analysis of a neutralization site of HSV-1 glycoprotein D. Virology 174:375-387. [DOI] [PubMed] [Google Scholar]
- 33.Nicola, A. V., C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1997. Antigenic structure of soluble herpes simplex virus (HSV) glycoprotein D correlates with inhibition of HSV infection. J. Virol. 71:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicola, A. V., M. Ponce de Leon, R. Xu, W. Hou, J. C. Whitbeck, C. Krummenacher, R. I. Montgomery, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1998. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J. Virol. 72:3595-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 37.Reymond, N., J. Borg, E. Lecocq, J. Adelaide, G. Campadelli-Fiume, P. Dubreuil, and M. Lopez. 2000. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene 255:347-355. [DOI] [PubMed] [Google Scholar]
- 38.Reymond, N., S. Fabre, E. Lecocq, J. Adelaide, P. Dubreuil, and M. Lopez. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276:43205-43215. [DOI] [PubMed] [Google Scholar]
- 39.Rux, A. H., S. H. Willis, A. V. Nicola, W. Hou, C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J. Virol. 72:7091-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruyechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satoh-Horikawa, K., H. Nakanishi, K. Takahashi, M. Miyahara, M. Nishimura, K. Tachibana, A. Mizoguchi, and Y. Takai. 2000. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J. Biol. Chem. 275:10291-10299. [DOI] [PubMed] [Google Scholar]
- 42.Sodora, D. L., G. H. Cohen, M. I. Muggeridge, and R. J. Eisenberg. 1991. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J. Virol. 65:4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, K., H. Nakanishi, M. Miyahara, K. Mandai, K. Satoh, A. Satoh, H. Nishioka, J. Aoki, A. Nomoto, A. Mizoguchi, and Y. Takai. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J. Cell Biol. 145:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gH/gL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitbeck, J. C., M. I. Muggeridge, A. H. Rux, W. Hou, C. Krummenacher, H. Lou, A. van Geelen, R. J. Eisenberg, and G. H. Cohen. 1999. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J. Virol. 73:9879-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, G., and B. Roizman. 2002. Cation-independent mannose 6-phosphate receptor blocks apoptosis induced by herpes simplex virus 1 mutants lacking glycoprotein D and is likely the target of antiapoptotic activity of the glycoprotein. J. Virol. 76:6197-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, G., and B. Roizman. 2001. The domains of glycoprotein D required to block apoptosis depend on whether glycoprotein D is present in the virions carrying herpes simplex virus 1 genome lacking the gene encoding the glycoprotein. J. Virol. 75:6166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, Y., B. Doray, A. Poussu, V. P. Letho, and S. Kornfield. 2001. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292:1716-1718. [DOI] [PubMed] [Google Scholar]