Abstract
The hepatitis C virus (HCV) serine protease is necessary for viral replication and represents a valid target for developing new therapies for HCV infection. Potent and selective inhibitors of this enzyme have been identified and shown to inhibit HCV replication in tissue culture. The optimization of these inhibitors for clinical development would greatly benefit from in vitro systems for the identification and the study of resistant variants. We report the use HCV subgenomic replicons to isolate and characterize mutants resistant to a protease inhibitor. Taking advantage of the replicons' ability to transduce resistance to neomycin, we selected replicons with decreased sensitivity to the inhibitor by culturing the host cells in the presence of the inhibitor and neomycin. The selected replicons replicated to the same extent as those in parental cells. Sequence analysis followed by transfection of replicons containing isolated mutations revealed that resistance was mediated by amino acid substitutions in the protease. These results were confirmed by in vitro experiments with mutant enzymes and by modeling the inhibitor in the three-dimensional structure of the protease.
Despite the introduction of blood-screening tests ∼10 years ago, hepatitis C virus (HCV) is still the major cause of blood-borne chronic hepatitis, with nearly 200 million infected people worldwide. HCV infection often evolves into a chronic disease, which can lead to liver dysfunction and hepatocellular carcinoma. Current therapeutic regimens based on alpha interferon (IFN-α) and the nucleoside analog ribavirin are only partially effective and are limited by the adverse effects of both agents (50). Given the high prevalence of this disease, developing new treatments is a major public health objective. Similarly to human immunodeficiency virus (HIV) research, most efforts to develop antiviral agents for HCV have focused on the inhibition of key viral enzymes, serine protease, helicase, and polymerase (2).
The most extensively studied HCV target has been the NS3-4A serine protease, a heterodimeric enzyme comprising the N-terminal domain of the NS3 protein (amino acids 1 to 180) and the small hydrophobic NS4A protein (3). This protease cleaves the viral polyprotein at four junctions (NS3/NS4A, NS5A/NS5B, NS4A/NS4B, and NS4B/NS5A), and its activity is necessary for viral replication (24). Although the NS3 protease domain possesses enzymatic activity, the 54-amino-acid NS4A protein is required for cleavage at the NS3/NS4A and NS4B/NS5A sites and increases cleavage efficiency at the NS4A/NS4B and NS5A/NS5B junctions (4, 14, 28, 47). X-ray crystallography (20, 35, 51) and nuclear magnetic resonance (NMR) spectroscopy (1, 36) have shown that the NS3-4A structure is similar to that of other chymotrypsin-like serine proteases, with two domains, both composed of a β-barrel and two short α-helices. The catalytic triad comprises histidine 57, aspartate 81, and serine 139 and is located between the two domains. The central region of NS4A is an integral part of the amino-terminal domain and forms the seventh strand of an eight-stranded β-barrel. Comparison of the NS3 protease structures in the presence and in the absence of NS4A suggested that NS4A stabilizes the N-terminal domain of the protease, thus optimizing the orientation of the catalytic triad. A characteristic feature of NS3 is the presence of a structural zinc ion that is coordinated tetrahedrally by three cysteines and a histidine residue at a site located opposite the active site (12, 20, 35, 51). Though the NS3 protease domain is covalently attached to an RNA helicase possessing ATPase activity, the helicase domain is not required for optimal protease activity. Both domains can be expressed in isolation as fully active and stably folded proteins. In line with functional studies, the X-ray structure of the full-length NS3 protein showed that the protease and helicase domains are segregated and connected by a single strand (52).
NS3-4Ap specificity has been defined by identification (17, 44) and mutagenesis (5, 23, 49, 53) of the natural cleavage sites and selection of optimized cleavage sites using peptide libraries (21, 41). The NS3/NS4A junction is cleaved in cis and tolerates substitutions at all positions except P1, where a threonine residue is found in all isolates. The other three junctions are cleaved in trans and contain a cysteine residue at the P1 position. Efficient in vitro cleavage requires a peptide substrate of at least 10 residues spanning P6 to P4′ and, besides P1, residues at positions P6, P3, P2, P1′, and P4′ contribute to efficient substrate recognition. This requirement for large peptide substrates has been rationalized on the basis of structural information and modeling. Compared to other serine proteases, NS3-4Ap lacks several surface loops that form the N-terminal substrate-binding cleft, which consequently is shallow and solvent exposed. The binding energy for the substrate is derived from a series of weak interactions that are distributed along an extended contact surface. Only the shallow and hydrophobic S1 specificity pocket favorably accommodates the P1 cysteine. In addition, a hydrophobic cavity formed by NS4A and the NS4A binding region can accommodate the hydrophobic amino acid at the P4′ position of the substrate.
The development of potent competitive inhibitors of NS3-4Ap has been hampered by the structure of its active site: the absence of well-defined P2, P3, and P4 binding sites made the design of low-molecular-weight competitive inhibitors a daunting job (for a review, see reference 46). Nevertheless, the discovery of peptide inhibitors some 5 years ago opened a new avenue for drug discovery. These inhibitors were identified based on the observation that NS3-4Ap was competitively inhibited by the N-terminal cleavage products, which had affinities for the enzyme higher than those of the corresponding substrates (31, 45). These inhibitors have been subsequently optimized by progressive trimming of the original hexapeptides to tetra- or tripeptides, in parallel with replacing the side chains with unnatural substituents that significantly increased affinity for the enzyme (19, 29, 30, 38; M. D. Bailey, J. Bordeleau, C. Brochu, D. Cameron, M. Cartier, J.-S. Duceppe, A.-M. Faucher, G. Fazal, E. Ghiro, V. Gorys, N. Goudreau, S. Goulet, C. Grand-Maitre, T. Halmos, S. LaPlante, M. Marquis, R. Maurice, M. Poirier, M.-A. Poupart, J. Rancourt, B. Simoneau, D. Thibeault, Y. Tsantrizos, D. Wernic, D. Lamarre, and M. Llinas-Brunet, Abstr. Pap. Am. Chem. Soc. 220, abstr. MEDI-088, 2000). More recently, further structure-activity studies have yielded potent and selective inhibitors that begin to have properties suitable for a drug (48). This progress has fostered the hope that these inhibitors may in the near future be used to treat HCV-infected patients. In fact, clinical trials with an NS3-4Ap inhibitor, presumably derived from these peptidic leads, have recently been announced. Nonetheless, the successful clinical use of protease inhibitors still has to overcome the serious hurdle of the emergence of resistant HCV variants. Based on the HIV experience and given the high mutation rate observed for HCV, it is highly likely that viruses expressing a mutant protease with decreased inhibitor sensitivity could emerge during therapy. Because of this potential limitation, it is important that the preclinical development of NS3-4Ap inhibitors be accompanied by in vitro studies of their resistance profiles.
In this study, we describe for the first time the in vitro selection and characterization of HCV variants resistant to compound 1, a tripeptide inhibitor of NS3-4Ap (Fig. 1) (Bailey et al., Abstr. Pap. Am. Chem. Soc. 220). In the absence of an efficient tissue culture infection system for HCV, we have used a subgenomic HCV RNA (replicon) containing the neomycin phosphotransferase gene (Neo) in place of the viral structural gene. Lohmann and colleagues first demonstrated that these replicons replicated in Huh-7 cells, rendering the host cell resistant to neomycin sulfate (G418) and allowing the selection of G418-resistant cell clones (34). We have exploited this feature to select replicons resistant to compound 1. Characterization of these replicons indicated that specific mutations in the NS3 protease domain were important in conferring resistance to this compound.
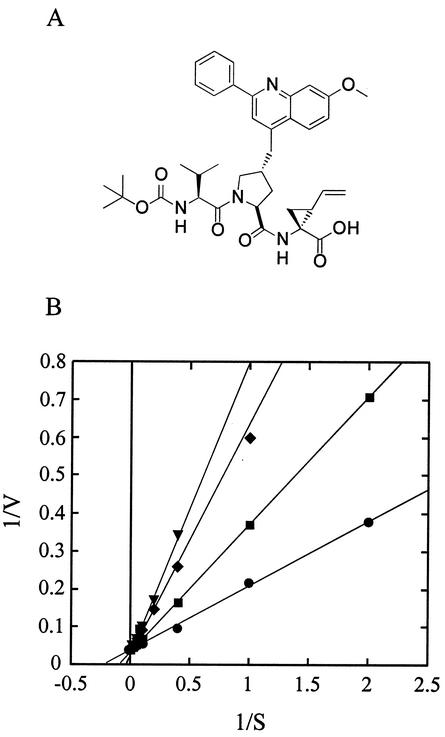
FIG. 1.
Structure (A) and mode of inhibition (B) of compound 1. (B) Reactions were performed as described in Materials and Methods with various substrate concentrations in the absence (•) or in the presence of 5 (▪), 10 (♦), and 20 (▾) nM compound 1. The data were fitted to a competitive mechanism. Plotting the slopes of the lines of the double-reciprocal plot yielded a Ki of 6.5 nM.
MATERIALS AND METHODS
Manipulation of nucleic acids and construction of recombinant plasmids.
Plasmids were assembled by standard restriction digestion protocols or by PCR amplification of the sequence of interest with primers containing the desired mutations. Plasmid DNA was prepared from overnight culture in Luria-Bertani broth using Qiagen 500 columns according to the manufacturer's instructions. The sequences of all plasmids were verified by automated sequencing using an ABI instrument. Plasmid pHCVNeo17.wt (37) was assembled by several subcloning steps and contains the cDNA coding for an HCV bicistronic replicon identical to replicon I377neo/NS3-3′/wt described by Lohmann et al. (34) (EMBL-GenBank no. AJ242652) under the control of a T7 promoter. All other plasmids for in vitro transcription were identical to pHCVNeo17.wt but contained the following mutations: (i) pHCVNeo17.A, insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nucleotides [nt] 4840 to 4842) coding for valine 67 in NS5A; (ii) pHCVNeo17.B, replacement of triplet GAA (nt 2329 to 2331) coding for glutamic acid 176 in NS3 protein with GGA coding for glycine and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; (iii) pHCVNeo17.C, replacement of triplet GAA (nt 2329 to 2331) coding for glutamic acid 176 in NS3 protein with GGA coding for glycine; (iv) pHCVNeo17.RRX1, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GCC coding for alanine; (v) pHCVNeo17.RRX2, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with TAC coding for tyrosine; (vi) pHCVNeo17.RRX3, substitution of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GTC coding for valine; (vii) pHCVNeo17.RRA1, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GCC coding for alanine and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; (viii) pHCVNeo17.RRA2, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with TAC coding for tyrosine and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; (ix) pHCVNeo17.RRA3, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GTC coding for valine and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; (x) pHCVNeo17.RRB1, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GCC coding for alanine, replacement of triplet GAA (nt 2329 to 2331) coding for glutamic acid 176 in NS3 protein with AAA coding for lysine, and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; (xi) pHCVNeo17.RRB2, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with TAC coding for tyrosine, replacement of triplet GAA (nt 2329 to 2331) coding for glutamic acid 176 in NS3 protein with GGA coding for glycine, and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A; and (xii) pHCVNeo17.RRB3, replacement of triplet GAC (nt 2305 to 2307) coding for aspartic acid 168 in NS3 protein with GTC coding for valine, replacement of triplet GAA (nt 2329 to 2331) coding for glutamic acid 176 in NS3 protein with GGA coding for glycine, and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842) coding for valine 67 in NS5A.
Plasmids pETNS3/4A(K) (16) and pETNS3/4C.wt contain the sequence coding for the full-length NS3-NS4A polypeptide precursor (amino acids 1027 to 1711) of the BK and Con1 strains of HCV, respectively, cloned between the NdeI and HindIII restriction sites of pET14b expression vector (Novagen). Cysteine 54 of NS4A was changed to glycine, and the sequence coding for three lysine residues was added downstream of this residue. Furthermore, a cysteine-to-serine mutation was introduced at position 428 of NS3 to prevent autolytic cleavage in the NS3 helicase domain. Plasmids pET3/4C.D168Y and pET3/4C.D168V are identical to pET3/4C.wt, except that the triplet GAC coding for aspartate 168 of the NS3 protein (amino acid 168 of the HCV polyprotein) was changed to TAC, coding for tyrosine, or GAC, coding for valine.
In vitro transcription and RNA transfection.
Huh-7 cells were kindly provided by Ralf Bartenschlager (University of Mainz, Mainz, Germany) and grown in Dulbecco's modified minimal essential medium (DMEM; Gibco BRL) supplemented with 10% fetal calf serum (FCS). HBI10 cells were cultured as described above, but the medium was supplemented with 0.8 mg of G418 (Gibco BRL)/ml. For routine work, cells were passaged twice a week at a dilution of 1 to 5, using 1× trypsin-EDTA (Gibco BRL). pHCVNeo17 plasmids were digested with the ScaI endonuclease (New England Biolabs) and transcribed in vitro with the T7 Megascript kit (Ambion). Transcription mixtures were treated with DNase (0.2 U/ml) to completely remove template DNA, extracted and precipitated as described previously (34), and resuspended with phosphate-buffered saline (PBS). RNA transfection was performed as described previously (34).
Sequence analysis of replicon RNAs.
Total RNA was extracted from parental and inhibitor-resistant cells using the RNeasy miniKit (Qiagen) according to the instructions. Replicon RNA was retrotranscribed using the antisense oligonucleotide HCVG36 (5′-GACCTTTCACAGCTAGCCGT-3′) and the Superscript II reverse transcriptase (Gibco BRL) according to the manufacturer's instructions and subsequently digested with 2 U of RNase H (Gibco BRL)/ml. The cDNA coding for the NS3-4A region was amplified by PCR using the oligonucleotides CITE3 (5′-TGGCTCTCCTCAAGCGTATTC-3′) and B52 (5′-CAGGTCCTCGGTGGAGGGCATCT-3′) and the LA Taq DNA polymerase (Takara Laboratories, Otsu, Japan). The cDNA coding for the entire nonstructural (NS) region was amplified as described above with the oligonucleotides CITE3 and RBA11 (5′-TCAGCTTACAGGCTTCCTCCAC-3′). To distinguish replicon mutations from those that could have arisen from the amplification procedure, at least eight independent reverse transcription-PCRs were performed for each cell clone, and the PCR products were pooled. Amplified cDNAs were purified using the QIAquick PCR purification kit (Qiagen), and the nucleotide sequences were determined by automated sequencing using an ABI instrument. The deduced amino acids sequences were aligned using VectorNTI software.
Antibodies.
The 10E5/24 anti-NS3 mouse monoclonal antibody (MAb) and the 55VI anti-NS5A rabbit antiserum were described previously (37).
Immunoblotting.
Samples were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (0.3 M Tris-Cl [pH 8.8], 2.5% SDS, 0.1 M dithiothreitol [DTT], 0.001% bromophenol blue), heated for 15 min at 55°C, and loaded on SDS-10% PAGE. Proteins were electrophoretically transferred to nitrocellulose filters, which were then incubated with primary antibodies diluted in the blocking buffer, followed by alkaline phosphatase-conjugated secondary antibodies. After being washed, bound antibodies were detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate.
Metabolic labeling and immunoprecipitation.
Cells (106) were plated in 10-cm-diameter tissue culture dishes. After 24 h, the cells were starved for 1 h with medium lacking methionine and cysteine and supplemented with 1% dialyzed fetal bovine serum and labeled with 35S-labeled amino acids (Easytag; Amersham, Little Chalfont, United Kingdom) for 30 min at 37°C. For pulse-chase labeling, cells were plated and labeled as described above for 15 min at 37°C and then chased for the appropriate time with DMEM supplemented with 10% FCS, 10× amino acids, and 50 μg of cycloheximide/ml. The cell monolayers were washed once with PBS and lysed by incubating them for 5 min at room temperature with 0.4 ml of immunoprecipitation buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA) supplemented with 1% SDS. The lysates were collected with the help of a disposable scraper, passed through a Qiashredder (Qiagen) to reduce viscosity, heated for 5 min at 95°C, diluted with immunoprecipitation buffer, and supplemented with Triton X-100 to adjust the final detergent concentration to 0.25% SDS-1% Triton X-100. The lysates were then clarified by centrifugation for 10 min at top speed in a refrigerated microcentrifuge. Immunoprecipitation was performed as described previously (40), and samples were analyzed on SDS-10% PAGE. The gel was exposed to X-ray film or to a phosphorimaging screen for quantitative analysis with the Molecular Dynamics system. The data reported (see Fig. 3, bottom) were calculated by dividing the signal of each band by the sum of the signals of all the bands in the same lane and multiplying the result by 100.
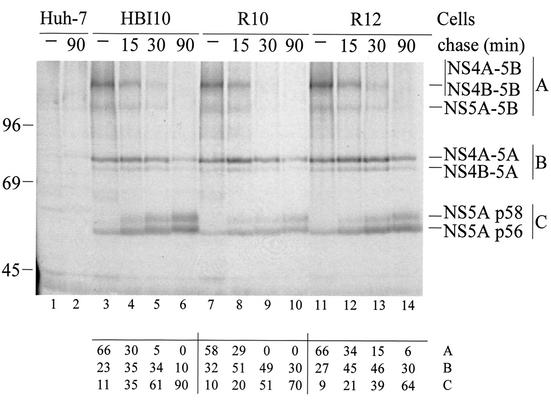
FIG. 3.
Kinetics of polyprotein processing in parental cells and clones resistant to compound 1. Huh-7 (lanes 1 and 2), HBI10 (lanes 3 to 6), R10 (lanes 7 to 10), and R12 (lanes 11 to 14) cells were pulse-labeled with 35S-labeled amino acids for 15 min at 37°C and then chased for the time indicated above each lane. The labeled proteins were immunoprecipitated with the 55IV anti-NS5A antiserum and analyzed by SDS-10% PAGE. The quantitative data below each lane were generated by PhosphorImager analysis as indicated in Materials and Methods and are expressed as percents of the total radioactivity counted in viral proteins. A is the sum of the NS4A-5B, NS4B-5B, and NS5A-5B precursors; B is the sum of the NS4A-5A and NS4B-5A precursors; C is the sum of the p56 and p58 forms of NS5A. The positions of molecular mass standards (in kilodaltons) and NS5A and uncleaved precursors are indicated.
Cell ELISA.
Cells were monitored for expression of the NS3 protein by enzyme-linked immunosorbent assay (ELISA) with the anti-NS3 MAb 10E5/24. Compounds were dissolved and serially diluted in dimethyl sulfoxide in such a way that the final dimethyl sulfoxide concentration was 1%. For the standard assay, cells were seeded in 96-well plates at a density of 104 per well in a final volume of 0.1 ml of DMEM-10% FCS. Two hours after the cells were plated, 50 μl of DMEM-10% FCS containing a 3× concentration of inhibitor was added, and the cells were incubated for 96 h at 37°C. The plates were then fixed for 10 min with ice-cold isopropanol, washed twice with PBS, blocked with 5% nonfat dry milk in PBS-0.1% Triton X-100-0.02% SDS (PBSTS), and then incubated overnight at 4°C with the 10E5/24 MAb diluted in milk-PBSTS. After being washed five times with PBSTS, the cells were incubated for 3 h at room temperature with anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Sigma) and diluted in milk-PBSTS. After being washed again as described above, the reaction was developed with p-nitrophenyl phosphate disodium substrate (Sigma), and the absorbances at 405 and 620 nm were read at intervals. Each condition was tested in triplicate, and average absorbance values were used for calculations, using data sets in which samples incubated without inhibitors had absorbance values between 0.8 and 1.5. The inhibitor concentration that reduced by 50% the expression of NS3 (IC50) was calculated with Kaleidagraph software by fitting the data to the Hill equation: fraction inhibition = 1 − (Ai − b)/(A0 − b) = [I]n/([I]n + IC50), where Ai is the absorbance value of HBI10 cells supplemented with the appropriate inhibitor (I) concentration, A0 is the absorbance value of HBI10 cells incubated without inhibitor, b is the absorbance value of Huh-7 cells plated at the same density in the same microtiter plates and incubated without inhibitor, and n is the Hill coefficient.
For transient-transfection assays, 10AIFN cells were transfected by electroporation as described above and seeded in 96-well plates at densities of 5 × 104 (plates A) or 1 × 104 (plates B) per well in a final volume of 0.1 ml of DMEM-10% FCS. Plates A were used to monitor transfection efficiency. Immediately after transfection, the samples in plates A were supplemented with IFN-α2b (Schering-Plough) at a final concentration of 100 IU/ml to block HCV replication. Plates A were fixed and processed for cell ELISA 24 h after transfection. The transfection efficiency was determined by dividing the absorbance value of the sample transfected with a given RNA by the absorbance value of mock-transfected cells. The samples in plates B were supplemented with the inhibitor and processed for cell ELISA 96 h after transfection to determine replication competence and sensitivity to the effect of the inhibitor. Replication competence was determined by dividing the absorbance value of the sample transfected with a given RNA by the absorbance value of mock-transfected cells. The values were normalized to the transfection efficiency using the data generated with the corresponding plate A. The effect of the inhibitor was determined by fitting the data to the Hill equation as described above, except that absorbance values from mock-transfected 10AIFN cells were used as background.
Northern blot hybridization.
HBI10 cells were seeded in six-well plates at a density of 5 × 105 per well in a final volume of 1 ml of DMEM-10% FCS. Twenty-four hours after the cells were plated, 1 ml of DMEM-10% FCS containing a 2× concentration of inhibitor was added, and the cells were incubated for 48 h at 37°C. Total cellular RNA was extracted as described above. Two micrograms of total RNA was electrophoresed through a 1% agarose gel containing 2.2 M formaldehyde, transferred to a Hybond+ nylon membrane (Amersham Pharmacia Biotech), and immobilized by alkaline cross-linking. Hybridization was carried out with [32P]UTP-labeled in vitro-transcribed RNA in a solution containing 50% deionized formamide, 5× SSC (750 mM sodium chloride plus 75 mM sodium citrate), 5× Denhardt's solution, 0.1% SDS, and 100 μg of sheared denatured herring sperm DNA/ml for 16 h at 65°C. The membranes were washed twice in 1× SSC-0.1% SDS for 10 min at 65°C and twice in 0.1× SSC-0.1% SDS for 30 min at 65°C. The membranes were exposed to X-ray film or to a phosphorimaging screen for quantitative analysis with the Molecular Dynamics system. The amount of HCV replicon RNA was determined with respect to serial dilutions of an in vitro transcript loaded in parallel onto the gel (not shown). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) hybridization signal was used to correct for different amounts of total RNA loaded in each lane of the gel. The resulting values were used for calculation of the IC50 with the Hill equation (see above).
Preparation of cells cured of endogenous replicon.
HBI10 cells were cultured for 11 days in the presence of 100 IU of recombinant human IFN-α2b (intron A; Schering-Plough)/ml and subsequently for 4 days in the absence of interferon. At the end of this procedure, HCV proteins and RNA could no longer be detected.
Compounds.
The synthesis of compound 1 (32), compounds 2 and 3 (38, 39), and compound 4 (10) has been described previously.
Selection of compound 1-resistant clones.
HBI10 cells were plated in 15-cm-diameter tissue culture dishes at a density of 3 × 103/cm2 and cultured in the presence of 1 mg of G418/ml and 12 μM compound. The majority of cells remained resistant to G418 and duplicated normally for ∼7 days. This lasting resistance to G418 was likely due to persistence of the neomycin phosphotransferase protein. The cells became confluent after 10 days of culture and were passed 1 to 10. Approximately 15 days after selection began, G418 resistance declined, most cells died, and small colonies of cells resistant to the inhibitor and the antibiotic became visible. Resistant colonies were isolated, expanded, and characterized.
Expression and purification of the NS3-4A complex from bacteria.
All protease complexes were produced in Escherichia coli BL21(DE3) cells and purified as described previously (16) with minor modifications. The final yield was 1 to 2 mg/liter of culture with a purity of ≥50%, as judged by SDS-PAGE. Protein stocks were stored at a concentration of 10 to 20 μM in 25 mM HEPES (pH 7.4)-1 mM EDTA-20% glycerol-3 mM DTT-0.1% dodecyl-maltoside-1 M NaCl at −80°C after being shock frozen in liquid nitrogen. The enzyme concentration was determined by active-site titration experiments using a hexapeptide α-ketoacid (38).
Peptides and high-performance liquid chromatography protease assays.
The peptide substrate NS5A-NS5B, having the sequence H-EAGDDIVPC/SMSYTWTGA-OH, was purchased from Anaspec. The concentration of stock peptide aliquots was determined by quantitative amino acid analysis performed on HCl-hydrolyzed samples. Cleavage assays were performed in 60 μl of a buffer containing 50 mM HEPES (pH 7.5), 0.15 M NaCl, 0.1% Triton X-100, 15% glycerol, and 5 mM DTT (protease activity buffer). Reactions were started by addition of the substrate to the final desired concentration. Incubation times at 23°C were adjusted to obtain <10% conversion. The reactions were stopped by the addition of 40 μl of 1% trifluoroacetic acid to the reaction mixture. Cleavage of peptide substrates was quantified by high-performance liquid chromatography using a Merck-Hitachi chromatograph equipped with an autosampler, as described previously (16). The kinetic parameters were calculated from a nonlinear least-squares fit of initial rates as a function of the substrate concentration with the aid of Kaleidagraph software, assuming Michaelis-Menten kinetics.
Structural analyses and modeling.
The structural data for the complex of compound 1 and the NS3 protease domain-NS4A cofactor NS4A were taken from the solution structure of the corresponding complex in the absence of NS4A (9). Additional information was taken from the cocrystal structures of NS3-NS4A with ketoacid inhibitors (13) and the NMR structure of tripeptide inhibitors published by the Boehringer group (26, 27). All calculations were carried out with the program BatchMin and the molecular modeling package InsightII/Discover (Biosym Technologies Inc., San Diego, Calif.).
RESULTS
Activity of compound 1.
Compound 1 is a tripeptide containing a C-terminal carboxylic acid, a vinyl-cyclopropyl substituent in P1, a 2-phenyl-8-methoxyquinoline substituent on the P2 hydroxyproline, a valine residue in P3, and an N-terminal tert-butyl carbamate capping group (Fig. 1A). It is the result of the extensive optimization of NS3-4Ap product inhibitors reported by Llinas-Brunet and colleagues and inhibits the enzyme both in vitro and in a surrogate cell-based assay (30, 32; Bailey et al., Abstr. Pap. Am. Chem. Soc. 220). Thus, it is an ideal candidate for resistance studies.
The ability of this compound to inhibit NS3-4Ap in vitro was confirmed using purified, recombinant full-length NS3-4A proteases derived from the HCV isolates Con1 and BK and a peptide substrate corresponding to the sequence of the NS5A-5B cleavage site. Both enzymes contained the cysteine 428-to-serine mutation (C428S) to prevent autolytic cleavage in the NS3 helicase domain. The Con1 enzyme also contained a replacement of glutamate 176 with glycine (E176G) that is required to enhance replication in cultured cells (see below). The Con1 enzyme showed activity indistinguishable from that of the previously described BK protease (16): for both enzymes, the Km for the substrate was 4 μM and the kcat was 30 min−1, suggesting that the E176G mutation had no effect on protease activity. Compound 1 inhibited both the BK and the Con1 enzymes, with IC50s of 12 and 10 nM, respectively. To confirm the predicted mechanism of inhibition, the activities of compound 1 on the Con1 protease were determined at different substrate concentrations. A double-reciprocal plot of the data demonstrated that compound 1 was competitive with the substrate, with a Ki of 6.5 nM (Fig. 1B).
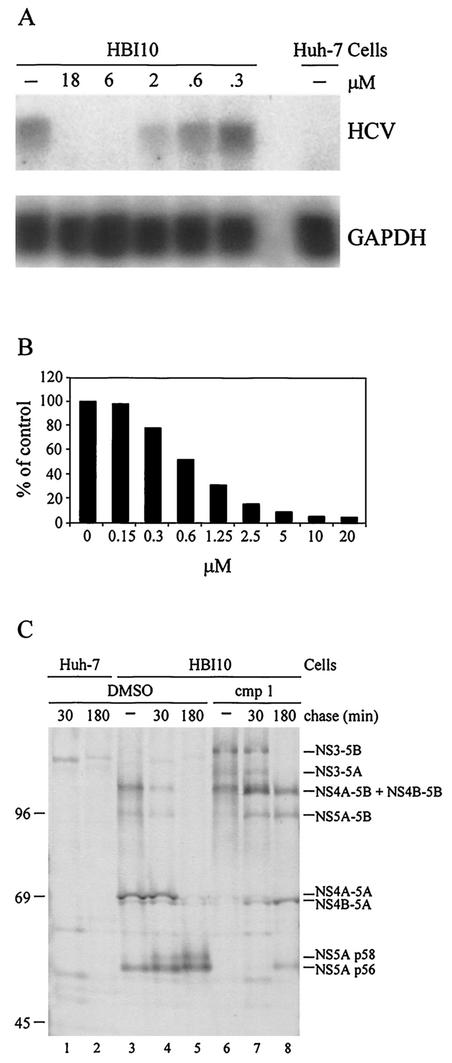
To determine the effect of compound 1 on viral replication, we used the cell clone HBI10 (37), a Huh-7 derivative hosting a selectable HCV replicon identical to replicon I377neo/NS3-3′/wt (34). The effect of compound 1 on viral replication was monitored by measuring replicon RNA by Northern blotting and measuring the NS3 protein by an ELISA performed directly on cells grown in 96-well microtiter plates (cell ELISA). Due to the different half-lives of viral RNA and proteins (reference 43 and data not shown), the cells were incubated with compound 1 for 48 h for Northern blotting experiments and for 96 h for cell ELISAs. Incubation with compound 1 resulted in a dose-dependent reduction of both viral RNA (Fig. 2A) and NS3 protein (Fig. 2B) and had no effect on the expression of the GAPDH mRNA (Fig. 2A). Quantitative analysis of these experiments indicated that compound 1 inhibited replication with an IC50 of ∼1 μM in both assays. Cytotoxicity assays and [14C]thymidine incorporation experiments showed that compound 1 was not toxic and had no effect on the cell growth rate at concentrations up to 30 μM (data not shown), thus indicating that the decrease of viral RNA and protein reflected a direct effect on viral replication. The 100-fold-reduced activity observed on viral replication compared to inhibition of the protease in vitro was not unexpected due to the peptidic nature of compound 1, and it probably reflects poor cell penetration. In addition, the level of protease activity required to sustain viral replication is not known, and therefore, it is difficult to predict the correlation between inhibition of protease and viral replication.
FIG. 2.
Effects of compound 1 on replication and polyprotein processing. (A) Viral (HCV) RNA levels present in cells treated with the indicated concentrations of compound 1 for 48 h were determined by Northern blot hybridization with a plus-strand-specific RNA probe. Hybridization with a probe specific for a cellular mRNA (GAPDH) served as a control. Quantitative data were generated by PhosphorImager analysis as indicated in Materials and Methods. (B) NS3 protein expression in cells treated with the indicated concentrations of compound 1 for 96 h was determined by cell ELISA. (C) Huh-7 (lanes 1 and 2) or HBI10 (lanes 3 to 8) cells were starved for 1 h, pulse-labeled with 35S-labeled amino acids for 15 min at 37°C, and then chased for the time indicated above each lane. Where indicated, starvation, labeling, and chasing were performed in the presence of 50 μM compound (cmp) 1. The labeled proteins were immunoprecipitated with the 55IV anti-NS5A antiserum and analyzed by SDS-10% PAGE. The positions of molecular mass standards (in kilodaltons) and NS5A and uncleaved precursors are indicated.
To investigate whether the effect of compound 1 on viral replication was exerted by inhibition of NS3-4p, in vivo processing of the HCV polyprotein was analyzed by pulse-chase labeling in the absence or in the presence of compound 1, followed by immunoprecipitation with an anti-NS5A antibody. In the absence of inhibitor, mature NS5A protein was released from the polyprotein in a time-dependent fashion with kinetics identical to those already reported (Fig. 2C) (40, 43). After the pulse, only the p56 form of NS5A was detected, and most of the protein was still contained in several partially processed precursors, the most abundant of which was NS4A-5A (Fig. 2C, lane 3). The amount of p56 increased with chase time in parallel with the appearance of the p58 hyperphosphorylated form of NS5A (Fig. 2C, lanes 4 and 5) (40). The increase of p56 and p58 was paralleled by the disappearance of the precursor proteins. In the presence of compound 1, only high-molecular-weight bands, corresponding to uncleaved (NS3-5B) and partially cleaved (NS3-5A, NS4A-5B, and NS4B-5B) precursor proteins were observed after the pulse (Fig. 2C, lane 6). These precursors were slowly converted to other incompletely processed proteins, and p56 became barely visible only after 3 h of chase. This strong delay in the processing of the polyprotein indicated that compound 1 inhibited NS3-4Ap almost completely. Interestingly, the accumulation of the NS3-5B full-length precursor polyprotein indicated that compound 1 inhibited cleavage at all NS3-4Ap-processed junctions, including the NS3/4A site, which is cleaved only intramolecularly. A similar delay in processing was observed when the biogenesis of the NS3 protein was examined (data not shown).
Selection of replicons resistant to compound 1 and identification of resistance mutations.
The ability of the HBI10 cells to survive in the presence of G418 hinges on replication of the HCV subgenomic RNAs expressing the resistance gene. Consequently, inhibitors of viral replication are expected to abolish resistance to G418. It is thus conceivable to select replicons resistant to compound 1 by culturing the cells in the presence of inhibitor and G418. Under these conditions, the majority of HBI10 cells are expected to succumb to the toxic effect of the antibiotic, and only cells containing mutant replicons with decreased sensitivity to compound 1 are predicted to survive and give rise to subclones.
In fact, clones resistant to compound 1 were selected by culturing HBI10 cells in the presence of 1 mg of G418/ml and 12 μM inhibitor. Less than 100 doubly resistant clones were obtained starting from 106 HBI10 cells. However, the frequency at which resistant clones emerged could not be estimated, since it was necessary to pass the cells once during selection. Three resistant clones (R7, R10, and R12) were expanded and studied in detail. These clones duplicated at the same rate as parental cells both in the absence and in the presence of G418 and expressed HCV RNA and proteins at levels comparable to those of the parental cells (Fig. 3 and data not shown). As expected, replication of the cognate replicons was partially resistant to compound 1: the inhibitor displayed IC50s for all tested clones at least 50 times higher than that for HBI10 cells (Table 1). Selected clones were also tested for sensitivity to IFN-α, which has been shown to inhibit replication of subgenomic HCV RNA (15, 18). All clones were as sensitive to the cytokine as the parent cells, indicating that resistance was specific for compound 1 (Table 1).
TABLE 1.
Effects of compound 1 and IFN-α on replication of parental and resistant replicons
| Clone | IC50a
|
|
|---|---|---|
| Compound 1 (μM) | IFN-α (IU/ml) | |
| HBI10A | 1 | 1 |
| R7 | >50 | 1.4 |
| R10 | >50 | 0.9 |
| R12 | >50 | 1.3 |
Data are averages of at least two separate experiments.
To ascertain if resistance was due to a distinctive replicon mutation(s), we determined the sequences of the NS3 (R7) or entire NS (R10 and R12) coding regions of replicons extracted from resistant clones and compared them to those of replicons extracted from parental HBI10 cells. This comparison revealed the presence of 2 mutations common to parental and resistant cells, namely, replacement of glutamate 176 of NS3 (residue 1202 of the polyprotein) with either lysine (E176K) or glycine (E176G) and insertion of a lysine residue after valine 67 of NS5A (residue 2039 of the polyprotein) (Table 2, K @67). Both mutations had already been identified in HBI10 cells and in other replicon clones and had been shown to enhance replication in cultured cells (8, 25, 33, 42).
TABLE 2.
Sequence analysis of replicons extracted from parental and resistant clones
| Clone | Mutation(s)
|
||||||
|---|---|---|---|---|---|---|---|
| NS3 (protease) | NS3 (helicase) | NS5A | |||||
| HBI10A | E176G | K@67 | |||||
| R10 | D168Y | E176G | P264L | K@67 | |||
| R12 | D168V | E176G | G394S | I446V | K@67 | E438K | |
| R7a | P67S | D168A | E176K | R512K | |||
Only the sequence of the NS3-4A region was determined for this clone.
Replicons isolated from resistant clones contained several other mutations not observed in HBI10 cells (Table 2). Notably, no mutations were found in the NS3-4Ap cleavage sites and only 1 mutation was common to all clones, namely, replacement of aspartate 168 of NS3 (residue 1194 of the polyprotein) with either alanine (D168A), tyrosine (D168Y), or valine (D168V). The fact that this substitution was the only mutation in the NS3 protease domain suggested that it was responsible for the resistance phenotype. Thus, we assembled replicon constructs containing several combinations of the D168A/Y/V, E176G/K, and K@67 mutations and tested them for replication competence and sensitivity to compound 1 in transient-transfection experiments, monitoring replication by cell ELISA (Table 3). For this experiment, we used as the recipient 10AIFN cells, a derivative of the HBI10 clone that supported HCV replication much more efficiently than parental Huh-7 cells (data not shown). These cells were obtained by treating HBI10 cells with IFN-α to remove the endogenous replicons (11). As already reported by other groups (8, 25), the replicon without mutations (wild type [wt]) did not replicate appreciably in transient transfection. Conversely, the replicons containing the E176G (C) or K@67 (A) mutation replicated at low and medium levels, respectively. The replicon containing both E176G and K@67 (B) replicated very efficiently, indicating that these mutations enhanced replication synergistically. Replication of replicons A, B, and C was sensitive to compound 1, and the IC50 for replicon B was comparable to that observed for HBI10 cells, indicating that the E176G and K@67 substitutions did not affect sensitivity to the inhibitor. Replicons containing only a replacement of aspartate 168 with alanine (RRX1), tyrosine (RRX2), or valine (RRX3) did not show visible replication. Replicons containing replacement of D168 in combination with K@67 (RRA1, RRA2, and RRA3) or with E176G and K@67 (RRB1, RRB2, and RRB3) replicated to the same extent as the corresponding replicons lacking mutations of aspartate 168, indicating that replacement of this residue had no measurable effect on replication competence. The RRA replicons were clearly less sensitive than replicon A to compound 1, though we did not determine the IC50s because of the low replication efficiency. More importantly, the IC50s measured for RRB replicons were similar to those observed in the R7, R10, and R12 clones, thus demonstrating that replacement of aspartate 168 was sufficient to fully recapitulate the resistance phenotype displayed by selected clones.
TABLE 3.
Replication competences and sensitivities to compound 1 of mutant replicons
| Replicon pHCVNEO17. | Mutation(s)
|
Replication efficiencya,b | Compound 1 IC50 (μM)b | ||
|---|---|---|---|---|---|
| NS3 | NS5A | ||||
| wt | 0.9 | NA | |||
| A | K@67 | 3.0 | NA | ||
| B | E176G | K@67 | 15.5 | 0.7 | |
| C | E176G | 1.5 | NA | ||
| RRX1 | D168A | 1.0 | NA | ||
| RRX2 | D168Y | 0.9 | NA | ||
| RRX3 | D168V | 0.9 | NA | ||
| RRA1 | D168A | K@67 | 1.9 | NA | |
| RRA2 | D168Y | K@67 | 1.6 | NA | |
| RRA3 | D168V | K@67 | 2.8 | NA | |
| RRB1 | D168A | E176K | K@67 | 12.9 | >50 |
| RRB2 | D168Y | E176G | K@67 | 12.1 | >50 |
| RRB3 | D168V | E176G | K@67 | 14.4 | >50 |
Replication efficiency is expressed in arbitrary units and was calculated as described in Materials and Methods.
Data are averages of at least two separate experiments. NA, not applicable.
To investigate if the mutations found in resistant replicons affected the ability of NS3-4Ap to process the polyprotein, we analyzed the biogenesis of the NS5A protein by pulse-chase-immunoprecipitation experiments (Fig. 3). In both the R10 and R12 clones, NS5A was released from the precursor polyprotein with kinetics similar to those observed in HBI10 cells, although quantitative analysis revealed minor differences. While the disappearances over time of the NS4A-5B, NS4B-5B, and NS5A-5B precursors were similar in parental and resistant cells, the NS4A-5A and NS4B-5A precursors of resistant clones showed half-lives slightly longer than that of HBI10 cells. Consistently in the two resistant clones, the appearance of the p56 and p58 forms of NS5A was also slightly delayed compared to parental cells. Overall, these results suggested that the mutant proteases encoded by resistant replicons cleaved the NS3-4A, NS4A-4B, and NS5A-5B junctions with efficiencies comparable to that of the wt enzyme but were to some extent impaired in processing the NS4B-5A site.
Characterization of mutant enzymes.
The demonstration that a single amino acid substitution in the NS3 protease domain rendered HCV replication resistant to compound 1 strongly suggested that resistance was due to a reduced sensitivity of the mutant proteases to inhibition by compound 1. To confirm this hypothesis, we expressed and purified recombinant Con1 proteases containing the D168Y (D168Yp) or D168V (D168Vp) substitution and compared them to the wt enzymes. D168Yp and the D168Vp showed affinity for the NS5A-5B substrate and catalytic efficiencies comparable to those of the Con1 and BK enzymes (Table 4). Most importantly, compound 1 was ∼1,000-fold less active on D168Yp and D168Vp than on the Con1 and BK enzymes, thus confirming that substitution of aspartate 168 was responsible for resistance to compound 1 and that resistance was due to reduced affinity of the mutant proteases to the inhibitor.
TABLE 4.
Activities and sensitivities to compound 1 of recombinant proteasesa
| Enzyme | Strain | Mutation(s)
|
Substrate
|
Compound 1
|
||||
|---|---|---|---|---|---|---|---|---|
| Km (μM) | Kcat (min−1) | IC50 (μM) | Ki (μM) | |||||
| BK | BK | C428S | 4 | 29 | 0.012 | |||
| Con1 | Con1 | E176G | C428S | 4 | 32 | 0.010 | 0.006 | |
| D168Yp | Con1 | D168Y | E176G | C428S | 8 | 35 | 10 | 8.5 |
| D168Vp | Con1 | D168V | E176G | C428S | 7 | 16 | 17 | 8.5 |
Data are averages of at least two separate experiments.
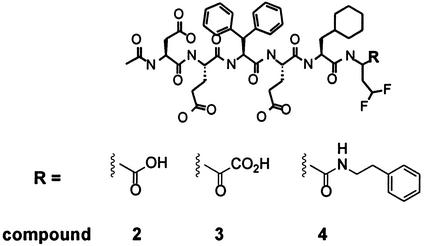
As an initial effort toward elucidating of the mechanism of resistance to compound 1, we tested resistant proteases for cross-resistance to three hexapeptide inhibitors. These compounds have the same structure, with acidic residues in P6, P5, and P3, diphenylalanine in P4, cyclohexylalanine in P2, and difluoroamino butyric acid as a cysteine mimetic in P1 and differ only in the substituent on the P1 C-alpha position (Fig. 4). Similarly to compound 1, compound 2 has an alpha carboxylic acid and is a well-characterized product inhibitor (38, 39). In compound 3, the P1 carboxylic acid is replaced by an alpha ketoacid, resulting in a covalent inhibitor of the protease (38). In compound 4, the alpha carboxylic acid is replaced by phenethyl-amide (10). In contrast to compound 1, mutations of aspartate 168 reduced the potency of compound 2 only four- to eightfold and did not seem to affect the activities of compounds 3 and 4 at all (Table 5). The lack of cross-resistance suggests that replacement of aspartate 168 influences interaction with the side chains rather than the backbone of the inhibitor. In addition, comparison of the three hexapeptides indicates that P1 alpha carboxylic acid has only a minor role in the specificity of resistance.
FIG. 4.
Structures of compounds 2, 3, and 4.
TABLE 5.
Activities of compounds 2, 3, and 4 on wt and mutant proteases
| Enzyme | IC50 (μM)a
|
||
|---|---|---|---|
| Compound 2 | Compound 3 | Compound 4 | |
| Con1 | 1.8 | 0.13 | 8 |
| D168Yp | 7.4 | 0.09 | 9 |
| D168Vp | 12 | 0.04 | 14 |
Data are averages of at least two separate experiments.
Modeling.
To gain some understanding of the mechanism of resistance induced by replacement of aspartate 168, compound 1 was modeled in the three-dimensional structure of the enzyme. The modeling was based on the NMR structures of product inhibitors bound to the protease (9, 26) and the crystal structures of alpha ketoacids covalently bound to the catalytic serine (13). According to this model, and consistent with the structure-activity studies reported for tripeptide inhibitors (Bailey et al., Abstr. Pap. Am. Chem. Soc. 220), the major determinants for inhibitor binding appear to be the P1 residue, both through its side chain and through its alpha carboxylate, and the bulky P2 substituent (Fig. 5). The P1 cyclopropyl seats favorably in the S1 specificity pocket, while the carboxylate is in the oxyanion hole at hydrogen bond distance with the protonated ɛ-N of the catalytic histidine 57 and the side chain of lysine 136. The P2 phenylmethoxy quinoline substituent lies on the enzyme surface and partially covers a chain of residues with alternating charges that starts with catalytic histidine 57 and aspartate 81 and continues with arginine 155, aspartate 168, and arginine 123. The P3 valine and the tert-butyl capping group also establish hydrophobic interactions with the surface of the enzyme. The tert-butyl group is in contact with valine 158 and is near aspartate 168, but it is not in direct contact with this residue. Comparison with compounds 2, 3, and 4, which have the bulky diphenylalanine in the position corresponding to the capping group and which do not show a significant change in potency upon mutation of aspartate 168, suggests that the capping group of compound 1 is not responsible for resistance. Thus, initial evidence indirectly points to the P2 residue of compound 1 as the major determinant of resistance. However, additional experiments are necessary to support this hypothesis.
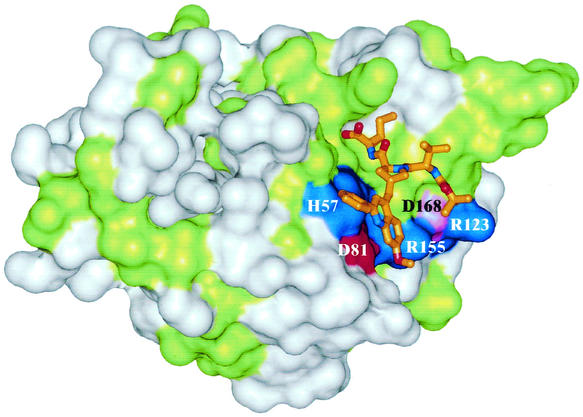
FIG. 5.
Model of compound 1 in the active site of the NS3-4A protease. Conserved residues are shown in green, and nonconserved residues are white. Charged residues are colored according to their charges, with basic residues in blue, acidic residues in red, and aspartate 168 in magenta. Compound 1 is presented as a stick model.
DISCUSSION
Although the development of antiviral agents to treat HCV infection is less advanced than that for HIV, potent inhibitors of the HCV serine protease and RNA-dependent RNA polymerase have been identified, and the most advanced are being evaluated in clinical trials (6). The long-term clinical success of these molecules depends on their abilities to suppress drug-resistant viral variants. In fact, given the high mutation rate observed in vivo for HCV, it is likely that the virus can accumulate in its enzymes specific mutations that could lead to decreased sensitivity to inhibitors. Thus, it is important that the optimization of antiviral drugs for HCV be accompanied by the study of their resistance profiles. These studies should help in developing inhibitors active against a broad range of viral variants and facilitate the identification of inhibitors with distinct resistance profiles that could be used in combination therapy, thus increasing the chances to completely suppress viral replication.
In the case of HIV, the possibility of selecting and characterizing in vitro viruses resistant to antiviral agents has complemented resistance studies in patients receiving therapy and greatly aided the development of compounds with different resistance profiles.
For a long time, the absence of efficient in vitro infection systems for HCV prevented the assessment of the efficacy of enzyme inhibitors in infected cells and the performance of resistance analysis. The advent of a robust cell culture HCV replication system based on subgenomic replicons has changed this scenario, offering the first tractable means of testing antiviral compounds in live cells (34). Subsequently, the identification of adaptive variants that replicated very efficiently made possible the use of replicons for reverse-genetics studies (8, 25, 33).
In this paper, we show for the first time that HCV subgenomic replicons can also be used for conventional virus genetic studies, and more specifically for the selection and characterization of mutants with decreased sensitivity to inhibitors of viral enzymes. Indeed, our data indicate that in vitro selection of HCV subgenomic replicons with an inhibitor of the NS3-4A protease leads to the accumulation of resistant replicons containing specific protease mutations which make the enzyme less sensitive to inhibition. Replacement of aspartate 168 in the NS3 protease domain is sufficient to make the enzyme about 3 orders of magnitude less sensitive to compound 1, with no effect on catalytic efficiency. Accordingly, in tissue culture, the same substitution shows only minor effects on polyprotein processing and is not detrimental to replication of partially or fully adapted replicons. Although the effects of specific mutations on the replication of an authentic HCV cannot be assessed at present, our data suggest that mutations that induce antiviral resistance, and specifically replacement of aspartate 168, can be well tolerated by the virus and have little if any impact on virus fitness. Interestingly, aspartate 168 is one of the few active-site residues not entirely conserved among HCV genotypes, and it is replaced by glutamate and glutamine in isolates belonging to genotypes 5 and 3, respectively. The glutamine substitution has no effect on enzyme activity and does not change the enzyme sensitivity to hexapeptide inhibitors (7). Although the effects of these natural substitutions of aspartate 168 on the activity of compound 1 have not been investigated, their occurrence suggests that virus mutants resistant to this inhibitor may exist in the natural population.
The mechanisms by which replacements of aspartate 168 induce resistance to compound 1 are not obvious, and our data provide only hints as to the roles of the different substituents of compound 1 as resistance determinants. Modeling of compound 1 in the structure of the protease suggests that aspartate 168 does not establish direct contact with the inhibitor. Thus, replacement of this residue seems to affect affinity for the inhibitor indirectly. Aspartate 168 is part of an alternating chain of positively and negatively charged residues that neutralize each other (Fig. 5). Presumably, neutralizing aspartate 168 increases the excess positive charge of the ligand binding site, leading the neighboring arginine residues (155 and 123) to become more strongly solvated. This increases the energy required to desolvate these residues upon binding of the phenylmethoxy quinoline substituent of compound 1. We speculate that the increase in the desolvation energy, especially for arginine 155 in the S2 site, upon binding of the lipophilic P2 side chain of compound 1 is responsible for the loss of activity on the mutated enzyme. This effect would be less pronounced for compounds 2, 3, and 4, with smaller P2 side chains. In addition, an unfavorable change in intramolecular electrostatic interactions may also play a role. We have shown that electrostatic interactions between the charged residues in the ligand binding site of NS3 increase upon ligand binding due to the reduction of electrostatic screening when water is replaced by a low-dielectric ligand. Whereas this effect increases interactions between oppositely charged residues in wt enzyme and is thus favorable (22), it would clearly be unfavorable in the case of the mutant. This analysis suggests that the P2 residue of compound 1 is the major factor responsible for the loss of affinity for the resistance mutation through a combination of desolvation and electrostatic effects. Thus, we suggest that one possible way to overcome resistance is the design of compounds with altered P2 side chains. Clearly, additional experiments are necessary to support this hypothesis.
The compound used for this study is a micromolar inhibitor of the NS3-4A protease in replicon cells; however, it is expected that the selection procedure used is also applicable to more potent compounds and to inhibitors of other viral functions. Furthermore, the selection strategy described in this paper can also be used to map mutations responsible for resistance to replication inhibitors with unknown mechanisms, thus facilitating the identification of the molecular target.
Conceptually, the selection method described in this paper is similar to the procedures conventionally used to select resistant mutants with authentic viruses. The major difference is that replicons are selected together with host cells, posing the question of whether the resistance phenotype results from replicon mutations or from changes in the selected cells. In the case of clones resistant to compound 1, the mechanism of resistance could be easily identified by sequencing the replicons extracted from resistant clones. A limited number of mutations were found in each clone, and the mutation responsible for resistance was the same in the three cell clones analyzed, making the subsequent reverse-genetic analysis rather simple. This may not be the case with other inhibitors, and in addition to direct sequencing, other approaches may be required to discriminate between the cellular and the replicon contributions.
In conclusion, our results with compound 1 may be predictive of the resistance patterns that will probably be observed in vivo with structurally related compounds, and they should contribute to the identification and clinical development of resistance-repellent inhibitors.
Acknowledgments
We thank Riccardo Cortese for continuous support and Janet Clench for editing the manuscript.
REFERENCES
- 1.Barbato, G., D. O. Cicero, M. C. Nardi, C. Steinkuhler, R. Cortese, R. De Francesco, and R. Bazzo. 1999. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol. 289:371-384. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R. 1997. Candidate targets for hepatitis C virus-specific antiviral therapy. Intervirology 40:378-393. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6:165-181. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., L. Ahlborn-Laake, K. Yasargil, J. Mous, and H. Jacobsen. 1995. Substrate determinants for cleavage in cis and in trans by the hepatitis C virus NS3 proteinase. J. Virol. 69:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaulieu, P. L., and M. Llinas-Brunet. 2002. Therapies for hepatitis C infection: targeting the non-structural proteins of HCV. Curr. Med. Chem. 1:163-176. [Google Scholar]
- 7.Beyer, B. M., R. Zhang, Z. Hong, V. Madison, and B. A. Malcolm. 2001. Effect of naturally occurring active site mutations on hepatitis C virus NS3 protease specificity. Proteins 43:82-88. [PubMed] [Google Scholar]
- 8.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Cicero, D. O., G. Barbato, U. Koch, P. Ingallinella, E. Bianchi, M. C. Nardi, C. Steinkuhler, R. Cortese, V. Matassa, R. De Francesco, A. Pessi, and R. Bazzo. 1999. Structural characterization of the interactions of optimized product inhibitors with the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein by NMR and modelling studies. J. Mol. Biol. 289:385-396. [DOI] [PubMed] [Google Scholar]
- 10.Colarusso, S., C. Gardelli, B. Gerlach, S. Harper, U. Koch, V. G. Matassa, E. Muraglia, F. Narjes, O. J. M. Ontoria, A. Petrocchi, S. Ponzi, I. Stansfield, and V. Summa. 2002. Peptides and their use as inhibitors of hepatitis C virus ns3 protease. WO patent 0279234.
- 11.De Francesco, R., G. Migliaccio, and G. Paonessa. 2002. Hepatitis C virus replicons and replicon enhanced cells. WO patent 0259321.
- 12.De Francesco, R., A. Urbani, M. C. Nardi, L. Tomei, C. Steinkuhler, and A. Tramontano. 1996. A zinc binding site in viral serine proteinases. Biochemistry 35:13282-13287. [DOI] [PubMed] [Google Scholar]
- 13.Di Marco, S., M. Rizzi, C. Volpari, M. A. Walsh, F. Narjes, S. Colarusso, R. De Francesco, V. G. Matassa, and M. Sollazzo. 2000. Inhibition of the hepatitis C virus NS3/4A protease: the crystal structures of two protease-inhibitor complexes. J. Biol. Chem. 275:7152-7157. [DOI] [PubMed] [Google Scholar]
- 14.Failla, C., L. Tomei, and R. De Francesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 16.Gallinari, P., C. Paolini, D. Brennan, C. Nardi, C. Steinkuhler, and R. De Francesco. 1999. Modulation of hepatitis C virus NS3 protease and helicase activities through the interaction with NS4A. Biochemistry 38:5620-5632. [DOI] [PubMed] [Google Scholar]
- 17.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingallinella, P., S. Altamura, E. Bianchi, M. Taliani, R. Ingenito, R. Cortese, R. De Francesco, C. Steinkuhler, and A. Pessi. 1998. Potent peptide inhibitors of human hepatitis C virus NS3 protease are obtained by optimizing the cleavage products. Biochemistry 37:8906-8914. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. Y., K. W. Park, Y. J. Lee, S. H. Back, J. H. Goo, O. K. Park, S. K. Jang, and W. J. Park. 2000. In vivo determination of substrate specificity of hepatitis C virus NS3 protease: genetic assay for site-specific proteolysis. Anal. Biochem. 284:42-48. [DOI] [PubMed] [Google Scholar]
- 22.Koch, U., G. Biasiol, M. Brunetti, D. Fattori, M. Pallaoro, and C. Steinkuhler. 2001. Role of charged residues in the catalytic mechanism of hepatitis C virus NS3 protease: electrostatic precollision guidance and transition-state stabilization. Biochemistry 40:631-640. [DOI] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., E. V. Agapov, and C. M. Rice. 1994. Specificity of the hepatitis C virus NS3 serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J. Virol. 68:7525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaPlante, S. R., N. Aubry, P. R. Bonneau, G. Kukolj, D. Lamarre, S. Lefebvre, H. Li, M. Llinas-Brunet, C. Plouffe, and D. R. Cameron. 2000. NMR line-broadening and transferred NOESY as a medicinal chemistry tool for studying inhibitors of the hepatitis C virus NS3 protease domain. Bioorg. Med. Chem. Lett. 10:2271-2274. [DOI] [PubMed] [Google Scholar]
- 27.LaPlante, S. R., D. R. Cameron, N. Aubry, S. Lefebvre, G. Kukolj, R. Maurice, D. Thibeault, D. Lamarre, and M. Llinas-Brunet. 1999. Solution structure of substrate-based ligands when bound to hepatitis C virus NS3 protease domain. J. Biol. Chem. 274:18618-18624. [DOI] [PubMed] [Google Scholar]
- 28.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llinas-Brunet, M., M. Bailey, R. Deziel, G. Fazal, V. Gorys, S. Goulet, T. Halmos, R. Maurice, M. Poirier, M. A. Poupart, J. Rancourt, D. Thibeault, D. Wernic, and D. Lamarre. 1998. Studies on the C-terminal of hexapeptide inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett. 8:2719-2724. [DOI] [PubMed] [Google Scholar]
- 30.Llinas-Brunet, M., M. Bailey, G. Fazal, E. Ghiro, V. Gorys, S. Goulet, T. Halmos, R. Maurice, M. Poirier, M. A. Poupart, J. Rancourt, D. Thibeault, D. Wernic, and D. Lamarre. 2000. Highly potent and selective peptide-based inhibitors of the hepatitis C virus serine protease: towards smaller inhibitors. Bioorg. Med. Chem. Lett. 10:2267-2270. [DOI] [PubMed] [Google Scholar]
- 31.Llinas-Brunet, M., M. Bailey, G. Fazal, S. Goulet, T. Halmos, S. Laplante, R. Maurice, M. Poirier, M. A. Poupart, D. Thibeault, D. Wernic, and D. Lamarre. 1998. Peptide-based inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett. 8:1713-1718. [DOI] [PubMed] [Google Scholar]
- 32.Llinas-Brunet, M., M. D. Bailey, D. Cameron, A.-M. Faucher, E. Ghiro, N. Goudreau, T. Halmos, M.-A. Poupart, J. Rancourt, Y. S. Tsantrizos, D. M. Wernic, and B. Simoneau. 2000. Preparation of hepatitis C inhibitory tripeptides. WO patent 0009543.
- 33.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 35.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 36.McCoy, M. A., M. M. Senior, J. J. Gesell, L. Ramanathan, and D. F. Wyss. 2001. Solution structure and dynamics of the single-chain hepatitis C virus NS3 protease NS4A cofactor complex. J. Mol. Biol. 305:1099-1110. [DOI] [PubMed] [Google Scholar]
- 37.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 38.Narjes, F., M. Brunetti, S. Colarusso, B. Gerlach, U. Koch, G. Biasiol, D. Fattori, R. De Francesco, V. G. Matassa, and C. Steinkuhler. 2000. Alpha-ketoacids are potent slow binding inhibitors of the hepatitis C virus NS3 protease. Biochemistry 39:1849-1861. [DOI] [PubMed] [Google Scholar]
- 39.Narjes, F., K. F. Koehler, U. Koch, B. Gerlach, S. Colarusso, C. Steinkuhler, M. Brunetti, S. Altamura, R. De Francesco, and V. G. Matassa. 2002. A designed P(1) cysteine mimetic for covalent and non-covalent inhibitors of HCV NS3 protease. Bioorg. Med. Chem. Lett. 12:701-704. [DOI] [PubMed] [Google Scholar]
- 40.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacini, L., A. Vitelli, G. Filocamo, L. Bartholomew, M. Brunetti, A. Tramontano, C. Steinkuhler, and G. Migliaccio. 2000. In vivo selection of protease cleavage sites by using chimeric Sindbis virus libraries. J. Virol. 74:10563-10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, F. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizzi, E., A. Tramontano, L. Tomei, N. La Monica, C. Failla, M. Sardana, T. Wood, and R. De Francesco. 1994. Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc. Natl. Acad. Sci. USA 91:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinkuhler, C., G. Biasiol, M. Brunetti, A. Urbani, U. Koch, R. Cortese, A. Pessi, and R. De Francesco. 1998. Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 37:8899-8905. [DOI] [PubMed] [Google Scholar]
- 46.Steinkuhler, C., U. Koch, F. Narjes, and V. G. Matassa. 2001. Hepatitis C virus protease inhibitors: current progress and future challenges. Curr. Med. Chem. 8:919-932. [DOI] [PubMed] [Google Scholar]
- 47.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsantrizos, Y. S., D. R. Cameron, A.-M. Faucher, E. Ghiro, N. Goudreau, T. Halmos, and M. Llinas-brunet. 2000. Preparation of macrocyclic peptides active against the hepatitis C virus. WO patent 0059929.
- 49.Urbani, A., E. Bianchi, F. Narjes, A. Tramontano, R. De Francesco, C. Steinkuhler, and A. Pessi. 1997. Substrate specificity of the hepatitis C virus serine protease NS3. J. Biol. Chem. 272:9204-9209. [DOI] [PubMed] [Google Scholar]
- 50.Wright, M., J. Main, and H. C. Thomas. 2001. Treatment of chronic viral hepatitis. Antivir. Chem. Chemother. 12:201-212. [DOI] [PubMed] [Google Scholar]
- 51.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Struct. Fold Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, R., J. Durkin, W. T. Windsor, C. McNemar, L. Ramanathan, and H. V. Le. 1997. Probing the substrate specificity of hepatitis C virus NS3 serine protease by using synthetic peptides. J. Virol. 71:6208-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]