Abstract
Molecular-phylogenetic sequence analyses have provided a new perspective on microbial communities by allowing the detection and identification of constituent microorganisms in the absence of cultivation. In this study we used broad-specificity amplification of ribosomal DNA (rDNA) genes to survey organisms present in the human outer ear canal. Samples were obtained from 24 individuals, including members of three extended families, in order to survey the resident microbiota and to examine microbial population structures in individuals related by familial or household associations. To examine the stability of the microbial populations, one individual was sampled four times and another twice over a 14-month period. We found that a distinct set of microbial types was present in the majority of the subjects sampled. The two most prevalent rDNA sequence types that were identified in multiple individuals corresponded closely to those of Alloiococcus otitis and Corynebacterium otitidis, commonly thought to be associated exclusively with infections of the middle ear. Our results suggest, therefore, that the outer ear canal may serve as a reservoir for normally commensal microbes that can contribute to pathogenesis upon introduction into the middle ear. Alternatively, culture analyses of diseases of the middle ear may have been confounded by these contaminating commensal organisms.
The indigenous microbiota of humans plays a variety of roles in the maintenance of health in the host. Commensal microbes of the gastrointestinal tract, for instance, stimulate the immune system, aid mineral and vitamin absorption by the intestine, synthesize nutrients used by the host, and ward off colonization by potentially pathogenic organisms (16). By analogy, similar functions likely are assumed by the microbiota indigenous to other sites of the human body, such as the skin or genital tract. Despite the many contributions of commensal microorganisms to human health, relatively little is known about the identities of these organisms. Traditionally, the identification and enumeration of microbial species has depended entirely upon pure-culture techniques. However, the difficulty with which some types of organisms are cultured, particularly fastidious and/or anaerobic microbes, means that the more easily grown species in a mixed microbial community are likely to be overrepresented by cultivation and plate counts. In many complex natural environments, for example, <1% of the viable microbes present can be cultured under standard conditions (24).
Recently, culture-independent molecular methods of microbial identification and characterization have been developed and applied in the context of microbial ecology. Several of these techniques involve the use of rRNA gene sequences as tools for species identification by means of phylogenetic sequence analysis. rRNA genes can be amplified by PCR directly from mixed-community DNA preparations (e.g., representative of both host and resident microbes in the case of clinical samples) and cloned, and individual clones can be sequenced. The occurrence of a particular rRNA gene sequence in a clone library indicates that the organism that carries this sequence is present in the sampled community. On the basis of rRNA sequence comparisons, species-specific DNA or RNA hybridization probes can subsequently be designed and used to enumerate the various types of organisms present in a sample (9). Several laboratories have used rRNA-based molecular techniques to identify and characterize human pathogens and commensals (20, 25, 30, 34, 36). These studies have identified a plethora of microbes associated with humans, many of which represent novel genera that previously were undescribed at the molecular level.
Although a diverse collection of microbes has been associated with the outer ear canal over the years based on the results of cultivation studies, none of these microbes are reported to be highly prevalent in the human population (18). We now report the results of a molecular survey of the microbes present in the outer ear canals of healthy humans. The goal of this study was to survey the commensal microbes resident within human ear canals in order to ascertain whether this environment is host to distinct, stable populations of microbes. For this purpose, outer ear swabs were obtained from 24 individuals, and the contents were subjected to molecular phylogenetic analyses. Samples were obtained from three extended families in order to correlate the occurrence of microbial types within household and family units.
MATERIALS AND METHODS
Subjects and sample collection.
Outer ear canal samples were collected from 24 individuals with sterile BBL CultureSwabs (Becton Dickson). An initial sample population was identified among employees of the Department of Molecular, Cellular, and Developmental Biology at the University of Colorado, Boulder. Participants were chosen based on the willingness of additional family members to enroll in the study. All adult participants and guardians signed informed consent forms. Individuals were not compensated for their participation in the study. The protocol of this study was approved by the University of Colorado, Boulder.
Adults collected samples from their own right outer ear canals by gentle swabbing. Samples from children were collected by their guardians. The samples were collected under the supervision of one of the authors (D.N.F.), who first demonstrated to the study participants sterile technique for swabbing the outer ear canal while avoiding contact with the auricle, hair, face, etc. None of the study participants had been swimming the day of sampling or had bathed within 4 h of sample collection. Although the sampled material consisted mainly of cerumen, epithelial cells and skin-associated microbes probably also were included. All subjects reported themselves to be healthy at the time of sampling. Samples collected in the laboratory were immediately frozen in the absence of culture medium and stored at −20°C (for individuals of households A, B, C, and K). For samples collected in the field (from individuals of households D, E, F, G, and H), the heads of the swabs were cut away from the handles with isopropanol-washed scissors, immersed in one milliliter of 100% isopropanol in a sterile 2-ml screw-cap microcentrifuge tube, stored frozen, and transported to the laboratory in a frozen state. Seven negative control swabs were collected during the sampling process (four were collected in the field and three were collected in the laboratory) and processed identically to the sample swabs in all procedures. Samples prepared from individual B1 produced nearly identical results, regardless of the sample storage method (see Table 3 and data not shown). Samples were frozen from 24 h to 3 months prior to DNA extraction.
TABLE 3.
Familial distribution of ear canal phylotypes
| Parameter | Valuea
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I
|
II
|
III
|
IV
|
|||||||||||||||||||||
| A
|
B
|
C
|
D
|
E
|
F
|
G
|
H
|
J
|
K
|
|||||||||||||||
| A1 | A2 | A3 | B1a | B1b | B1c | B1d | B2 | C1 | C2 | D1 | D2 | E1 | E2 | F1 | F2 | F3 | G1 | H1a | H1b | H3 | H4 | J1 | K1 | |
| Phylotypeb | ||||||||||||||||||||||||
| A. otitis | 80 | 97 | 70 | 70 | 55 | 61 | 77 | 38 | 100 | 31 | 79 | 35 | 41 | 12 | 67 | 47 | 59 | 100 | 97 | 92 | 95 | |||
| C. otitidis | 13 | 10 | 2 | 30 | 28 | 22 | 33 | 40 | 22 | 4 | 52 | 52 | 84 | 24 | 50 | 33 | ||||||||
| S. auricularis | 32 | 10 | 6 | 19 | 12 | 11 | 11 | 5 | 3 | 23 | 60 | 2 | 5 | 1 | ||||||||||
| C. auris | 44 | 31 | 2 | |||||||||||||||||||||
| Corynebacterium sp. strain 61720 | 19 | 1 | ||||||||||||||||||||||
| S. epidermidis | 1 | 1 | 1 | 3 | ||||||||||||||||||||
| P. acnes | 1 | 2 | 4 | 5 | ||||||||||||||||||||
| Streptococcus gordonii | 3 | 1 | ||||||||||||||||||||||
| Fusobacterium periodonticum | 1 | 1 | 3 | |||||||||||||||||||||
| Pseudomonas lanceolata | 1 | 2 | 1 | |||||||||||||||||||||
| Enterobacter sp. strain B509 | 1 | 2 | ||||||||||||||||||||||
| Uncultured bacterium strain BPC009 | 1 | 1 | ||||||||||||||||||||||
| Enterobacter agglomerans | 1 | 1 | ||||||||||||||||||||||
| Tilletiara anomala | 43 | 5 | 7 | 1 | 1 | 6 | 2 | 1 | 2 | 4 | 1 | |||||||||||||
| M. obscurus fungus | 5 | 2 | 4 | |||||||||||||||||||||
| Trhypochthanius tectorum | 1 | 6 | 3 | |||||||||||||||||||||
| P. pubis fungus | 2 | 1 | ||||||||||||||||||||||
| Sclerotium sp. | 2 | 1 | ||||||||||||||||||||||
| Homo sapiens (mitochondrion) | 1 | 4 | ||||||||||||||||||||||
| Other | 1 | 1 | 4 | 26 | 1 | 3 | 2 | 1 | 1 | 3 | 4 | 1 | ||||||||||||
| No. of clones | 88 | 87 | 93 | 54 | 89 | 89 | 94 | 84 | 84 | 95 | 89 | 90 | 93 | 87 | 73 | 91 | 84 | 80 | 91 | 81 | 89 | 88 | 83 | 87 |
| PCR primersc | U | U | U | B | U | U | U | B | U | U | U | U | U | U | U | U | U | U | B | U | U | U | U | B |
| DNA extractiond | S | S | S | C | S | S | S | C | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | C |
| Collection sitee | L | L | L | L | L | L | F | L | L | L | F | F | F | F | F | F | F | F | F | F | F | F | F | L |
| Elapsed timef | 0 | 4 | 12 | 13 | 0 | 14 | ||||||||||||||||||
| Sexg | M | F | F | M | F | M | F | M | F | M | F | F | F | M | M | M | F | F | M | M | ||||
| Ageh | 32 | 32 | 5 | 35 | 35 | 65 | 62 | 66 | 61 | 35 | 31 | 23 | 1 | 55 | 28 | 39 | 1 | 4 | 66 | 57 | ||||
Column headers indicate family (roman numerals), household (letters), and individuals (letters plus numbers). Multiple samples for an individual are designated by lowercase letters.
Nearest relative based on Blastn search. Data are presented as percentages of the numbers of clones screened for each library.
U, Universal primers 515F-1391R; B, bacterial primers 8F-805R.
S, simple boiling preparation; C, chemical, enzymatic, mechanical extraction of DNA.
L, laboratory; F, field.
Months elapsed from initial sampling for individuals sampled on multiple occasions.
M, male; F, female.
Age in years at time of sampling.
DNA sample preparation.
Prior to extraction of DNA from isopropanol-preserved samples, the microcentrifuge tubes, along with the swab tips, were subjected to centrifugation (16,100 × g) for 20 min in order to pellet any material that had dissociated from the swabs. Approximately 750 μl of isopropanol was removed from the microcentrifuge tubes, and the swabs, along with the pellets, were allowed to dry for 3 h in a vacuum chamber. For samples that were stored dry (for individuals of households A, B, C, and K), the heads of the swabs were severed from their handles with isopropanol-washed scissors and placed in sterile 2-ml screw-cap microcentrifuge tubes.
Two protocols were used to prepare mixed-community genomic DNAs for PCR analysis. For both protocols, samples were processed by adding 500 to 750 μl of buffer A (200 mM NaCl, 200 mM Tris-Cl [pH 8.0], 20 mM EDTA) plus 0.1% NP-40 nonionic detergent (Igepal CA630; Sigma) to the dried swab heads in 2-ml sterile screw-cap tubes. The majority of samples (see Table 3) were then lysed by a simple boiling method in which they were incubated twice for 10 min at 95°C and vortexed for 30 s during the interval. The lysed samples were either placed on ice or stored at −20°C until they were used for PCR. At the outset of this study, a more rigorous solvent-and-grinding DNA extraction protocol was used to process some of the samples (libraries IB1a, IB2a, and IVK1). In addition to buffer A, lysozyme was added to the mixture at a final concentration of 5 mg/ml. Samples were incubated for 30 min at 37°C. Proteinase K (final concentration, 2 mg/ml) and sodium dodecyl sulfate (final concentration, 0.3%) were added, and the samples were incubated for 30 min at 50°C. The sodium dodecyl sulfate concentration was adjusted to 5%, and 500 μl of phenol-chloroform and 0.5 g of zirconium beads were added to the mixture. The samples were agitated in a Mini Beadbeater-8 (Biospec Products) on high for 2 min and then subjected to centrifugation (16,100 × g) for 5 min. The aqueous phase was reextracted two times with phenol-chloroform and once with chloroform. DNA was precipitated by the addition of NaCl (final concentration, 280 mM) and 2.5 volumes of ethanol followed by centrifugation (16,100 × g) for 30 min. The DNA pellets were washed once with 70% ethanol, allowed to dry in a laminar flow hood, and resuspended in 50 μl of sterile Tris-EDTA (10 mM Tris-Cl [pH 8.0], 1 mM EDTA).
All DNA extraction and PCR amplification were conducted in the laboratory of N.R.P. under the supervision of D.N.F. Although samples were not processed simultaneously, frozen aliquots of reagents were used for DNA extractions and PCR amplifications in order to minimize sample-to-sample variation.
PCR, cloning, and sequence analysis.
Small-subunit (SSU) rRNA genes were amplified from swab DNA samples by PCR with one of two sets of primers (22, 35): (i) bacterial genes were amplified with the primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 805R (5′-GACTACCAGGGTATCTAAT), and (ii) all SSU rRNA genes in a sample were surveyed with the universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA) and 1391R (5′-GACGGGCGGTGWGTRCA). Each 30-μl PCR mixture included 3 μl of 10× PCR buffer, 2.5 μl of deoxynucleoside triphosphate mix (2.5 mM each deoxynucleoside triphosphate), 1.5 μl of 50 mM MgCl2, 75 ng of each primer, 3 μl of 10-mg/ml bovine serum albumin (Sigma), 1 U of Taq polymerase, and 1 μl of genomic DNA lysate (following the manufacturers' protocols). Bovine serum albumin was included in all PCR mixtures in order to suppress PCR inhibitors (19). PCR reagents from Bioline (Biolase polymerase) and Eppendorf (MasterTaq polymerase) produced indistinguishable results. Thirty cycles of amplification (92°C for 30 s, 52°C for 60 s, and 72°C for 90 s) usually were sufficient to obtain a product of the appropriate length that was visible in ethidium bromide-stained agarose gels (NucleoTech Inc.). Additional cycles of PCR were required for a subset of the samples in order to produce sufficient product for purification and cloning. Samples IA1, IA2, IA3, and IB1b were amplified for 32 cycles, and samples IB1a, IB2, IIIH1a, and IVK1 were amplified for 35 cycles (negative control reactions were run in parallel with these extended PCR amplifications). Two negative control PCRs, one with lysate from control swab extractions and the other with sterile H2O serving as a template, were performed for each set of samples processed in order to assess whether contamination of reagents had occurred. Positive control PCRs, which used an environmental genomic DNA sample as a template, were also performed for each set of samples processed. For PCR inhibition controls, equal quantities of positive control DNA templates were added to each of two PCR mixtures set up in parallel, one of which was supplemented with ear swab DNA.
DNA fragments were excised from agarose gels (1% agarose gel in Tris-borate-EDTA) and purified with the QIAquick gel extraction kit (Qiagen). A portion of each PCR product was cloned into the pCR4-TOPO vector of the Invitrogen TOPO TA cloning kit. For each clone library that was constructed, 96 transformants were grown overnight at 37°C in a 96-well culture plate filled with 1.5 ml of 2× yeast extract-tryptone medium per well. In order to screen for positive transformants, 20 μl of each overnight culture was added to 20 μl of 10 mM Tris-Cl (pH 8.0), heated for 10 min at 95°C, and centrifuged for 10 min at 2,000 × g in a 96-well plate centrifuge (Eppendorf). One microliter of culture supernatant was used as a template in a 30-μl PCR mixture (38 cycles of the program described above) with vector-specific primers (T7 and T3 sites). Five microliters of PCR product were separated by agarose gel electrophoresis in order to screen for cloned inserts. The remainder of the PCR product was digested with 2 U each of MspI and HinPI restriction enzymes for 3 h at 37°C. The restriction fragments were separated by gel electrophoresis on 4% GenePure HiRes agarose (ISC BioExpress). Restriction fragment length polymorphism (RFLP) types were sorted by visual inspection of digitized gel images (NucleoTech Inc.).
In the rare instances in which some signal was observed in the negative control PCRs (always at a level well below reactions with ear swab samples), clone libraries were also constructed from this material and screened along with sample libraries. Sequence analyses of contaminant libraries indicated that cross-contamination was not a problem, since contaminant sequence types were not found in ear swab sample libraries (data not shown).
Representative RFLP types were sequenced on a Licor automated DNA sequencer with the ThermoSequenase cycle-sequencing kit (U.S. Biochemicals), following the manufacturers' directions. The majority of both strands of DNA was sequenced in all instances. Initial microbial species identifications were made by a batch BlastN search (National Center for Biotechnology Information [NCBI]) using the client application BlastCl3 (NCBI). Blast results were filtered through the program Extract (D. N. Frank, unpublished). SSU rRNA gene sequences were aligned to an existing database of rRNA gene sequences (23) using the computer application ARB (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps). Phylogenetic analyses, including phylogenetic-tree estimations, utilized ARB, as well as PAUP* (32). The percent sequence identities (see Table 2) are uncorrected. The type sequences used for analysis (see Table 2) were downloaded from GenBank (accession numbers X59765 [Alloiococcus otitis], X73976 [Corynebacterium otitidis], and D83358 [Staphylococcus auricularis]).
TABLE 2.
Summary of sequence distances between prevalent ear canal clones and type species
| Phylotype | Distance (%)
|
No. of Clones:
|
ng | |||
|---|---|---|---|---|---|---|
| Meand | Maximum | Minimum | <1%e | <3%f | ||
| A. otitisa | 1.0 (0.5) | 2.4 | 0.1 | 18 | 33 | 33 |
| C. otitidisb | 1.3 (0.6) | 2.9 | 0.4 | 5 | 21 | 21 |
| S. auricularisc | 0.7 (0.4) | 1.6 | 0.1 | 27 | 33 | 33 |
Accession no. X59765.
Accession no. X73976.
Accession no. D83358.
Mean uncorrected evolutionary distance (standard deviation) of all sequenced clones in the phylotype compared to reference sequence.
Number of clones with <1% evolutionary distance to reference sequence.
Number of clones with <3% evolutionary distance to reference sequence.
Sample size.
Nucleotide sequence accession numbers
The nucleotide sequence data presented in this paper have been deposited in the GenBank database, with accession numbers as follows: for A. otitis-like clones, AY189528 to AY189560; for C. otitidis-like clones, AY189561 to AY189581; for S. auricularis-like clones, AY189582 to AY189614; and for other clones, AY189615 to AY189674.
RESULTS
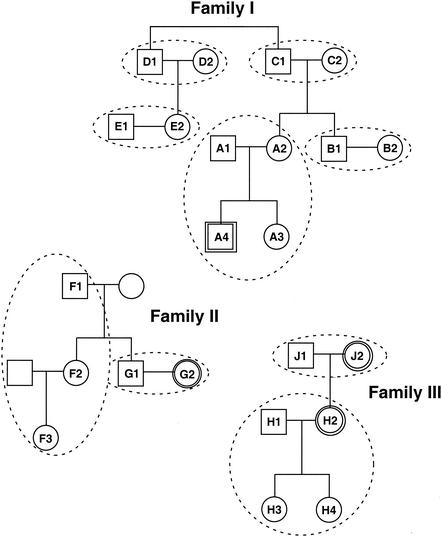
Twenty-four healthy subjects, ranging in age from 1 to 66 years, were chosen to participate in this study. The study population was split approximately evenly between female (n = 13) and male (n = 11) subjects. All participants were Caucasian long-term residents of the United States, specifically of the states of Colorado, Illinois, Ohio, North Carolina, and South Carolina. In order to trace potential familial correlations in microbial populations, we obtained ear swab samples from three families, each of which extended for three generations. Nine multisubject households (i.e., groups of individuals that share homes, regardless of familial association) were sampled in this study. Familial pedigrees and household groupings are summarized in Fig. 1. Ear swab samples were collected over a 2-year period. The sampled material was predominantly cerumen but also was expected to include epithelial cells and skin-associated microbes.
FIG. 1.
Pedigrees of families included in this study. Individuals from whom cerumen samples were collected are labeled with numbers and letters. Samples that were SSU PCR negative are designated by double lines. Individuals who lived in the same household are grouped by dashed lines.
Mixed-community genomic DNAs (i.e., DNAs of the host and associated organisms) were prepared from each sample either by simply boiling the collected swabs in a solution of 10 mM Tris-EDTA-0.1% NP-40 nonionic detergent or by a more rigorous chemical, enzymatic, and mechanical extraction protocol (detailed in Materials and Methods). Little difference in clone library complexity or composition was noted when the two DNA extraction procedures were applied to samples collected from one individual and the results were compared. Consequently, the simpler, more rapid boiling protocol was used for the majority of the collected samples (see Table 3 and Materials and Methods) in order to maximize the DNA yield and minimize potential DNA contamination from enzymes, solvents, and other reagents.
SSU rRNA genes were amplified by PCR from swab DNA samples using either universal SSU ribosomal DNA (rDNA) primers (515F-1391R; Escherichia coli numbering) capable of recognizing all known SSU rRNA genes or Bacteria-specific primers (8F-805R; E. coli numbering), which recognize most known bacterial SSU rRNA genes (22, 35). Of the 24 samples collected, only 4 failed to produce PCR amplification products (three adult females, IIG2, IIIH2, and IIIJ2, and one 2-year-old male, IA4) (Fig. 1). Additional samples obtained from individuals IA4 and IIIH2 similarly failed to amplify. PCR inhibition controls, in which positive control PCR mixtures were spiked with ear canal DNA lysates, indicated that the failure of the samples from these four individuals to amplify was not a consequence of PCR inhibitors. Rather, it is likely that low sample biomass and/or poor DNA recovery from these samples prevented PCR amplification.
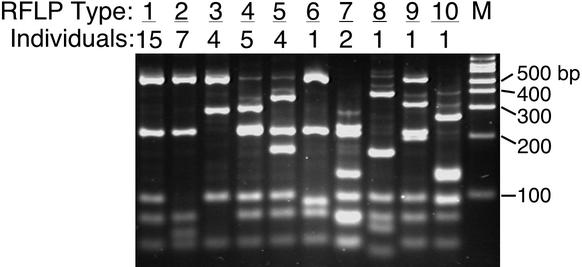
To isolate and analyze individual gene sequences, amplicons were cloned and bacterial transformants were screened for recombinants by PCR of cultures with vector-specific primer sets. Ninety-six transformants were screened for each PCR library (5). To simplify sequence analysis, an RFLP assay was used to sort rDNA clones into relatedness groups, designated RFLP types (4, 5, 17, 34). The DNA sequence of at least one example of each RFLP type was determined. Sequence analysis of multiple examples of the more prevalent RFLP types (e.g., 11, 9, and 13 clones from the three most prevalent RFLP types, members of the A. otitis, C. otitidis, and S. auricularis phylotypes, respectively), selected from different individuals, confirmed the accuracy of the sorting process. Although nonidentical DNA sequences often occur among independent clones belonging to a particular RFLP type, high rDNA sequence identities (>97%) were observed for each RFLP type (i.e., there were no false-positive examples of RFLP sorting). In contrast, several examples of false-negative RFLP sorting (i.e., different RFLP types that showed high-percentage sequence identities) were observed (Fig. 2).
FIG. 2.
Diversity of RFLP types associated with A. otitis 16S rRNA clones. The RFLP analysis is described in Materials and Methods. The number of individuals who exhibited each RFLP pattern is indicated. A total of 15 individuals in whom A. otitis was detected by universal SSU rRNA PCR were included in this analysis. Lane M, molecular size marker.
Analysis of the cloned sequences by either BlastN search (NCBI) or phylogenetic reconstruction identified the phylogenetic affiliations of the organisms present in the ear canal samples. A small percentage of sequences were not those of SSU rRNA genes, indicating that some mispriming events occurred during PCR; these sequences were excluded from further analysis. As described above, in several instances, different RFLP types were found to be closely related by analysis of multiple selected sequences (Fig. 2). These RFLP types were collapsed into groups, designated “phylotypes,” based on similarity of the sequences and the common nearest known relative, as determined by the BlastN score. Table 1 lists the closest relative of each phylotype analyzed, ranges of the BlastN similarity scores of the sequences compared to their closest known relatives, and the overall frequency with which each phylotype was observed in the study as a whole. In many instances, the sequences obtained from the samples were closely related (BlastN scores of >97%) to the SSU rDNA sequences of previously analyzed microbes and thus allowed provisional identification of organisms at the species level. A cutoff value of 97% was chosen based on the correlation of 16S rRNA sequence distances with DNA-DNA reassociation rates by Stackebrandt and Goebel, which suggests that SSU rRNA sequences that share <97% nucleotide identities constitute distinct species (28). Exact sequence matches between SSU rRNA gene isolates and those of known organisms are rare, indicating considerable biological diversity within the sampled populations. Indeed, 16 of the 45 phylotypes detected were composed solely of clones with BlastN scores of <97% in comparison to their closest known relatives. Thus, these phylotypes likely represent novel microbial species that have not previously been characterized at the molecular level (28).
TABLE 1.
Diversity of phylogenetic types associated with human ear canals
| Nearest relativea | Cladeb | % IDc | No. of clonesd | % of total |
|---|---|---|---|---|
| A. otitis | Low G + C | 96-98 | 1,221 | 56.71 |
| C. otitidis | Actinobacteria | 96-99 | 440 | 20.44 |
| S. auricularis | Low G + C | 98-99 | 212 | 9.85 |
| C. auris | Actinobacteria | 98 | 67 | 3.11 |
| Tilletiaria anomala | Eucaryote | 94-95 | 65 | 3.02 |
| P. acnes | Actinobacteria | 99 | 35 | 1.63 |
| Corynebacterium sp. strain 61720 | Actinobacteria | 97 | 17 | 0.79 |
| M. obscurus fungus | Eucaryote | 94-97 | 10 | 0.46 |
| Stephanoascus ciferrii | Eucaryote | 98 | 8 | 0.37 |
| Trhypochthonius tectorum | Eucaryote | 91 | 8 | 0.37 |
| S. epidermidis | Low G − C | 95 | 6 | 0.28 |
| Fusobacterium periodonticum | Fusobacteria | 98 | 4 | 0.19 |
| Homo sapiens (mitochondrion) | Plastid | 97 | 4 | 0.19 |
| Pseudomonas lanceolata | γ Proteobacteria | 99 | 4 | 0.19 |
| Streptococcus gordonii | Low G + C | 99 | 4 | 0.19 |
| Aspergillus fumigatus | Eucaryote | 96 | 3 | 0.14 |
| P. pubis fungus | Eucaryote | 99 | 3 | 0.14 |
| Sclerotium sp. strain BSC-97 | Eucaryote | 96 | 3 | 0.14 |
| Enterobacter sp. strain B509 | γ Proteobacteria | 99 | 3 | 0.14 |
| Staphylococcus hominis | Low G − C | 90 | 3 | 0.14 |
| Kocuria erythromyxa | Actinobacteria | 94 | 2 | 0.09 |
| Uncultured bacterium strain BPC009 | Actinobacteria | 95-98 | 2 | 0.09 |
| Enterobacter agglomerans | γ Proteobacteria | 99 | 2 | 0.09 |
| Bacillus licheniformis | Low G − C | 98 | 2 | 0.09 |
| Streptococcus sp. oral strain B5SC | Low G − C | 98 | 2 | 0.09 |
| Alpha proteobacterium strain VUN10077 | α Proteobacteria | 99 | 1 | 0.05 |
| Uncultured bacterium 16S rRNA | β Proteobacteria | 98 | 1 | 0.05 |
| Acinetobacter sp. | γ Proteobacteria | 98 | 1 | 0.05 |
| Haemophilus paraphrophilus | γ Proteobacteria | 96 | 1 | 0.05 |
| Brevibacterium avium | Actinobacteria | 98 | 1 | 0.05 |
| Brevibacterium sp. | Actinobacteria | 98 | 1 | 0.05 |
| Corynebacterium sp. strain X82493 | Actinobacteria | 95 | 1 | 0.05 |
| Dietzia maris | Actinobacteria | 95 | 1 | 0.05 |
| Gram-positive bacterium strain Wuba45 | Actinobacteria | 97 | 1 | 0.05 |
| Human oral bacterium strain AC1 | Actinobacteria | 98 | 1 | 0.05 |
| Synechococcus sp. strain ATCC 700246 | Cyanobacteria | 95 | 1 | 0.05 |
| Tetranychus urticae | Eucaryote | 95 | 1 | 0.05 |
| Abiotrophia defectiva | Low G + C | 96 | 1 | 0.05 |
| Carnobacterium sp. strain LV62LW1 | Low G + C | 92 | 1 | 0.05 |
| Staphylococcus piscifermentans | Low G + C | 96 | 1 | 0.05 |
| Streptococcus mitis | Low G + C | 99 | 1 | 0.05 |
| Streptococcus sp. oral clone BW009 | Low G + C | 99 | 1 | 0.05 |
| Veillonella sp. oral clone X042 | Low G + C | 98 | 1 | 0.05 |
| Uncultured bacterium strain WCHA2-01 | OP11 | 100 | 1 | 0.05 |
| Uncultured bacterium 16S rRNA, strain BD7-4 | OP11 | 87 | 1 | 0.05 |
| Total | 2,150 | 100.00 |
Based on BlastN search.
Kingdom (eucaryote)- or division (bacteria)-level phylogenetic affiliation.
BlastN score. ID, identity.
Total number of clones screened in all libraries.
It is evident from Table 1 that the ear canal SSU clone libraries were dominated numerically by only a few types of SSU rRNA genes, mainly representing bacteria closely related to A. otitis, C. otitidis, and S. auricularis. These accounted for 86% of the SSU rDNA clones recovered in this study. Overall, the two gram-positive divisions of Bacteria, the Actinobacteria and the low-G+C group, were the most prevalent higher-level phylogenetic groupings observed (Table 1). As discussed below, A. otitis and C. otitidis are commonly thought to be exclusive pathogens of the middle ear.
The BlastN scores reported in Table 1 only approximate the sequence similarities between the ear canal clones and their closest known relatives. To more precisely determine the relationships within the three most prevalent phylotypes (i.e., the A. otitis, C. otitidis, and S. auricularis relatedness groups), we constructed multiple sequence alignments of all sequenced clones that belong to these groups (obtained from multiple individuals) along with the SSU rRNA sequences of the type species. Uncorrected evolutionary distances were then calculated for each ear canal sequence paired with its closest known relative (Table 2). This analysis revealed that the 33 cloned sequences that were assigned to the A. otitis phylotype were on average 99.0% identical (range, 97.6 to 99.9%) to the published SSU rRNA sequences of A. otitis. Similarly, the C. otitidis-like clones averaged 98.7% identity (range, 97.1 to 99.6%) to C. otitidis SSU rRNA sequences. The S. auricularis-like clones were also closely related (99.3% mean identity; range, 98.4 to 99.9%) to published S. auricularis SSU rRNA sequences. These high sequence similarities suggest that the organisms represented by the ear canal clones correspond to the species A. otitis, C. otitidis, or S. auricularis (see Discussion).
Eucaryotes represented approximately 5% of the clones sequenced. However, because of the greater length of eucaryal 18S SSU rRNA genes compared to bacterial and plastid SSU genes and the bias of PCR toward amplifying shorter DNA fragments, eucaryotes possibly were underrepresented in this survey. We note that sequences from two of the eucaryal phylotypes were initially identified by BlastN searches as being related to the metazoans Phthirus pubis (pubic louse; unpublished GenBank submission AF139485) and Magmatodrilus obscurus (an annelid; unpublished GenBank submission AF310699). More detailed phylogenetic analysis of the P. pubis and M. obscurus 18S SSU genes obtained from GenBank revealed that these sequences, along with the related ear canal-associated sequences isolated in this study, clearly represent members of the basidiomycetous fungi (http://pacelab.colorado.edu/publications). Presumably, the genomic DNA preparations from which the putative P. pubis and M. obscurus 18S rRNA genes originally were isolated were contaminated with fungal DNA.
Table 3 shows the distribution of clones observed for each phylotype, sorted by individual, household, and extended family. The frequencies with which rDNA phylotypes were observed in the PCR libraries provide semiquantitative estimates of the relative abundances of organisms in the samples and thus provide for comparison of microbial population structures between individuals. The relative numbers of rDNA phylotypes that were observed do not precisely reflect the relative numbers of individual organisms, in part because rRNA gene copy numbers vary between microbial species. To simplify presentation, only phylotypes that were observed in at least two individuals are included in Table 3 (the entire data set is available at http://pacelab.colorado.edu/publications). For all but two subjects (IA1 and IIG1) (Fig. 1), either the A. otitis or C. otitidis phylotype was the predominant group of sequences detected in the individual swab samples. In two instances, IC2 and IIIH3, examples of the A. otitis phylotype were observed in 100% of the SSU rDNA clones that were screened (n = 95 and 89, respectively). A. otitis-like sequences were not detected in only three subjects, IA1, IIF3, and IIG1. It is noteworthy that the individuals that lacked the A. otitis phylotype also exhibited the highest proportions of S. auricularis-like clones (23 to 60% of the clones analyzed) of any of the individuals examined. This suggests that S. auricularis and A. otitis may compete with one another for a niche within the outer ear canal.
Examples of the C. otitidis phylotype were not detected in seven individuals, including the entirety of extended family III (IB2, IC2, IIIH1, IIIH3, IIIH4, IIIJ1, and IVK1). In general, those subjects in which the C. otitidis phylotype was not detected exhibited few occurrences of S. auricularis-like sequences (the exception being subject IB2, who had the fourth highest percentage of S. auricularis-like sequences in the study population). Rather, clones of the A. otitis phylotype were found to be relatively more abundant in these individuals than the average. The sole exception to this observation was subject IIIH1, in whom the C. otitidis phylotype seemed to be replaced by the Corynebacterium auris phylotype.
In the course of testing various methods of sample collection and DNA extraction, we sampled two individuals multiple times over a 14-month period. These samples provided an opportunity to examine the temporal stabilities of microbial populations within the outer ear. Despite the use of different PCR primer sets (i.e., bacterial versus universal) and different methods of sample collection and processing, the results indicate that the major microbial types identified in the samples remained relatively constant over the sampling period (Table 3, individuals IB1 and IIIH1). Thus, the two most highly represented phylotypes that were observed for each individual (A. otitis, C. otitidis, and/or C. auris phylotypes) remained the same at each time point. Any variability in population structure was evident only among the less prevalent organisms. This variation may reflect the transience of particular microbial species within the ear canal samples or systematic sampling or PCR errors.
DISCUSSION
Molecular phylogenetic sequence analyses have revolutionized the characterization of complex microbial communities that previously were accessible only through relatively inefficient and often biased culture methodologies (9). In some clinical microbiology laboratories, the analysis of SSU rRNA gene sequences of clinical isolates has become a standard means of species identification. However, the use of PCR to obtain rRNA genes also permits the amplification and characterization of rDNA sequences directly from mixed communities, thus allowing the identification of microbes in the absence of cultivation. Several laboratories have begun to use these techniques to explore the population structures of human microbial pathogens and commensal organisms (1, 20, 25, 30, 33, 34, 36). In this study, we used broad-range amplification of rDNA genes to survey organisms present in the healthy human outer ear. Samples were obtained from 24 individuals, including members of three extended families, in order to survey the resident microbiota and to ascertain whether microbial population structures were influenced by familial or household associations. Characterization of multiple samples collected from two of the study participants over a 14-month period demonstrated both the reproducibility of the community-based molecular phylogenetic methodology and the temporal stability of cerumen-associated microbial populations within select individuals (Table 3). Indeed, similar results were obtained regardless of the method of sampling, the DNA extraction protocol, or the selection of rDNA PCR primers.
Microbes of the human outer ear generally are described as idiosyncratic in occurrence, i.e., no microbe is “commonly found in ears” (18). In contrast, we observed relatively stable microbial populations, the most prevalent members of which occur in multiple subjects regardless of age, sex, or geographical location (within the United States). Microbes closely related to A. otitis, C. otitidis, and S. auricularis were detected in high abundance in the rDNA clone libraries of multiple individuals (approximately 85% of the 2,150 rDNA clones surveyed). Because the distribution of SSU rRNA clones in a library can be skewed by a number of factors, including differential cell lysis and DNA extraction, interspecies differences in gene copy number, and biased PCR amplification, the actual abundances of microbes in ear canal samples must be confirmed by other techniques, such as in situ hybridization. In a preliminary experiment (following the methods of Campos et al. [3]), cultivation of microbes from the ear canal of individual IB1 (obtained at the same time as sample IB1d) revealed growth on tryptic soy agar and blood agar plates of S. auricularis (45 of 95 colonies), C. otitidis (44 of 95 colonies), C. auris (3 of 95 colonies), A. otitis (1 of 95 colonies), Staphylococcus epidermidis (1 of 95 colonies), and Propionibacterium acnes (1 of 95 colonies). Except for the paucity of A. otitis isolates, these results corroborate those of the molecularly based microbial survey and provide further evidence that S. auricularis and C. otitidis are numerically dominant constituents of the healthy human ear canal microbiota. Because A. otitis is a fastidious microbe (7, 8, 12, 15, 21), it is likely that cultivation underestimates the abundance of this organism in ear canal specimens.
In contrast to the more abundant microbes, the less prevalent species rarely were observed in more than one individual. In these cases, we cannot distinguish organisms that are indigenous to the ear canals of particular individuals from those that are transient. Fluctuation due to sampling of low-abundance organisms probably also accounts for some of the observed differences. Regardless of the precise quantitation of microbial species in the ear canal samples, the A. otitis, C. otitidis, and S. auricularis relatedness groups constituted the most widely distributed phylotypes, with representative clones detected in 17, 13, and 12 individuals, respectively, out of the 20 total individuals from whom clone libraries were constructed. The widespread distribution of the most prevalent groups of microbes indicates that they are indigenous to the outer ear canals of healthy humans. The human outer ear canal, therefore, is host to a well-defined, relatively simple population of commensal organisms. Delineation of the constituents of this microbiota provides an essential base of information for analyses of microbes in pathogenic states.
The lists of microbial species that are presented in Tables 1 and 3 minimize the actual extent of organism diversity that was evident in our molecular survey of ear canal samples. Few of the SSU rRNA sequences that were determined were 100% matches to one another or to those of previously characterized microbial species (Table 1, %ID). Thus, rather than describing distinct, monolithic species, this collection of sequence data suggests the existence of clusters of closely related microbial types, a theme that is emerging in microbial ecology (20, 25, 26, 30, 31, 34, 36). Qualitatively, the range of sequence diversity within a particular phylotype (e.g., the A. otitis group) can be observed in the variety of RFLP types associated with that phylotype. Figure 2 highlights the common RFLP patterns for the A. otitis group. Note that several of the A. otitis RFLP types were observed in multiple individuals and that some individuals were characterized by multiple A. otitis RFLP types. RFLP type 1, for instance, was observed in all individuals in which examples of the A. otitis phylotype were present and was the numerically dominant RFLP type within the A. otitis group (956 of 1,221 clones). Together, these results suggest that the divergence in SSU rRNA sequences within a particular phylotype reflects real subpopulations of microbial types that are distributed both across multiple hosts and within individual hosts. In general, SSU rRNA sequence similarities within each phylotype were too great to accurately differentiate subgroups within a phylotype. Other more rapidly evolving molecules (e.g., ribosomal spacer sequences) undoubtedly will be useful in elucidating the population biology of the prevalent outer ear microbes.
Despite the relatively small sample population of this study, analysis of the data in Table 3 revealed several distinct trends. Both the age and sex of individuals influenced the diversity of microbial types identified in this study. Females, for instance, exhibited less complex populations than did males. Indeed, we were unable to detect microbes in the cerumen samples of three adult females (IIG2, IIIJ2, and IIIH2 [Fig. 1]). Similarly, the microbial populations of children's (1 to 5 years) outer ears were less complex, on average, than those of older individuals (23 to 66 years). Whether these correlations are due to biological or sociological (e.g., gender- or age-specific grooming practices) determinants has yet to be resolved. In three of the four children analyzed, A. otitis-like clones represented >97% of the clones analyzed, suggesting that organisms belonging to this relatedness group are early colonizers of the outer ear. Because attempts to sample the ears of younger children were not successful, we were unable to test this hypothesis. Additional sampling of the children included in this study over the course of several years will be of interest in order to determine whether changes in microbial community structures occur with age.
In several instances, pairs of individuals who had cohabitated for several years, or even decades, exhibited quite different patterns of microbes (e.g., IA1 and IA2, IC1 and IC2, IIF1 and IIF3). Thus, simple household associations are unlikely to greatly influence the colonization of the outer ear canals. Moreover, the suite of microbes detected in a child could not necessarily be predicted by inspection of their parents' microbial populations (e.g., IIF3 versus IIF1 and IIIH1 versus IIIH3 and IIIH4). The occurrence of the most prevalent microbial species, however, was not random. For example, the relatively abundant C. otitidis phylotype was absent from all members of family III. The absence of C. otitidis-like microbes in members of this family was accompanied by high frequencies of the A. otitis and/or C. auris phylotype, suggesting competition between these microbes for a particular niche.
From a clinical standpoint, it is important to address the question of whether the organisms that are represented by the SSU rRNA clones identified in this study are constituents of known species or are novel species related to known species. Although no SSU rRNA sequence similarity value can be used to unequivocally differentiate microbial species, Stackebrandt and Goebel (28) have proposed a value of 97% as a cutoff for assigning phylotypes to particular species. By this criterion, many of the phylotypes identified in the ear canals of healthy individuals that have BlastN scores of <97% represent species, or even genera, of microbes that have not previously been characterized by molecular or culture techniques. Conversely, a 97% cutoff would indicate that all of the clones assigned to the A. otitis, C. otitidis, and S. auricularis phylotypes are members of the named species. Even considering a more stringent cutoff of 99% nucleotide identity (within the range of intraspecific SSU rRNA evolutionary distances observed among strains of E. coli), a substantial number of clones would be classified as being representatives of the species A. otitis, C. otitidis, or S. auricularis (55, 24, and 82% of sequenced clones, respectively). Furthermore, 14 of the 15 A. otitis clones that were amplified with the primers 8F-805R and subsequently sequenced encoded exact matches to three A. otitis species-specific oligonucleotides that have been used for clinical detection of the species (6, 14, 15). These data strongly suggest, but do not prove formally, that the SSU rRNA clones that were isolated in this study indicate the presence of strains of the species A. otitis, C. otitidis, and S. auricularis in numerous healthy individuals.
The prominence of both A. otitis and C. otitidis in the outer ear canals of multiple healthy individuals was surprising because these organisms commonly are cited as associated solely with pathogenic states. A. otitis, for example, was first cultured from the middle ear fluids of children with chronic otitis media with effusion (7). Several subsequent studies, which have used both molecular and culture identification techniques, have reported high incidences of A. otitis in middle ear fluids of otitis media with effusion patients (1, 2, 13). Similarly, C. otitidis was initially identified in cultures of middle ear fluids from patients with chronic and acute otitis media (10, 11, 27). Based on these findings, A. otitis and C. otitidis are commonly considered to be exclusively associated with middle ear infections (8).
Several recent studies, however, have reported the isolation or detection of A. otitis and/or C. otitidis in the outer ear canals and nasopharynges of healthy individuals (6, 12, 29). Using a PCR assay specific for A. otitis, Durmaz et al. (6) detected the presence of this microbe in 4 of 50 (8%) nasopharynx swabs and 3 of 95 (3%) outer ear canal swabs. Overall, six of the seven positive swabs were collected from children with chronic otitis media. Significantly, one of the seven positive swabs was obtained from a child with no symptoms of chronic otitis media (1 of 23 healthy individuals), indicating that A. otitis is resident in the ear canals of healthy individuals (6). Stroman et al. (29) subjected cerumen and outer ear canal swabs, collected from 164 healthy individuals, to cultivation in order to survey the normal microbiota of the ear. Of the 624 total organisms that were characterized, 48 (8%) A. otitis and 74 (12%) C. otitidis isolates were identified (29). The distribution of these organisms throughout the study population was not reported. However, assuming at most one isolate of a particular species per individual, these data would indicate that a maximum of 29% of the individuals were culture positive for A. otitis and 45% were culture positive for C. otitidis. In contrast, 85% of the individuals from whom SSU rRNA libraries were characterized in this study were positive for A. otitis and 65% were C. otitidis positive. Thus, our results not only corroborate the finding of A. otitis and C. otitidis in healthy individuals but also demonstrate a much wider distribution of these species in the general population. Our results, together with those of previously published studies (6, 12, 29), clearly indicate that A. otitis and C. otitidis are commensal inhabitants of the human outer ear canal. The culture of these organisms from middle ear fluids of otitis media patients may simply reflect contamination by cerumen-associated microbes during the sampling process. Alternatively, organisms that are commensals in the context of the outer ear canal may contribute to, or even cause, pathology when present in inappropriate sites such as the middle ear, a putatively sterile environment under normal circumstances. How microbes such as A. otitis and C. otitidis might be introduced into the middle ear remains an open question. Although A. otitis has been detected in nasopharyngeal swabs of some individuals with otitis media (6), broad-range molecular phylogenetic surveys of oral microbes have failed to detect either A. otitis or C. otitidis in the mouth (20, 25). Prior ruptures of the tympanic membrane, through tympanocentesis, for example, might also provide an avenue for entry into the middle ear for prominent inhabitants of the outer ear. In any case, the identification of A. otitis and C. otitidis as prevalent commensals of the healthy human outer ear canal indicates a wider niche for these microbes than was previously thought. Consequently, reevaluation of the roles of these two species in the pathogenesis of the middle ear is warranted.
Acknowledgments
This work was supported by grants from the NIH.
We thank members of the Pace laboratory and anonymous reviewers for many helpful criticisms of the manuscript.
REFERENCES
- 1.Beswick, A. J., B. Lawley, A. P. Fraise, A. L. Pahor, and N. L. Brown. 1999. Detection of Alloiococcus otitis in mixed bacterial populations from middle-ear effusions of patients with otitis media. Lancet 354:386-389. [DOI] [PubMed] [Google Scholar]
- 2.Bosley, G. S., A. M. Whitney, J. M. Pruckler, C. W. Moss, M. Daneshvar, T. Sih, and D. F. Talkington. 1995. Characterization of ear fluid isolates of Alloiococcus otitidis from patients with recurrent otitis media. J. Clin. Microbiol. 33:2876-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos, A., A. Arias, L. Betancor, C. Rodriguez, A. M. Hernandez, D. L. Auago, and A. Sierra. 1998. Study of common aerobic flora of human cerumen. J. Laryngol. Otol. 112:613-616. [DOI] [PubMed] [Google Scholar]
- 4.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durmaz, R., L. H. Ozerol, M. T. Kalcioglu, S. Oncel, B. Otlu, S. Direkel, and P. H. Hendolin. 2002. Detection of Alloiococcus otitidis in the nasopharynx and in the outer ear canal. Microbiologica 25:265-268. [PubMed] [Google Scholar]
- 7.Faden, H., and D. Dryja. 1989. Recovery of a unique bacterial organism in human middle ear fluid and its possible role in chronic otitis media. J. Clin. Microbiol. 27:2488-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraise, A. P., A. L. Pahor, and A. J. Beswick. 2001. Otitis media with effusion: the role of Alloiococcus otitidis. Ann. Med. 33:1-3. [DOI] [PubMed] [Google Scholar]
- 9.Frank, D. N., and N. R. Pace. 2001. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr. Opin. Gastroenterol. 17:52-57. [DOI] [PubMed] [Google Scholar]
- 10.Funke, G., G. E. Pfyffer, and A. von Graevenitz. 1993. A hitherto undescribed coryneform bacterium isolated from patients with otitis media. Med. Microbiol. Lett. 2:183-190. [Google Scholar]
- 11.Funke, G., S. Stubbs, M. Altwegg, A. Carlotti, and M. D. Collins. 1994. Turicella otitidis gen. nov., sp. nov., a coryneform bacterium isolated from patients with otitis media. Int. J. Syst. Bacteriol. 44:270-273. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Hernando, C., C. Toro, M. Gutierrez, A. Enriquez, and M. Baquero. 1998. Isolation of Alloiococcus otitidis from the external ear in children. Eur. J. Clin. Microbiol. Infect. Dis. 18:69-80. [DOI] [PubMed] [Google Scholar]
- 13.Hendolin, P. H., U. Karkkainen, T. Himi, A. Markkanen, and J. Ylikoski. 1999. High incidence of Alloiococcus otitis in otitis media with effusion. Pediatr. Infect. Dis. J. 18:860-865. [DOI] [PubMed] [Google Scholar]
- 14.Hendolin, P. H., A. Markkanen, J. Ylikoski, and J. J. Wahlfors. 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J. Clin. Microbiol. 35:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendolin, P. H., L. Paulin, and J. Ylikoski. 2000. Clinically applicable multiplex PCR for four middle ear pathogens. J. Clin. Microbiol. 38:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 17.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isenberg, H. D., and R. F. D'Amato. 1995. Indigenous and pathogenic microorganisms of humans, p. 5-18. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 19.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaClaire, L. L., and R. R. Facklam. 2000. Comparison of three commercial rapid identification systems for the unusual gram-positive cocci Dolosigranulum pigrum, Ignavigranum ruoffiae, and Facklamia species. J. Clin. Microbiol. 38:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 25.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappe, M. S., M. T. Suzuki, K. L. Vergin, and S. J. Giovannoni. 1998. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl. Environ. Microbiol. 64:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonet, M., D. D. Briel, I. Boucot, R. Minck, and M. Veron. 1993. Coryneform bacteria isolated from middle ear fluid. J. Clin. Microbiol. 31:1667-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 29.Stroman, D. W., P. S. Roland, J. Dohar, and W. Burt. 2001. Microbiology of normal external auditory canal. Laryngoscope 111:2054-2059. [DOI] [PubMed] [Google Scholar]
- 30.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki, M. T., M. S. Rappe, Z. W. Haimberger, H. Winfield, N. Adair, J. Strobel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford, D. L. 1999. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 33.Tanner, M. A., C. L. Everett, and D. C. Youvan. 2000. Molecular phylogenetic evidence for noninvasive zoonotic transmission of Staphylococcus intermedius from a canine pet to a human. J. Clin. Microbiol. 38:1628-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner, M. A., D. Shoskes, A. Shahed, and N. R. Pace. 1999. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and “nonbacterial” prostatitis. J. Clin. Microbiol. 37:1863-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]