Abstract
Shigella sonnei isolates from southwestern Korea during the epidemic periods of 1998 to 2000 were genetically related. The antimicrobial susceptibilities of the outbreak-related isolates changed annually. All isolates carried class 2 integrons, and the outbreak-related isolates from 1999 also carried class 1 integrons. The antimicrobial susceptibilities of S. sonnei isolates are readily changed by antibiotic selective pressures, and integrons are responsible for resistance to antimicrobial agents commonly used to treat shigellosis.
Bacillary dysentery caused by Shigella species is an important cause of acute diarrheal disease in both developing and industrialized countries (11). Shigellosis is a serious public health problem in Korea, because large outbreaks of S. sonnei infections in many parts of this country during the period 1998 to 2000 were reported. The annual incidence of shigellosis was estimated at about 10 cases before 1997 but explosively increased to about 1,000 to 2,500 cases during the period 1998 to 2000 (8). The frequency of S. sonnei during epidemic periods was over 90%. Ampicillin or trimethoprim combined with sulfamethoxazole has been used to treat shigellosis during the last 2 decades. However, a high proportion of S. sonnei isolates from Korea were resistant to these antimicrobial agents (2, 4). Integrons are known to be a novel mechanism for dissemination of resistance genes among gram-negative bacteria (1). The horizontal transfer of integrons may account for the dissemination of resistance genes and the emergence of multiresistant strains. The aim of this study is to determine the epidemiological relationships among, antimicrobial susceptibilities of, and presence of integrons in S. sonnei isolates from southwestern Korea during the epidemic periods of 1998 to 2000.
Sixty-seven S. sonnei isolates, 59 isolates from three defined outbreaks and 8 isolates from sporadic cases, were collected from stool samples in southwestern Korea (Chonnam province) during the period 1998 to 2000 (Table 1). One isolate (91NH14) was included for the comparison of epidemiological relationships. Biotyping was performed by the method of Nastasi et al. (6). The MICs of the antimicrobial agents were determined by means of the agar dilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (7). The antimicrobial agents included were ampicillin, cefoxitin, cefotaxime, nalidixic acid, ciprofloxacin, chloramphenicol, kanamycin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim. For pulsed-field gel electrophoresis (PFGE), genomic DNA was digested with XbaI for 20 h and separated in a 1% agarose gel with a contour-clamped homogeneous electric field apparatus (CHEF-DR2; Bio-Rad). The conditions for electrophoresis were 6 V/cm for 21 h, with the pulse time increasing from 5 to 40 s. PFGE patterns were interpreted by using the criteria established by Tenover et al. (9). PCR amplification of internal fragments of integrase genes intI1, intI2, and intI3 was performed to determine the presence of integrons (2, 10). Gene cassette regions of class 1 integrons were amplified (10) and sequenced. To determine dfr genes, PCR was performed as described previously (3). The presence of dfrA1 downstream of intl2 was analyzed by PCR using specific primers for intl2 and the 5′ end of dfrA1 (2).
TABLE 1.
Characterization of S. sonnei isolates used in this study
| Group | PFGE pattern | Presence of integrase gene:
|
Antibiograma | No. of isolates in:
|
||||
|---|---|---|---|---|---|---|---|---|
| intI1 | intI2 | intI3 | 1998 | 1999 | 2000 | |||
| S1 | A | − | + | − | TcSmSuTpNa | 27 | 4b,c | 1b |
| S2 | B | − | + | − | TcSmSuTpNaKm | 1b | 0 | 0 |
| S3 | A | − | + | − | SmTpNa | 1 | 0 | 0 |
| S4 | A | + | + | − | TcSmSuTpNaAp | 0 | 10 | 0 |
| S5 | A | − | + | − | TcSmSuTpNaAp | 1b | 0 | 0 |
| S6 | C | − | + | − | TcSmSuTpNaApKm | 2b | 0 | 0 |
| S7 | D | − | + | − | TcSmSuTpApKm | 0 | 0 | 20 |
Abbreviations of antimicrobial agents: Tc, tetracycline; Sm, streptomycin; Su, sulfamethoxazole; Tp, trimethoprim; Na, nalidixic acid; Km, kanamycin; Ap, ampicillin.
Eight isolates were collected from sporadic cases.
Of the four isolates, three were isolated from sporadic cases and one was isolated from the outbreak.
Sixty-seven S. sonnei isolates were investigated for epidemiological relationships on the basis of biotype, PFGE profile, presence of class 1 integrons, and antibiogram (Table 1). All isolates showed identical biotypes: all were positive for o-nitrophenyl-β-d-galactopyranoside and negative for fermentation of rhamnose and fermentation of xylose. S. sonnei isolates showed four different PFGE patterns with one to three band differences (Fig. 1); this indicates that they are genetically related by the criteria of Tenover et al. (9). The class 1 integrons from 10 outbreak-related isolates from 1999 were amplified. Based on antimicrobial susceptibility, six different antibiograms with 32, 1, 1, 11, 2, and 20 isolates were defined. Overall, the 67 S. sonnei isolates were classified into seven groups.
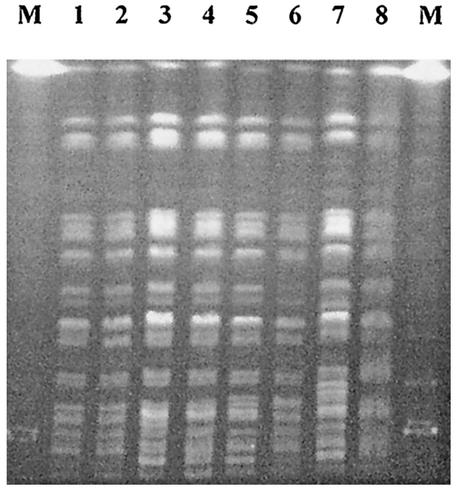
FIG. 1.
PFGE patterns of XbaI-cleaved genomic DNA of S. sonnei isolates. Lanes: M, lambda phage molecular weight ladder; 1, outbreak-related isolate from 1998; 2, sporadic isolate from 1998; 3, outbreak-related isolate from 1998; 4, outbreak-related isolate from 1999; 5, sporadic isolate from 1998; 6, sporadic isolate from 1998; 7, outbreak-related isolate from 2000; 8, 91NH14 from 1991. The PFGE patterns of lanes 1, 3, 4, 5, and 8 were identical and were classified pattern A. The PFGE patterns of lanes 2 and 6 were classified patterns B and C, respectively. The pattern of the 20 outbreak-related isolates from 2000 (lane 7) was classified pattern D.
The S. sonnei isolates were all resistant to streptomycin and trimethoprim, 99% were resistant to tetracycline and sulfamethoxazole, 70% were resistant to nalidixic acid, 49% were resistant to ampicillin, and 34% were resistant to kanamycin. No isolates were resistant to chloramphenicol, cefoxitin, cefotaxime, or ciprofloxacin. All outbreak-related isolates from 1998 except one were resistant to tetracycline, streptomycin, sulfamethoxazole, trimethoprim, and nalidixic acid. Ten isolates among the 11 outbreak-related isolates from 1999 were resistant to ampicillin, and the outbreak-related isolates from 1999 were also resistant to the five antimicrobial agents mentioned above for the 1998 outbreak-related isolates. The outbreak-related isolates from 2000 were resistant to tetracycline, streptomycin, sulfamethoxazole, trimethoprim, ampicillin, and kanamycin but were susceptible to nalidixic acid. Trimethoprim resistance was mediated by both dfrA1 and dfrA12. intI2 was amplified in all S. sonnei isolates, while intI3 was not found. A 1,650-bp class 1 integron for dfrA12, truncated orfF, and aadA2 from the 10 outbreak-related isolates from 1999 was amplified. Southern hybridization analyses revealed that intl1 and intl2 were located in plasmids and chromosomes, respectively. All S. sonnei isolates were found to carry dfrA1 downstream of intl2.
This study showed that one epidemic clone of S. sonnei has been spread in southwestern Korea during the epidemic periods of 1998 to 2000. The PFGE profile of 91NH14 was identical or very similar to those of the isolates from 1998 to 2000 (Fig. 1). These findings suggested that the isolate from 1991 and the isolates from 1998 to 2000 originated from a common ancestral clone.
The outbreak-related isolates from 1998 were susceptible to ampicillin, while the outbreak-related isolates from 1999 and 2000 were resistant to ampicillin. S. sonnei isolates from Korea between 1992 and 1997 were all susceptible to ampicillin (2). This result indicates that resistance to ampicillin in Korean S. sonnei isolates increased sharply over time. Ampicillin was the most commonly prescribed antimicrobial agent to treat shigellosis during the last decade in Korea. This may account for the emergence of resistance to ampicillin and the selection of ampicillin-resistant strains. All isolates from 1998 and 1999 were resistant to nalidixic acid, while the outbreak-related isolates from 2000 were susceptible to nalidixic acid. We previously reported that most S. sonnei isolates from 1980 to 1997 were susceptible to nalidixic acid (4). Nalidixic acid has not been an antimicrobial agent commonly used for bacterial infections during the last 2 decades. The limited use of nalidixic acid for a long time is responsible for the loss of resistance to nalidixic acid in the outbreak-related isolates from 2000, although the emergence of resistance to nalidixic acid in the isolates from 1998 and 1999 remains undefined. The prevalence of resistance to trimethoprim and sulfamethoxazole could be explained by the fact that trimethoprim combined with sulfamethoxazole has been used to treat shigellosis for a long time, thereby ensuring selection pressure for the maintenance of resistance.
Trimethoprim resistance was mediated by both dfrA1 and dfrA12. dfrA1 was located in class 2 integrons. Class 1 integrons, which comprised dfrA12, truncated orfF, and aadA2, were also detected in the outbreak-related isolates from 1999. These results indicate that integrons are responsible for resistance to trimethoprim. The same array of gene cassettes in class 1 integrons was first identified in Klebsiella pneumoniae from Korea and had a size of 1,911 bp (GenBank accession no. AF180731). Class 1 integron, which comprised dfrA12, orfF, and aadA2 gene cassettes, was also detected in urinary isolates of E. coli from the 1990s (3). The emergence of this type of class 1 integron in S. sonnei reflected the horizontal transfer of integrons between different species of Enterobacteriaceae. The horizontal transfer of class 1 integrons is responsible for the dissemination of antimicrobial resistance and alteration of genetic contents in bacteria. Therefore, characterization of class 1 integrons is a useful epidemiological tool for differentiation of the genetically related strains. In this study, S. sonnei isolates showing identical PFGE patterns and antibiograms were further discriminated on the basis of differences in class 1 integrons.
It has been recently reported that all S. sonnei biotype g isolates from Australia carried class 2 integrons and that two epidemic strains also carried class 1 integrons, which comprised dfrA12 and aadA2 gene cassettes (5). Biotypes and gene cassettes responsible for antimicrobial resistance were identical to those of the isolates from Korea. Furthermore, the majority of the isolates from Australia during the period 1997 to 2000 were resistant to streptomycin, sulfafurazole, tetracycline, and trimethoprim. The Australian epidemic isolates from 2000 were susceptible to kanamycin, while the Korean outbreak-related isolates from 2000 were resistant to kanamycin. Genotypic and phenotypic similarities of S. sonnei isolates from two countries raised the question of whether these were due to the spread of identical clones by travelers or to the same evolutionary changes in the bacteria caused by antimicrobial selective pressures.
Since the antimicrobial susceptibilities of S. sonnei isolates are readily changed by antibiotic selective pressures, an appropriate antimicrobial therapy is necessary to prevent the emergence of resistant strains and the dissemination of resistance genes.
Acknowledgments
This work was supported in part by the grant from Ministry of Health and Welfare in Korea (HMP-99-M-04-002).
REFERENCES
- 1.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 2.Kim, K. S., J. Y. Oh, Y. W. Jeong, J. W. Cho, J. C. Park, D. T. Cho, and J. C. Lee. 2002. Epidemiological typing and characterization of dfr genes of Shigella sonnei isolates in Korea during the last two decades. J. Microbiol. Biotechnol. 12:106-113. [Google Scholar]
- 3.Lee, J. C., J. Y. Oh, J. W. Cho, J. C. Park, J. M. Kim, S. Y. Seol, and D. T. Cho. 2001. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 47:599-604. [DOI] [PubMed] [Google Scholar]
- 4.Lee, J. C., J. Y. Oh, K. S. Kim, Y. W. Jeong, J. W. Cho, J. C. Park, S. Y. Seol, and D. T. Cho. 2001. Antimicrobial resistance of Shigella sonnei in Korea during the last two decades. APMIS 109:228-234. [DOI] [PubMed] [Google Scholar]
- 5.McIver, C. J., P. A. White, L. A. Jones, T. Karagiannis, J. Harkness, D. Marriott, and W. D. Rawlinson. 2002. Epidemic strains of Shigella sonnei biotype g carrying integrons. J. Clin. Microbiol. 40:1538-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nastasi, A., S. Pignato, C. Mammina, and G. Giammanco. 1993. rRNA gene restriction patterns and biotypes of Shigella sonnei. Epidemiol. Infect. 110:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 8.National Institute of Health in Korea. 2002. Epidemic characterization and prospect of recent shigellosis in Korea. Communicable Dis. Monthly Rep. 13:69-75. [Google Scholar]
- 9.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosini, F., P. Visca, I. Luzzi, A. M. Dionisi, C. Pezzella, and A. Petrucca. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila, J., J. Gascon, S. Abdalla, J. Gomez, F. Marco, A. Moreno, M. Corachan, and T. Jimenez de Anta. 1994. Antimicrobial resistance of Shigella isolates causing traveler's diarrhea. Antimicrob. Agents Chemother. 38:2668-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]