Abstract
Quantitative monitoring of human cytomegalovirus (HCMV) infection is helpful in determining appropriate antiviral management of transplant recipients. Quantitative PCR technologies have demonstrated accuracy in measuring systemic HCMV loads. A total of 298 consecutive whole-blood specimens submitted to the Clinical Virology Laboratory at Vanderbilt University Medical Center from 15 February to 31 October 1999 were included in the study. In addition to a qualitative colorimetric microtiter plate PCR assay (MTP-PCR) and a semiquantitative pp65 antigenemia assay, each specimen was measured for HCMV loads by a quantitative PCR assay performed on an ABI PRISM 7700 Sequence Detection System (TaqMan). Compared to results of the MTP-PCR, the sensitivity, specificity, positive predictive value, and negative predictive value were 70.5, 97.5, 87.8, and 92.8% for the antigenemia assay and were 96.7, 92.0, 75.6, and 99.1% for the TaqMan assay, respectively. There was a high correlation between antigenemia values and HCMV loads as determined by the TaqMan (r = 0.989; P < 0.001). Antigenemia values of 0, 1 to 10, 11 to 100, 101 to 1,000, and over 1,000 positive cells per 2 × 105 leukocytes corresponded to median HCMV loads measured by TaqMan of 125, 1,593, 5,713, 16,825, and 5,425,000 copies/ml, respectively. Corresponding to antigenemia values of 1 to 2, 10, and 50 positive cells per 2 × 105 leukocytes, HCMV viral loads of 1,000, 4,000, and 10,000 copies/ml are proposed as cutoff points for initiating antiviral therapy in patient groups with high, intermediate, and low risk of CMV diseases.
Human cytomegalovirus (HCMV) continues to be a significant cause of morbidity and mortality in immunocompromised patients, such as solid organ or bone marrow transplant recipients (21, 36). Effective preemptive antiviral therapy that includes ganciclovir and foscarnet depends on detecting HCMV at an early stage of infection (3, 14, 21, 37), but these drugs must be used carefully because of their side effects. The early and accurate diagnosis of CMV infection status is an essential part of the management of these patients (14, 36). However, because the primary HCMV infection results in a latent life-long infection and because many transplant patients secrete HCMV without any clinical disease, the mere detection of HCMV does not always indicate the need for treatment (3, 21, 36).
Quantitation of the systemic HCMV load may provide a highly sensitive and specific method to predict which patients will develop HCMV diseases. Several techniques have been used to monitor HCMV loads. The traditional methods, based on virus isolation as indicated by cytopathic effect, are time-consuming procedures, have poor reproducibility, and have only moderate sensitivity. The antigenemia assay, which detects the viral lower matrix protein pp65 (UL83) in blood leukocytes (4, 11, 13, 34), has been widely used as a tool to monitor HCMV infections. The number of pp65-positive cells has been shown to correlate with HCMV disease, although the threshold for HCMV disease may vary between patient groups (3, 23). This method, however, needs to be processed within 8 h of collection and suffers from its lack of standardization (3, 11, 23). Only virus associated with cells is detected, and free viruses in biological fluids are not detectable by this assay. The reading of the assay depends on technical skill and training and requires a moderate amount of technologist time. Furthermore, this assay cannot be carried out when a patient's leukocyte count is very low (4).
The wide application of in vitro amplification techniques for the detection of viral nucleic acids has become the most important innovation in laboratory diagnosis and clinical management of posttransplant HCMV infection. Multiple studies have shown that the real-time quantitative PCR for HCMV DNA in plasma and other biological samples is useful in the rapid diagnosis of HCMV infection and in effective monitoring of clinical course as well as response to therapy (7, 10, 11, 14, 15, 26, 27, 29, 32, 37). The real-time systems measure the accumulation of PCR products with a fluorogenic probe and instantaneous laser scanning (7, 15). Unlike regular PCR procedures performed by using conventional thermocyclers, the real-time PCR system is closed and therefore avoids the important concerns of carryover contamination with the previous product. In addition, the nucleic acid products can be monitored in real time after each cycle of amplification. As a result of shorter thermocycling and simultaneous product detection, the turnaround time of the assay is significantly shorter than that of prior quantitative assays.
In this study we developed such a real-time PCR assay that uses the 7700 ABI PRISM Sequence Detector (Applied Biosystems, Foster City, Calif.) (12) to quantitate HCMV loads in whole-blood specimens collected from transplant recipients. We compared this assay with an antigenemia assay that has been in use for years on which we had reliable threshold points for treatment in different patient populations.
(This study was presented in part at the 102nd General Meeting of the American Society for Microbiology, Salt Lake City, Utah, 19 to 23 May 2002.)
MATERIALS AND METHODS
Clinical specimens.
A total of 298 consecutive whole-blood specimens with sufficient volumes submitted for the HCMV antigenemia assay to the Clinical Virology Laboratory at Vanderbilt University Medical Center from 15 February to 31 October 1999 were included in the study. The study was approved by the Vanderbilt University Medical Center Institutional Review Board (no. 9912). The samples were from patients undergoing bone marrow or solid organ transplantation at the Vanderbilt University Transplantation Center. Further aliquots of EDTA blood specimens were prepared and stored at −70°C from those specimens with sufficient remaining volumes after routine antigenemia testing. In addition to the antigenemia assay, each specimen was tested for HCMV qualitatively and quantitatively by two PCR-based assays as described below.
Antigenemia assay.
The CMV Brite assay (Biotest Diagnostics Corp., Denville, N.J.) was performed according to the manufacturer's instructions (13). Briefly, leukocytes were separated from whole EDTA blood, enumerated, and centrifuged onto two 2-well slides per specimen. Cells on each slide were fixed and permeabilized, and one slide was stained with anti-CMV pp65 monoclonal antibodies; this was followed by staining with fluorescein isothiocyanate-labeled rabbit anti-mouse immunoglobulin G conjugate. A positive result consisted of one or more HCMV antigen-positive cells per set of duplicate wells. Results were expressed as the number of positive cells per 2 × 105 leukocytes. Approximate antigenemia thresholds used at our Vanderbilt University Medical Center were 1 to 2, 10, and 50 positive cells per 2 × 105 leukocytes for allogeneic bone marrow transplant and HCMV-seronegative and -seropositive recipients of solid organs, respectively (4, 23, 34).
DNA extraction.
Nucleic acid was extracted from 0.2 ml of EDTA-anticoagulated whole blood by using the Qiagen blood/tissue kit according to the manufacturer's instructions (7, 15). DNA was eluted from the Qiagen columns in a final volume of 50 μl of distilled water and was stored at −70°C until used. These extracted DNA samples were used for both qualitative and quantitative PCR assays.
Qualitative PCR.
A colorimetric microtiter plate PCR assay (MTP-PCR) was performed to detect HCMV DNA according to the procedure published previously (31). A primer set (CMV-1, 5′-GGG TGC TGT CCT GCT ATG TCT TA-3′; and CMV-2, 5′-CAT CAC TCT GCT CAC TTT CTT CC-3′) was designed to target the HCMV immediate-early antigen gene (30). After PCR, amplification products were identified by detecting digoxigenin-labeled PCR products as previously described (31). An HCMV-specific 5′-biotinylated capture probe (CMV-3, 5′-CGG CCT CTG ATA ACC AAG CCT G-3′) was applied to detect and confirm the amplification product. A similar format was used to amplify a housekeeping gene to assure the quality of extracted DNA samples. The primer set (5′-ATC ATG TTT GAG ACC TTC AAC-3′ and 5′-CAG GAA GGA AGG CTG GAA GAG-3′) defined a 438-bp target within the human β-actin gene (19). The 5′-biotinylated β-actin gene-specific probe (5′-GTA CCA CTG GCA TCG TGA TGG A-3′) was used to identify and confirm the amplification product as described above. Output signals were measured at optical densities of 450 (OD450) and 490 (OD490). A positive result was defined as an OD450-OD490 value greater than or equal to 0.1 (31).
Quantitative real-time PCR.
A quantitation standard curve was achieved by using six 10-fold serial dilutions of a plasmid standard (pCT-gpB) containing the primer-spanning region of the HCMV glycoprotein B gene. Sequences of the primer set (gpB-A), which yield a 254-bp product, were published previously (2). The amplicon was generated by PCR and was subsequently cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.). The plasmid standard DNA concentration was calibrated by spectrophotometry at 260 nm (27). Quantitative PCR was performed in a real-time format on the 7700 ABI Prism Sequence Detector (Applied Biosystems). An aliquot of 5 μl of the extracted nucleic acid was added to 20 μl of reaction mixture containing 0.8 μM concentrations of each primer and 0.4 μM fluorophore probe (final concentration) and was mixed with 25 μl of TaqMan Universal PCR Master Mix (Applied Biosystems) (12). The HCMV-specific fluorophore probe (5′-TTG CTG CCC AGC AGA TAA GTG GTG T-3′) was designed with a reporter dye, 6-carboxyfluorescein, and a quencher dye, 6-carboxytetramethylrhodomine, covalently linked to the 5′ and 3′ ends, respectively (Applied Biosystems) (12). The TaqMan cycling conditions were a 2-min degradation of the preamplified templates at 50°C and then 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 58°C for 60 s.
Statistics.
Data were entered into Microsoft Excel for further analysis. Pearson correlation coefficients of HCMV loads between antigenemia and TaqMan assays were calculated and tested for statistical significance with SSPS (version 10; SSPS Inc., Chicago, Ill.). Comparisons of sensitivities, specificities, and predictive values were performed with Epiinfo software (version 6; Centers for Disease Control and Prevention, Atlanta, Ga.). A reduced random effects model with two nesting levels (analysis of variance balanced design) was used for analysis of variance for the reproducibility data (SAS software, version 8.2; Chapel Hill, N.C.). A P value of ≤0.05 was considered statistically significant.
RESULTS
A total of 298 whole-blood specimens submitted to the Clinical Virology Laboratory at Vanderbilt University Medical Center from 15 February to 31 October 1999 was included in the study. A housekeeping gene, human β-actin, was detectable by MTP-PCR in all DNA samples extracted from whole EDTA blood, indicating that these DNA samples were free of amplification inhibitors.
All 298 specimens were tested for HCMV by three assays: the qualitative MTP-PCR, the semiquantitative antigenemia assay, and the quantitative TaqMan assay. The antigenemia assay detected from 1 to 5,700 HCMV-positive cells per 2 × 105 leukocytes in 49 specimens. The sensitivity, specificity, positive predictive value, and negative predictive value were 70.5, 97.5, 87.8, and 92.8% with the MTP-PCR as reference. There were 78 blood specimens yielding detectable copy numbers by the TaqMan with sensitivity, specificity, positive predictive value, and negative predictive value of 96.7, 92.0, 75.6, and 99.1%, respectively, compared to results of the MTP-PCR (Table 1). While the TaqMan assay demonstrated a significantly better sensitivity than the antigenemia assay (χ2M-H = 15.5, P < 0.001), the latter possessed a higher specificity (χ2M-H = 7.12, where the subscript M-H [Mantel-Haenszel] indicates an adjustment for the χ2 value; P < 0.01), which yielded a more than 12.2% difference in positive predictive values.
TABLE 1.
Results of the antigenemia and the quantitative TaqMan assays compared with those of MTP-PCR on 298 clinical whole-blood specimens
| Test | No. positive by MTP-PCR and:
|
No. negative by MTP-PCR and:
|
Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | ||
|---|---|---|---|---|---|---|---|---|
| Positive by indicated test | Negative by indicated test | Positive by indicated test | Negative by indicated test | |||||
| Antigenemia (≥1 detectable as positive) | 43 | 18 | 6 | 231 | 70.5 | 97.5 | 87.8 | 92.8 |
| TaqMan (detectable as positive) | 59 | 2 | 19 | 218 | 96.7 | 92.0 | 75.6 | 99.1 |
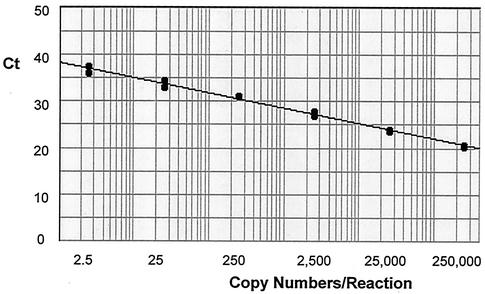
A quantitation standard curve was achieved by using six 10-fold dilutions of a plasmid standard containing the primer-spanning region of the HCMV glycoprotein B gene covering plasmid copies from 2.5 to 250,000 per reaction, which corresponded to 250 to 25,000,000 copies/ml of whole blood. The linear regression curve is shown in Fig. 1. The quantitative TaqMan assay detected from 48 to 5,425,000 copies of HCMV genomes/ml in 78 specimens. There was a high correlation between antigenemia values and HCMV loads determined by the quantitative TaqMan (r = 0.989, P < 0.001).
FIG. 1.
Standard curve generated by the quantitative TaqMan assay covering copy numbers from 2.5 to 250,000 per reaction. Duplicate samples were run for each copy number point. The standard curve possessed a correlation coefficient of 0.999 with a slope of −3.28 and a y-intercept value of 38.40. Ct, the threshold cycle (the cycle number at which the amount of amplified gene of interest reached a fixed threshold).
TaqMan reproducibility was determined on three blood samples (antigenemia values were 1, 83, and 650 positive cells per 2 × 105 leukocytes, respectively). These three samples were run in triplicate in one assay (on the same DNA extracts) and the same specimens in three different assays (on different DNA extracts) to assess the intra- and interassay variabilities. The variance of the HCMV load values within the assay was 0.026 among the triplicate tests for the same specimen. The interassay variances were 0.034 for the same specimens and 1.260 for different specimens. The interspecimen variance was about 37 times as large as the assay variance, indicating good reproducibility of the TaqMan assay. Also, there were significant differences between the underlying geometric mean HCMV load values for different specimens (P < 0.001).
The antigenemia values were used to determine several cutoff values of the TaqMan. Antigenemia values of 0 corresponded to a median TaqMan value of 125 copies/ml of whole blood (95% confidence limit; 146 to 233 copies/ml). Antigenemia values of 1 to 10 positive cells per 2 × 105 cells corresponded to a median HCMV of 1,593 copies/ml (95% confidence limit; 1,356 to 5,029 copies/ml). For antigenemia values of 11 to 100 positive cells per 2 × 105 cells, the corresponding median HCMV load measured by the TaqMan was 5,713 copies/ml (95% confidence limit; 3,232 to 12,827 copies/ml) (Table 2). HCMV loads of 1,000, 4,000, and 10,000 copies/ml were proposed as cutoff points corresponding to 1 to 2, 10, and 50 positive cells per 2 × 105 cells in the antigenemia tests.
TABLE 2.
Correlation of HCMV loads measured by the quantitative TaqMan and blood antigenemia assays
| Antigenemia values (positive cells/2 × 105 cells) | No. tested | HCMV median load (copies/ml) | 95% Confidence limit | Relative median ratio |
|---|---|---|---|---|
| 0 | 249 | 125 | 146-233 | 1 |
| 1-10 | 30 | 1,593 | 1,356-5,029 | 13 |
| 11-100 | 12 | 5,713 | 3,232-12,827 | 46 |
| 101-1,000 | 6 | 16,825 | 0-666,823 | 135 |
| >1,000 | 1 | 5,425,000 | NAa | 43,400 |
NA, not applicable.
We next studied whether leukocyte concentration had any effect on test efficacy. There were 271 specimens involved in this study that had total leukocyte count information available. As shown in Table 3, 13 (4.8%) specimens had leukocyte counts of less than 1,500/mm3. Discrepancies between the results of the antigenemia and TaqMan test were more common in the specimens with leukocyte counts of less than 1,500/mm3 (P < 0.005). All the discrepant results at low leukocyte numbers consisted of samples with negative antigenemia but detectable viral load by TaqMan (Table 3).
TABLE 3.
Antigenemia values and TaqMan HCMV loads in whole-blood specimens with different leukocyte counts
| Leukocyte level (per mm3) | No. tested | No. of samples with discrepant results
|
Total discrepancy rate (%) | |
|---|---|---|---|---|
| Antigenemia positive, TaqMan negative | Antigenemia negative, TaqMan positive | |||
| ≥1,500 | 258 | 6 | 25 | 12.0 |
| <1,500 | 13 | 0 | 6 | 46.2 |
| Total | 271 | 6 | 31 | 13.7 |
Several parameters of the three techniques described in this study, including material costs, minimal test turnaround time, technologist hands-on time, and test range, are listed in Table 4. While costs of materials and reagents were similar for these three tests, the hands-on time for performing the quantitative TaqMan assay was 2.5 min, which was only 18 and 13% of the hands-on time needed for the MTP-PCR and antigenemia assays, respectively. The quantitative TaqMan assay yielded a wide linear HCMV load range from ≤125 to 5,425,000 copies/ml within 3 h.
TABLE 4.
Comparison of assays for HCMV detection
| Test | Material cost/test ($) | Minimal test turnaround time (h) | Hands-on time (min) | Test range |
|---|---|---|---|---|
| MTP-PCR | 11.12 | 8 | 14.0 | Positive or negative (qualitative) |
| Antigenemia | 12.96 | 3 | 20.0 | 0-5,700 positives/2 × 105 leukocytes |
| Real-time TaqMan | 10.88 | 3 | 2.5 | ≤125-5,425,000 copies/ml of whole blood |
DISCUSSION
Quantitation of circulating HCMV has been demonstrated to be useful in clinical contexts, such as virological surveillance of transplant recipients and monitoring of antiviral therapy (3, 5, 9, 25). Higher viral loads measured by nucleic acid assays have been found to correlate with an increased likelihood of disease (1, 8, 16, 21), and nucleic acid quantitation can be used to track response to therapy (14, 15, 17, 33, 37). Due to the high sensitivity of these assays, HCMV is also detectable in a substantial number of patients with asymptomatic infection who never progress to disease (3, 23). Therefore, qualitative PCR-based assays are probably too sensitive for clinical purposes. The plus-minus answer they supply is overly simplistic for clinical purposes, as it only allows for a single threshold to initiate therapy and provides limited feedback on response to therapy (21, 28, 35).
The real-time PCR technology measures quantitative levels of HCMV DNA that are useful for predicting disease and monitoring response to antiviral therapy (7, 10, 11, 14, 15, 26, 27, 29, 32, 37). In our study, the HCMV DNA copy number determined with the quantitative TaqMan assay correlated with the number of pp65-positive cells, in agreement with the results of other studies of TaqMan-based assays (7, 10, 11, 15, 26, 29). An important test of the TaqMan was whether it could discriminate viral load as well as antigenemia (10, 11). The HCMV loads measured by our quantitative real-time PCR assay performed well in this regard. Antigenemia values of 0, 1 to 10, 11 to 100, 101 to 1,000, and over 1,000 positive cells per 2 × 105 leukocytes corresponded to median HCMV loads measured by TaqMan of 125, 1,593, 5,713, 16,825, and 5,425,000 copies/ml, respectively. We have been able to propose cutoff points of 1,000, 4,000, and 10,000 copies/ml as treatment thresholds for patients with high, intermediate, and low risk of CMV diseases corresponding to antigenemia values of 1 to 2, 10, and 50 positive cells per 2 × 105 leukocytes (4, 34).
Although HCMV is a cell-associated virus, HCMV quantitation can be performed in different acellular fractions of the blood, such as plasma and serum (14, 32). The optimal specimen depends on the test method and clinical setting; however, most studies show that the quantity of viral DNA in leukocytes is generally greater than that in plasma for both transplant recipients and subjects with AIDS (8, 38). The use of leukocytes may be more appropriate in situations where the viral DNA load is low (8) or when preemptive therapy is envisaged in a setting where there is rapid progression from the first detection to HCMV disease (4). However, testing on leukocytes requires immediate specimen processing and increases the test turnaround time significantly, thereby limiting the utility of this test. In this study we used whole EDTA blood in order to recover viral DNA from both cellular and acellular fractions. A housekeeping gene, β-actin, was successfully amplified from all DNA samples extracted by the Qiagen tissue/blood kit, indicating that nucleic acids extracted from whole EDTA blood were free of amplification inhibitors.
While real-time PCR assays have been successfully adapted for HCMV quantitation (2, 7, 10, 11, 14, 15, 26, 27, 29, 32, 37), it is important to point out that such assays may not be the tests of choice for qualitative purposes. In our present study, sensitivity and specificity of the TaqMan assay were 96.7 and 92.0%, respectively, compared to that of the MTP-PCR. This apparent reduction in sensitivity has been reported previously (18, 20, 27). In a study that compared HCMV load detection between the LightCycler real-time and the COBAS CMV Monitor systems in solid organ transplant recipients, higher HCMV DNA levels were observed with the latter system (22). The small volume of starting material used in real-time formats may contribute to a loss of sensitivity in the presence of low levels of virus. On the other hand, several specimens that were negative by MTP-PCR and antigenemia assays were found to have low but definite numbers of copies by the quantitative TaqMan assay (0.19 to 0.92 copies per reaction, corresponding to 48 to 245 copies/ml). This accounted for the positive predictive value of only 75.6% in our study. Discrepant results also occurred with higher cycling numbers when two gene targets were selected for the detection of varicella-zoster virus by using the LightCycler real-time system (6). While we are satisfied with our quantitation results by real-time formats, one should be careful in applying this method for qualitative purposes.
The most significant advance of the real-time systems comes from is its rapid thermocycling and simultaneous detection characteristics. Delays in sample processing can result in significant decrease in antigenemia levels (3), while blood samples for PCR testing may be stored for up to 72 h without deterioration (24, 28). While the costs of materials and reagents are similar to those of other tests in this study, the TaqMan system yielded test results within 3 h with only 2.5 min of a technologist's hands-on time for each specimen. The TaqMan system can simultaneously analyze 96 samples in a microwell format, which is especially beneficial for clinical laboratories handling large numbers of specimens. Additional advantages of the TaqMan system include easier handling and greater suitability to the automation of specimen processing. In a climate of continuous cost containment and decreasing reimbursement for clinical laboratory procedures, real-time PCR quantitation technologies, such as the TaqMan assay, have significant advantages for routine diagnostic testing.
In summary, we have addressed the appropriate cutoff points of HCMV loads determined by quantitative real-time PCR for the initiation of antiviral therapy. This assay yielded a wide linear quantitation range from <125 to 5,425,000 HCMV copies/ml. HCMV loads in whole-blood specimens correlated well with HCMV-positive cell counts in peripheral leukocytes tested by the antigenemia assay. This high correlation demonstrated between these two assays validates the quantitative real-time PCR as another rapid, accurate laboratory tool for monitoring HCMV infections in transplant recipients.
Acknowledgments
We thank Linda Carter, Melinda Caudill, Suzanne Fernandez, Valerie Hill, Elaine Mahaffey, Jill White-Abell, and Jannie Whitehurst for helping collect clinical specimens, Yuwei Zhu for assisting with statistical analysis, and Michael Kleines for providing plasmid pCT-gpB.
REFERENCES
- 1.Aitken, C., W. Barrett-Muir, C. Millar, K. Templeton, J. Thomas, F. Sheridan, D. Jeffries, M. Yaqoob, and J. Breuer. 1999. Use of molecular assays in diagnosis and monitoring of cytomegalovirus disease following renal transplantation. J. Clin. Microbiol. 37:2804-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, X., G. Hosler, B. B. Rogers, D. B. Dawson, and R. H. Scheuermann. 1997. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin. Chem. 43:1843-1849. [PubMed] [Google Scholar]
- 3.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation 64:108-113. [DOI] [PubMed] [Google Scholar]
- 5.Caliendo, A. M., R. Schuurman, B. Yen-Lieberman, S. A. Spector, J. Andersen, R. Manjiry, C. Crumpacker, N. S. Lurain, and A. Erice. 2001. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 39:1334-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espy, M. J., R. Teo, T. K. Ross, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Diagnosis of varicella-zoster virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:3187-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerna, G., M. Furione, F. Baldanti, and A. Sarasini. 1994. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J. Clin. Microbiol. 32:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths, P. D., A. V. Cope, A. F. Hassan-Walker, and V. C. Emery. 1999. Diagnostic approaches to cytomegalovirus infection in bone marrow and organ transplantation. Transplant. Infect. Dis. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 10.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J. H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 39:4362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guiver, M., A. J. Fox, K. Mutton, N. Mogulkoc, and J. Egan. 2001. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation 71:1609-1615. [DOI] [PubMed] [Google Scholar]
- 12.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landry, M. L., D. Ferguson, T. Stevens-Ayers, M. W. de Jonge, and M. Boeckh. 1996. Evaluation of CMV Brite kit for detection of cytomegalovirus pp65 antigenemia in peripheral blood leukocytes by immunofluorescence. J. Clin. Microbiol. 34:1337-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 15.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 38:2536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendez, J. C., M. J. Espy, T. F. Smith, J. A. Wilson, and C. V. Paya. 1998. Evaluation of PCR primers for early diagnosis of cytomegalovirus infection following liver transplantation. J. Clin. Microbiol. 36:526-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mutimer, D., A. Matyi-Toth, J. Shaw, E. Elias, K. O'Donnell, and P. Stalhandske. 1997. Patterns of viremia in liver transplant recipients with symptomatic cytomegalovirus infection. Transplantation 63:68-73. [DOI] [PubMed] [Google Scholar]
- 18.Najioullah, F., D. Thouvenot, and B. Lina. 2001. Development of a real-time PCR procedure including an internal control for the measurement of HCMV viral load. J. Virol. Methods 92:55-64. [DOI] [PubMed] [Google Scholar]
- 19.Ng, S. Y., P. Gunning, R. Eddy, P. Ponte, J. Leavitt, T. Shows, and L. Kedes. 1985. Evolution of the functional human beta-actin gene and its multi-pseudogene family: conservation of noncoding regions and chromosomal dispersion of pseudogenes. Mol. Cell. Biol. 5:2720-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicoll, S., A. Brass, and H. A. Cubie. 2001. Detection of herpes viruses in clinical samples using real-time PCR. J. Virol. Methods 96:25-31. [DOI] [PubMed] [Google Scholar]
- 21.Paya, C. V. 2001. Prevention of cytomegalovirus disease in recipients of solid-organ transplants. Clin. Infect. Dis. 32:596-603. [DOI] [PubMed] [Google Scholar]
- 22.Razonable, R. R., R. A. Brown, M. J. Espy, A. Rivero, W. Kremers, J. Wilson, C. Groettum, T. F. Smith, and C. V. Paya. 2001. Comparative quantitation of cytomegalovirus (CMV) DNA in solid organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 39:4472-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razonable, R. R., C. V. Paya, and T. F. Smith. 2002. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid-organ transplant recipients. J. Clin. Microbiol. 40:746-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts, T. C., R. S. Buller, M. Gaudreault-Keener, K. E. Sternhell, K. Garlock, G. G. Singer, D. C. Brennan, and G. A. Storch. 1997. Effects of storage temperature and time on qualitative and quantitative detection of cytomegalovirus in blood specimens by shell vial culture and PCR. J. Clin. Microbiol. 35:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saltzman, R. L., M. R. Quirk, and M. C. Jordan. 1992. High levels of circulating cytomegalovirus DNA reflect visceral organ disease in viremic immunosuppressed patients other than marrow recipients. J. Clin. Investig. 90:1832-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez, J. L., R. M. Kruger, S. Paranjothi, E. P. Trulock, J. P. Lynch, C. Hicks, W. D. Shannon, and G. A. Storch. 2001. Relationship of cytomegalovirus viral load in blood to pneumonitis in lung transplant recipients. Transplantation 72:733-735. [DOI] [PubMed] [Google Scholar]
- 27.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol. 38:4006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer, P., W. Tenschert, K. Gutensohn, and R. Laufs. 1997. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J. Clin. Microbiol. 35:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:455-462. [DOI] [PubMed] [Google Scholar]
- 30.Tang, Y. W., M. J. Espy, D. H. Persing, and T. F. Smith. 1997. Molecular evidence and clinical significance of herpesvirus coinfection in the central nervous system. J. Clin. Microbiol. 35:2869-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang, Y. W., P. N. Rys, B. J. Rutledge, P. S. Mitchell, T. F. Smith, and D. H. Persing. 1998. Comparative evaluation of colorimetric microtiter plate systems for detection of herpes simplex virus in cerebrospinal fluid. J. Clin. Microbiol. 36:2714-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tedder, R. S., U. Ayliffe, W. Preiser, N. S. Brink, P. R. Grant, K. S. Peggs, S. Mackinnon, F. Kreig-Schneider, S. Kirk, and J. A. Garson. 2002. Development and evaluation of an internally controlled semiautomated PCR assay for quantification of cell-free cytomegalovirus. J. Med. Virol. 66:518-523. [DOI] [PubMed] [Google Scholar]
- 33.Toyoda, M., J. B. Carlos, O. A. Galera, K. Galfayan, X. Zhang, Z. Sun, L. S. Czer, and S. C. Jordan. 1997. Correlation of cytomegalovirus DNA levels with response to antiviral therapy in cardiac and renal allograft recipients. Transplantation 63:957-963. [DOI] [PubMed] [Google Scholar]
- 34.van den Berg, A. P., W. van der Bij, W. J. van Son, J. Anema, M. van der Giessen, J. Schirm, A. M. Tegzess, and T. H. The. 1989. Cytomegalovirus antigenemia as a useful marker of symptomatic cytomegalovirus infection after renal transplantation-a report of 130 consecutive patients. Transplantation 48:991-995. [DOI] [PubMed] [Google Scholar]
- 35.Weber, B., U. Nestler, W. Ernst, H. Rabenau, J. Braner, A. Birkenbach, E. H. Scheuermann, W. Schoeppe, and H. W. Doerr. 1994. Low correlation of human cytomegalovirus DNA amplification by polymerase chain reaction with cytomegalovirus disease in organ transplant recipients. J. Med. Virol. 43:187-193. [DOI] [PubMed] [Google Scholar]
- 36.Whitley, R. J., M. A. Jacobson, D. N. Friedberg, G. N. Holland, D. A. Jabs, D. T. Dieterich, W. D. Hardy, M. A. Polis, T. A. Deutsch, J. Feinberg, S. A. Spector, S. Walmsley, W. L. Drew, W. G. Powderly, P. D. Griffiths, C. A. Benson, and H. A. Kessler. 1998. Guidelines for the treatment of cytomegalovirus diseases in patients with AIDS in the era of potent antiretroviral therapy: recommendations of an international panel. International AIDS Society-USA. Arch. Intern. Med. 158:957-969. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida, A., S. Hitomi, T. Fukui, H. Endo, Y. Morisawa, Y. Kazuyama, K. Osumi, S. Oka, and S. Kimura. 2001. Diagnosis and monitoring of human cytomegalovirus diseases in patients with human immunodeficiency virus infection by use of a real-time PCR assay. Clin. Infect. Dis. 33:1756-1761. [DOI] [PubMed] [Google Scholar]
- 38.Zipeto, D., S. Morris, C. Hong, A. Dowling, R. Wolitz, T. C. Merigan, and L. Rasmussen. 1995. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J. Clin. Microbiol. 33:2607-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]