Abstract
The recent description of the respiratory pathogen human metapneumovirus (hMPV) has highlighted a deficiency in current diagnostic techniques for viral agents associated with acute lower respiratory tract infections. We describe two novel approaches to the detection of viral RNA by use of reverse transcriptase PCR (RT-PCR). The PCR products were identified after capture onto a solid-phase medium by hybridization with a sequence-specific, biotinylated oligonucleotide probe. The assay was applied to the screening of 329 nasopharyngeal aspirates sampled from patients suffering from respiratory tract disease. These samples were negative for other common microbial causes of respiratory tract disease. We were able to detect hMPV sequences in 32 (9.7%) samples collected from Australian patients during 2001. To further reduce result turnaround times we designed a fluorogenic TaqMan oligoprobe and combined it with the existing primers for use on the LightCycler platform. The real-time RT-PCR proved to be highly reproducible and detected hMPV in an additional 6 out of 62 samples (9.6%) tested during the comparison of the two diagnostic approaches. We found the real-time RT-PCR to be the test of choice for future investigation of samples for hMPV due to its speed, reproducibility, specificity, and sensitivity.
The recent description of a new member of the family Paramyxoviridae, subfamily Pneumovirus, has provided an opportunity to define the extent of disease caused by this previously unknown human pathogen (12). Human metapneumovirus (hMPV) has been detected in, and isolated from, nasopharyngeal aspirates (NPA) taken from patients with respiratory disease (9, 12).
Disease caused by hMPV appears similar to that caused by human respiratory syncytial virus (hRSV), which is the most significant cause of viral lower respiratory tract disease in infants and children worldwide (2, 4, 5, 8, 10, 13). Patients show symptoms ranging from a wheeze to bronchiolitis and may require assisted ventilation.
The long-term presence of hMPV in the community has been confirmed by detection of anti-hMPV antibodies during retrospective seroepidemiological studies of stored Dutch sera (12). The virus appears to infect the majority of The Netherlands' population by the age of 5 years. Within our hospital population, the causative viral agent of respiratory tract disease remains unidentified in 38 to 40% of patients examined.
Because the virus is difficult to detect by cell culture, due to its selective, slow growth and mild cytopathicity and the absence of specific diagnostic reagents, the need for a reliable and rapid diagnostic method is obvious. During the course of routine culture to isolate respiratory viruses from these patients, cytopathicity is noticed. However, enzyme immunoassay, immunofluorescent assay, and reverse transcriptase PCR (RT-PCR) for common viral and bacterial agents of respiratory disease fail to identify the cause. The hMPV negative-sense RNA genome is only 20% homologous to the nucleocapsid (N) gene of hRSV and 52% homologous to avian pneumovirus (14), so it is sufficiently distinct that its sequences do not cross-react with existing nucleic acid amplification assays. Ideally, in addition to being specific, a new diagnostic method should be rapid, sensitive, and able to be incorporated into existing testing paradigms for the screening of respiratory tract disease.
We describe the development of novel conventional RT-PCR and real-time RT-PCR which utilizes the LightCycler platform to detect hMPV RNA in patient samples. Using these assays, we examined samples from patients with respiratory tract disease of unknown etiology.
MATERIALS AND METHODS
Samples and RNA purification.
Samples (n = 329) were collected from March to October 2001 from patients who tested negative for the common agents of respiratory disease. In addition, samples (n = 37) were included from patients with previously diagnosed microbial causes of respiratory disease for use in the specificity studies. These pathogens included Mycoplasma pneumoniae, adenovirus, influenza virus, parainfluenza virus I-III, and hRSV. These microbial diagnoses were made by validated cell culture or nucleic acid amplification assays. Nucleic acids were also extracted from a range of microbes for use in cross-reaction studies. These included Bacillus cereus, Bartonella henselae, Burkholderia cepacia, Campylobacter coli, Candida albicans, coronavirus 229E, Cryptococcus neoformans, Gardnerella vaginalis, Haemophilus influenzae, human rhinovirus, Klebsiella pneumoniae, M. pneumoniae, Neisseria lactamica, Neisseria meningitidis, Pseudomonas aeruginosa, Saccharomyces cerevisiae, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus hominis, Streptococcus pneumoniae, Streptococcus pyogenes, and Vibrio parahaemolyticus.
Nucleic acids were purified from NPA placed in viral transport medium (VTM). A spin column kit was used (High Pure viral nucleic acid kit; Roche Diagnostics, GmbH) according to the manufacturer's instructions. Purified nucleic acids and VTM were stored at −70°C.
Conventional PCR and nucleotide sequencing.
RT-PCR was performed using the SUPERSCRIPT One-Step system (Invitrogen, Melbourne, Australia). Each 25-μl reaction contained 3 μl of RNA, 3 mM MgSO4, 0.2 μM (each) primers (Table 1), and 5 μM digoxigenin-11-dUTP (Roche Molecular Biochemicals, GmbH). The reaction mixes were exposed to a 30-min, 50°C RT incubation and then 45 cycles of 94°C for 20 s, 55°C for 20 s, and 68°C for 20 s, with a final 68°C extension for 7 min. The amplification phase took approximately 2 h to complete and resulted in a 213-nucleotide (nt) PCR product (amplicon).
TABLE 1.
Nucleotide sequences of sense (01) and antisense (02) oligonucleotide primers and biotinylated (MPV P01) or fluorogenic endonuclease (MPV TM02) oligoprobesa
| Primer | Sequence |
|---|---|
| MPV01.2 | AAC CGT GTA CTA AGT GAT GCA CTC |
| MPV02.2 | CAT TGT TTG ACC GGC CCC ATA A |
| MPV P01 | Biotin-GAA GTT TGT TCA TTG AGT ATG G |
| MPV TM02 | FAM-CTT TGC CAT ACT CAA TGA ACA AAC-TAMRA |
Designs were based on the hMPV N gene sequence available from GenBank under accession number AF371337.
The amplicon was purified from solution using a spin column kit (QIAquick; Qiagen, Valencia, Calif.) according to the manufacturer's instructions and was then eluted into a final volume of 30 μl. Nucleotide sequencing reactions were prepared with the ABI PRISM BigDye cycle sequencing kit (Perkin Elmer Applied Biosystems Division, Norwalk, Conn.) according to the manufacturer's instructions. Sequences were determined by an Applied Biosystems 377 DNA sequencer.
Amplicon detection by ELAHA.
Modifications were made to our previously described enzyme-linked amplicon hybridization assay (ELAHA [7, 15]). These consisted of a reduction in the total volume of the mixture to be denatured and captured as well as a doubling of the length of the incubation of oligoprobe-target duplex in the streptavidin-coated microwells. The new method combined 5 μl of amplicon with 10 μl of biotinylated oligoprobe (MPV P01; 0.2 μM) (Table 1) and 30 μl of SSC (0.3 M NaCl, 0.15 M sodium citrate, pH 7.0). This mixture was incubated at 95°C for 5 min and then rapidly cooled to 4°C using a thermocycler. The oligoprobe-DNA complexes were captured onto the surfaces of streptavidin-coated microwells during a 40-min incubation at 37°C. The microwells were washed three times with SSC. Each well was then incubated for 20 min at 37°C with 50 μl of a peroxidase-labeled anti-digoxigenin-11-dUTP antibody (750 mU/ml; Roche Molecular Biochemicals, GmbH). Following another three washes, 50 μl of tetramethylbenzidine substrate (Astral Scientific, Gymea, New South Wales, Australia) was incubated in each well at room temperature for 10 min. The reaction was stopped by the addition of 50 μl of 1 M HCl per well. The optical density of each microwell was determined by spectrophotometer at a wavelength of 450 nm with a 690 nm reference filter (Murex Diagnostics, Dartford, United Kingdom). Samples that returned values of at least 0.2 optical density (OD) units were considered to contain hMPV RNA. This cutoff was determined after studying the OD values produced by amplification and detection of the 37 samples used for the specificity study. The final cutoff was calculated as the mean of these values plus 4 standard deviations and the result was rounded up to the first significant figure. The ELAHA took approximately 2 h to perform, depending on the number of samples.
Real-time RT-PCR.
Real-time RT-PCR was applied to the detection of viral RNA using a single-tube RT-PCR kit according to the manufacturer's instructions (RNA amplification kit for HybProbes; Roche Diagnostics, GmbH). The amplification and detection were performed in the LightCycler, a combined thermocycler and fluorimeter (Roche Diagnostics, GmbH). Each glass capillary contained a 10-μl reaction mix which included 3 μl of purified sample RNA, 1.0 μM sense primer (MPV01.2), 0.2 μM antisense primer (MPV02.2), 7 mM MgCl2, and 0.2 μM fluorogenic endonuclease probe (TaqMan; TIB-MOLBIOL, Berlin, Germany) labeled at the 5′ end with FAM (6-carboxy-fluorescein) and at the 3′ end with TAMRA (6-carboxy-tetramethyl-rhodamine) (Table 1). The reaction mixes were exposed to a 15-min, 50°C RT step, a 30-s denaturation at 95°C, and then 55 PCR cycles consisting of 94°C for 4 s, 55°C for 9 s, and 72°C for 12 s, using a channel setting of F1/F2.
A dual-round, single-tube, real-time RT-PCR was performed in two steps. The reaction mixes were created as described earlier, but they included a 0.2 μM symmetric primer mix and 10 μl of mineral oil (Nujol; Perkin Elmer). The capillaries were then capped, loaded into centrifuge adapters, and spun at 800 × g in a bench-top microcentrifuge. Next, the capillaries were returned to the preparation area and uncapped, and 0.5 μl of the previously described asymmetric primer mix was added. The capillaries were then capped, loaded into the LightCycler's carousel, and spun in the carousel centrifuge (Roche Diagnostics, GmbH). The first round of amplification incorporated reverse transcription, denaturation of the RT, and 15 cycles of PCR. The samples were cooled to 40°C and the carousel was removed, centrifuged, and replaced. The second round consisted of an additional 40 cycles of PCR.
To investigate the efficiency of the amplification reaction and the nature of the amplicon, fluorescence melting curve analysis was performed. At the completion of the amplification phase, the amplicon was denatured at 95°C for 0 s and then heated to 50°C and held for 30 s. Finally, the amplicon was heated to 95°C at 0.3°C/s while the fluorescent emissions were continuously monitored by the LightCycler and presented as the negative derivative of the fluorescence over time. The dual-round real-time RT-PCR took approximately 80 min to complete compared to 60 min for the single-round assay.
Immunofluorescent assay.
Aliquots of VTM which had been inoculated with NPA from the patients previously described were added to cultures of monkey kidney or human lung epithelial cells (LLC MK2 or A549, respectively) in the presence of 10% (vol/vol) fetal calf serum, 50 μM penicillin G, 50 μg of streptomycin sulfate per ml, and 0.02% trypsin (Invitrogen). Cultures were maintained for a week before screening of the supernatant and cells by real-time RT-PCR commenced. Mock-infected or hRSV-infected cultures were grown and screened in parallel. When cytopathic effects became apparent, culture supernatant and cells were used to infect fresh cell monolayers growing on chamber slides (Nunc, Rochester, N.Y.). Dried monolayers were probed using a diluted adult serum. Positive cells were detected after incubation with a fluorescein isothiocyanate-conjugated secondary antibody.
Nucleotide sequence accession numbers.
Sequences containing the amplicon described in this paper were deposited into the GenBank database under the accession numbers AF442513-6 and AF443830-4.
RESULTS
Primer and oligoprobe specificity.
A range of related and unrelated nucleic acid templates were subjected to PCR using primers MPV01.2 and MPV02.2, which are common to both assays. No signal was produced in the ELAHA or the real-time RT-PCR from any of these reactions (data not shown). To further examine the specificity of the ELAHA, a strongly positive hMPV amplicon was denatured in the presence of a series of biotinylated oligoprobes. The oligoprobes ranged in size from 23 to 28 nt and were designed for use in ELAHA to detect hRSV, influenza viruses, rhinoviruses, M. pneumoniae, parainfluenza viruses, adenoviruses, herpesviruses, human coronavirus, Candida species, Aspergillus species, measles virus, enteroviruses, the malarial parasites, Treponema pallidum, parvovirus B19, and Klebsiella species. No signal was produced using any of these templates, confirming that the hMPV assays are highly specific and that hybridization by ELAHA is a suitably stringent technique (data not shown).
Relative sensitivity of the ELAHA.
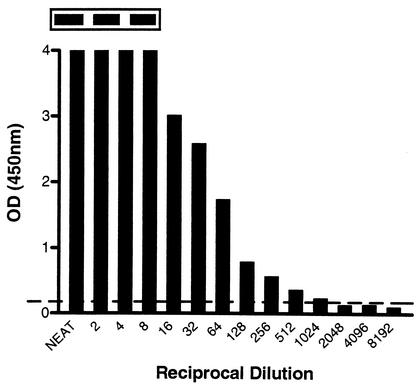
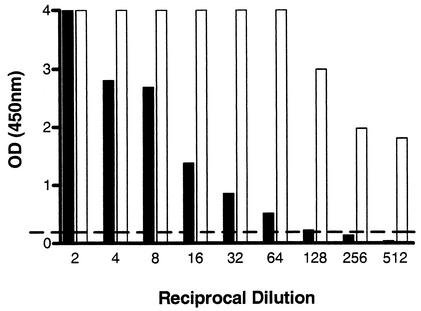
To investigate how sensitive the modified ELAHA was in comparison to a traditional amplicon detection method, we made serial twofold dilutions of an hMPV amplicon and examined 5 μl by agarose gel electrophoresis and ELAHA. The results revealed a 512-fold improvement in the sensitivity of the ELAHA compared to electrophoresis (Fig. 1). To examine the effect of an asymmetric PCR primer mix on the ELAHA, the antisense primer concentration was increased fivefold in an effort to make more of the strand to which the biotinylated oligoprobe will hybridize. This produced an increased signal, resulting in at least a fourfold increase in the sensitivity of amplicon detection (Fig. 2).
FIG. 1.
Titration of 5-μl aliquots of hMPV amplicon and detection by ELAHA. The inset panel displays the first three corresponding bands on an agarose gel. There is a faint band visible up to a dilution of 1:2, whereas the ELAHA is positive to a dilution of 1:1,024. The horizontal line represents the ELAHA cutoff value of 0.2 OD units.
FIG. 2.
Comparison of assay sensitivity using symmetric (filled bars) (0.2 μM sense and antisense primers) and asymmetric (open bars) (0.2 μM sense and 1.0 μM antisense primers) RT-PCR-ELAHA. The cutoff for a positive ELAHA result is shown as a broken, horizontal line.
Real-time RT-PCR.
To further reduce the turn-around time of the RT-PCR-ELAHA, we converted the conventional PCR to a real-time RT-PCR, which incorporated the same primers and included a fluorogenic oligoprobe. During this assay the LightCycler instrument continuously monitored amplicon accumulation by detecting the proportional increase in fluorescence emitted as the result of a loss of quenching of the FAM reporter fluorophore as the oligoprobe was hydrolyzed. The real-time assay was at least 180 min faster than the RT-PCR-ELAHA, taking approximately 60 min for 32 samples. In addition, there was no need to open the capillary to detect amplicon, thus significantly restricting the potential for carryover contamination.
Assay reproducibility, based on analysis of the fractional cycle number at which the fluorescence generated by the accumulating amplicon exceeds the baseline (threshold cycle [CT]), was determined after examining replicate samples, using different vials of RT-PCR master mix. Six replicates were assayed in each run, and this was performed by the same technician on different days. The assay gave highly reproducible results, with intra-assay coefficients of variation (CV) of 0.4 to 2% and an interassay CV of 3%.
Although the TaqMan and ELAHA oligoprobes hybridize with an almost identical sequence of the hMPV N gene, the target sequences are on opposite strands. This design was necessary in order to avoid a guanine on the template strand, which would occur adjacent to the FAM reporter molecule, resulting in nonspecific quenching.
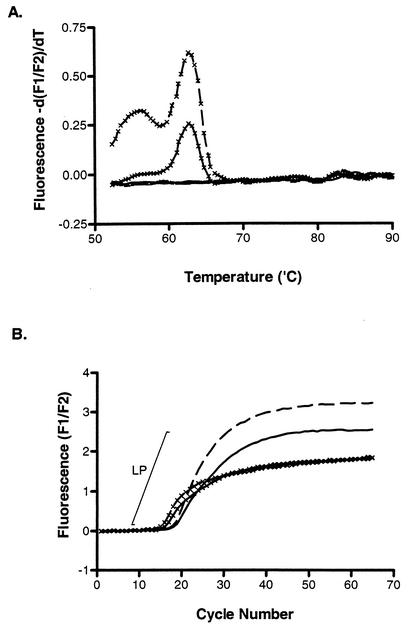
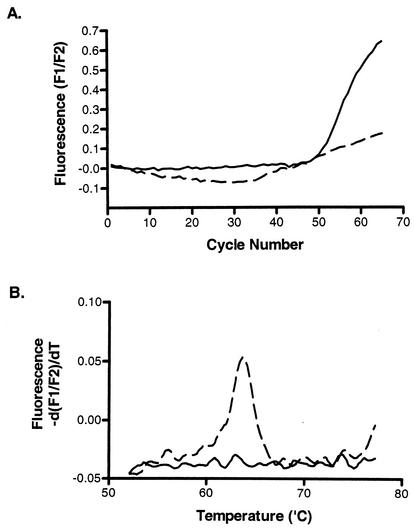
To study the efficiency of oligoprobe hydrolysis and, by extrapolation, the efficiency of the real-time PCR, we applied fluorescent melting curve analysis to the amplicon produced in the presence of an asymmetric primer mix. This resulted in a peak that was specific for the hMPV oligoprobe's nucleotide sequence and approximated its expected melting temperature (Tm) (Fig. 3A). When the kinetic curves were examined, it was apparent that the only reactions producing a melt curve were those containing a fivefold excess of the sense primer which produces the fluorogenic oligoprobe's target strand (Fig. 3B). The CT value decreased when using this primer mix; however, the kinetic curves reached a plateau earlier, resulting in short log-linear phases (LP). This is of concern if the assay is to be used quantitatively or if samples contain low levels of RNA template (Fig. 4). The product of reverse transcription of hMPV negative-sense RNA is positive-sense DNA, which is also the fluorogenic oligoprobe's target. We hypothesized that an asymmetric primer mix designed to produce an excess of the sense strand is not conducive, at least in the early cycles of PCR, to efficient exponential amplification of a sense-strand DNA due to the initial absence of negative-sense template DNA. We examined whether this was likely to reduce the PCR efficiency by developing a dual-round, real-time RT-PCR in which the first round of 15 cycles used a symmetric primer mix of MPV01.2 and MPV02.2 at 2 μM in order to allow the relatively unbiased reverse transcription and subsequent creation of antisense strands by PCR. The first round was separated from an asymmetric primer mix by a barrier of mineral oil. Prior to the second round of amplification, the capillaries were centrifuged, introducing the asymmetric primer combination, which contained a fivefold excess of the sense primer, to the PCR mix. While this format added approximately 15 min to the real-time assay, it retained the homogenous nature of real-time PCR. The dual-round assay produced reduced CT values for the detection of weakly positive RNA templates and improved PCR performance as gauged by the more pronounced linear phase of amplification seen in the resultant PCR curves of strongly positive templates (Fig. 4).
FIG. 3.
The effect of an asymmetric primer mix on the reaction efficiency of a strong hMPV-positive patient sample. Fluorescent melting-curve analysis (A) only produced peaks from the amplicon that was created in the presence of an excess of sense primer (0.5 μM MPV01.2-0.2 μM MPV02.2 or 1.0 μM MPV01.2-0.2 μM MPV02.2, indicated by a solid or broken line, respectively, with crosses), whereas amplification using an excess of antisense primer (0.2 μM MPV01.2-0.5 μM MPV02.2 or 0.2 μM MPV01.2-1.0 μM MPV02.2, indicated by a solid or broken line, respectively, without crosses) (B) produced a higher amplicon yield and extended the LP of amplification.
FIG. 4.
A weakly hMPV-positive patient sample was amplified by single-round (broken line) or dual-round (solid line) real-time RT-PCR. The second step of the reaction incorporated an asymmetric primer mix containing a fivefold excess of sense primer.
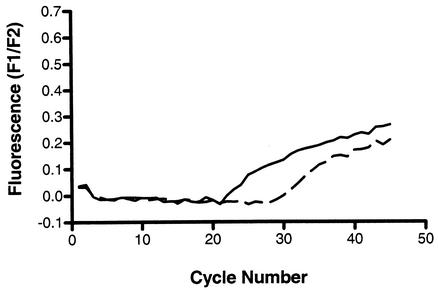
Rather than redesign the oligoprobe to hybridize to a location on the antisense strand, we investigated whether reversing the order of the asymmetric primer mix would improve the signal. Using 2 and 10 μM MPV01.2 and MPV02.2 primers, respectively, we were able to show improved amplification of the weakly positive RNA, producing a better quality of kinetic curve in the absence of a melt peak (Fig. 5).
FIG. 5.
Using the same weakly positive patient sample that was used for Fig. 4, hMPV RNA was amplified by single-round RT-PCR using asymmetric primers with a fivefold excess of the sense primer, MPV01.2 (broken line), or of the antisense primer, MPV02.2 (solid line). The kinetic curves indicate that the latter primer mix is superior (A) and this is confirmed by the melt analysis which shows incomplete oligoprobe hydrolysis when using more of the sense primer (B).
Specimen testing and comparative assay sensitivity.
Initially, the RT-PCR-ELAHA had been used to screen 329 NPA samples, detecting hMPV sequences in 32 (9.7%) of these.
To compare the ability of the real-time assay to detect template, fivefold dilutions of supernatant from an hMPV-positive culture were tested by both assays. The RT-PCR-ELAHA and the real-time RT-PCR were capable of detecting hMPV sequences to a dilution of 1:125, indicating equivalent analytical sensitivity (data not shown).
To evaluate the clinical sensitivity of these assays under routine conditions, a subset of 62 NPA samples was chosen for investigation using both assays. This subset contained 6 hMPV RNA-positive samples and 56 samples that did not contain hMPV RNA as determined by RT-PCR-ELAHA. The real-time assay detected hMPV sequences in an additional 6 of these specimens, bringing the total number of hMPV-positive samples to 12. This was equivalent to an improvement in the rate of detection of 9.6% within this population.
Sequencing of the amplicon.
To further ensure that the amplicon produced from these assays was due to the amplification of a portion of the hMPV N gene, nucleotide sequencing was performed.
Confirmation of active hMPV infection.
In order to confirm that our assays were detecting an active viral infection, cell cultures were incubated with VTM when sufficient volume was available (at least 200 μl) and provided that normal flora had not overgrown the sample during transport or specimen handling.
Virus was passaged several times and the status of virus replication monitored by real-time RT-PCR. In mock-infected or hRSV-infected control cell cultures, no PCR product was produced during any stage of the culture process. To further confirm that virus was actively replicating, immunofluorescence studies were performed on cells taken from RT-PCR positive cultures and probed using an adult serum. Positive, bright green cells were observed either singly or in adjacent clusters.
DISCUSSION
We have described the detection of hMPV from Australian children and infants by two newly developed RT-PCR assays. The need for rapid diagnosis of respiratory tract disease is acutely important during seasonal outbreaks of the causative agents (6).
Using these assays we were able to detect virus in 10% of local NPA samples collected during the period from March to October 2001, and subsequent isolation of hMPV was possible from RT-PCR positive VTMs stored at −70°C. These samples originated from patients presenting to the hospital with respiratory tract disease of an otherwise unknown microbial origin. Our rate of detection is identical to that reported by van den Hoogen et al. in their prospective studies of children who presented to a medical center with respiratory tract disease and were found to be RT-PCR positive for hMPV (14).
Neither of our assays cross-reacted with nucleic acids purified from the other common microbial agents of respiratory tract disease. This was examined both at the level of amplification by detection of the amplicon using agarose gel electrophoresis and at the level of detection by testing the specificity of the oligoprobes.
In our conventional RT-PCR assay, we replaced dTTP in the nucleotide mix with twice as much dUTP in order to allow the use of uracil-N-glycosylase pretreatment to control a possible contamination event. However, to date the careful separation of mix preparation, template addition, and postamplification areas as well as the use of dedicated lab coats, pipettes, and filtered pipette tips has protected this assay from contamination.
While optimizing the ELAHA, we applied an asymmetric primer mix designed to produce more of the strand to which the oligoprobe hybridized. We found that this improved the sensitivity of the assay at least twofold. This was in addition to a 512-fold improvement in amplicon detection compared to agarose gel electrophoresis.
Real-time PCR assays are ideal for use in studying the epidemiology of a newly described virus because the systems are homogeneous, combining nucleic acid amplification and detection in a single closed reaction vessel. This means that the amplicon can be discarded without ever exposing the environment to contamination, the preferred approach for a high-throughput molecular diagnostic laboratory. This feature is additionally important when studying a new virus, as amplicon contamination could prove disastrous to the process of virus characterization.
In order to accurately compare the conventional and real-time RT-PCR assays, we chose to target the same sequence with both oligoprobes. However, this was not possible due to a guanine nucleotide that would have been incorporated into the 5′ end of the fluorogenic oligoprobe, causing quenching of the reporter fluorophore (3). Therefore, we designed the fluorogenic oligoprobe's sequence to be the reverse complement of that used in the ELAHA, so that it would hybridize with the sense strand of the amplified DNA. In an attempt to increase the fluorescent signal from weakly positive samples, we used an asymmetric primer mix to create more of the DNA strand to which the fluorogenic oligoprobe hybridizes. However, a problem arose that manifested as an early plateau in amplicon detection. We developed a dual-round RT-PCR to further examine the effects of asymmetric PCR using this primer mix. The dual-round approach has been used for nested PCR using the LightCycler (1, 11). We modified this approach by separating two concentrations of primers rather than two different primer pairs. The results confirmed that the asymmetric primer mix we were using was unsuitable for the detection of low levels of hMPV RNA. Following reverse transcription, there is an abundance of sense strands, but no antisense strands, for the sense primer to hybridize with. In the presence of an excess of the antisense primer, however, the PCR can effectively reach exponential levels of amplification. Therefore, when an oligoprobe is used to detect a negative-sense RNA genome by single-tube RT-PCR, the inclusion of asymmetric primers needs to be carefully considered.
To investigate the efficiency of hydrolysis of the fluorogenic oligoprobe under asymmetric PCR conditions and, by extrapolation, the efficiency of the real-time PCR assay, we applied fluorescent melting-curve analysis to the amplicon. Melting peaks were obtained from reactions containing an excess of the sense primer but not from reactions with extra antisense primer. We believe these melting peaks are due to conformational changes occurring when intact hybridized oligoprobe is melted from its template. This reduces the “leaky” fluorescence which results from an incompletely quenched FAM molecule when it is maximally separated from TAMRA on an intact oligoprobe. As the oligoprobe returns to a relaxed state after it has melted from its template, the fluorophores are more likely to interact by molecular collision, increasing the extent of quenching. The entire process results in a decreased fluorescence, which is presented as a melting peak when the negative derivative of the rate of change in fluorescence is calculated by the LightCycler's software. The fact that this signal is only apparent from reaction mixes containing an excess of sense primer which achieve an early plateau of signal suggests that there is an excess of intact fluorogenic oligoprobe and that this may be a useful marker of the efficiency of a real-time PCR using this detection chemistry.
We discovered that although the two assays possessed equal analytical sensitivity, the real-time RT-PCR was more sensitive than the RT-PCR-ELAHA in clinical studies. This could be caused by differential effects due to the presence of inhibitors of the RT or PCR steps in the conventional assay or differences between the capabilities of the single-tube RT-PCR kits used for the two assays.
In conclusion, we have compared two rapid molecular diagnostic approaches for the amplification and detection of hMPV RNA. These assays can also be used to examine and define an improved model for culturing hMPV, which currently relies upon cell lines that show little to no effect of hMPV replication. The results show that although the analytical sensitivities of the assays were identical, upon testing the assays using clinical samples, the real-time RT-PCR had an improved sensitivity, was faster and more cost-effective, and greatly reduced the likelihood of environmental contamination by amplicon. Thus, it is the preferred choice for detecting the presence of a newly described viral pathogen in the routine laboratory, proving considerably more reliable than cell culture for diagnosing hMPV infection.
Acknowledgments
This work was supported by the Royal Children's Hospital Foundation grant number 922-034, which was sponsored by the Woolworth's “Care for Kids” campaign, and number R912-006, which supported this work.
We thank Andreas Nitsche for useful discussions and Katherine Arden for critical review of the manuscript.
REFERENCES
- 1.Berg, J., V. Nagl, G. Mühlbauer, and H. Stekel. 2001. Single-tube two-round polymerase chain reaction using the LightCycler instrument. J. Clin. Virol. 20:71-75. [DOI] [PubMed] [Google Scholar]
- 2.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1351. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 3.Crockett, A. O., and C. T. Wittwer. 2001. Fluorescein-labelled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem. 290:89-97. [DOI] [PubMed] [Google Scholar]
- 4.Domachowske, J. B., and H. F. Rosenberg. 1999. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 12:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holberg, C. J., A. L. Wright, F. D. Martinez, C. G. Ray, L. M. Taussig, and M. D. Lebowitz. 1991. Risk factors for respiratory syncytial virus-induced lower respiratory illnesses in the first year of life. Am. J. Epidemiol. 133:1135-1151. [DOI] [PubMed] [Google Scholar]
- 6.Lipson, S. M. 2002. Rapid laboratory diagnostics during the winter respiratory virus season. J. Clin. Microbiol. 40:733-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay, I. M., P. Metharom, T. P. Sloots, and M. Q. Wei. 2001. Quantitative PCR-ELAHA for the determination of retroviral vector transduction efficiency. Mol. Ther. 3:801-807. [DOI] [PubMed] [Google Scholar]
- 8.McClelland, L., M. R. Hilleman, V. V. Hamparian, A. Ketler, C. M. Reilly, and D. Cornfeld. 1961. Studies of acute respiratory illnesses caused by respiratory syncytial virus. N. Engl. J. Med. 264:1169-1175. [DOI] [PubMed] [Google Scholar]
- 9.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralph, D., M. McClelland, and J. Welsh. 1993. RNA fingerprinting using arbitrarily primed PCR identifies differentially regulated RNAs in mink lung (My1Lu) cells growth arrested by transforming growth factor β1. Proc. Natl. Acad. Sci. USA 90:10710-10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratge, D., B. Scheiblhuber, O. Landt, J. Berg, and C. Knabbe. 2002. Two-round rapid-cycle RT-PCR in single closed capillaries increases the sensitivity of HCV RNA detection and avoids amplicon carry-over. J. Clin. Virol. 24:161-172. [DOI] [PubMed] [Google Scholar]
- 12.Schutten, M., B. van den Hoogen, M. E. van der Ende, R. A. Gruters, A. D. M. E. Osterhaus, and H. G. M. Niesters. 2000. Development of a real-time quantitative RT-PCR for the detection of HIV-2 RNA in plasma. J. Virol. Methods 88:81-87. [DOI] [PubMed] [Google Scholar]
- 13.Selwyn, B. J. 1990. The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev. Infect. Dis. 12:S870-S888. [DOI] [PubMed] [Google Scholar]
- 14.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. M. Fouchier, and A. D. M. E. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiley, D. M., I. M. Mackay, and T. P. Sloots. 2001. Detection and differentiation of human polyomaviruses JC and BK by LightCycler PCR. J. Clin. Microbiol. 39:4357-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]