Abstract
The analytical performance of the NucliSens HIV-1 QT assay, a highly sensitive test based on nucleic acid sequence-based amplification technology, was evaluated in a multicenter trial. Assay specificity was evaluated with 502 plasma (EDTA) specimens from human immunodeficiency virus type 1 (HIV-1)-seronegative volunteer donors. No HIV-1 RNA was reported in any of the donor specimens. Analytical sensitivity and reproducibility were estimated with panels prepared from a high-titer well-characterized HIV-1 RNA stock (5.84 × 108 RNA copies/ml). The assay's dynamic range was linear from 106 to 101 HIV-1 RNA copies, with a lower detectable limit of 25 copies/ml and a 95% detection rate of 176 copies/ml. Sensitivity of the assay to detect HIV-1 RNA in clinical specimens from patients (n = 101) and in commercially available or prepared panels (n = 24) was compared with NASBA HIV-1 RNA QT (an earlier version of NucliSens HIV-1 QT) and with the Food and Drug Administration-approved standard and ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, assays. Detection of HIV-1 RNA was reproducible over a 5-log range (mean standard deviation = 0.15 log). The NucliSens and the standard AMPLICOR assays were equivalent in detection of HIV-1 RNA (concentration, 103 to 105 copies/ml) in 57 clinical specimens. The NucliSens assay was more sensitive in detecting HIV-1 RNA at lower concentrations (≤102 copies/ml) (44 of 44) than either the standard AMPLICOR test (12 of 19) or the NASBA assay (10 of 25). A 25% increase in HIV-1 RNA detection frequency with panels was observed with the NucliSens assay (23 of 24) compared with the standard AMPLICOR test (17 of 24). The new assay was highly specific and demonstrated good sensitivity with a broad linear dynamic range.
The measurement of human immunodeficiency virus type 1 (HIV-1) RNA levels in plasma (viral load) is a critical component in the management of persons with HIV-1 disease (20). Plasma HIV-1 RNA levels in conjunction with CD4+-T-cell counts enable clinicians and researchers to assess both the virologic and immunologic status of their patients and the success or failure of antiretroviral therapy regimens (14, 15, 20). The sequential monitoring of plasma viral load has aided investigators in understanding the nature of viral dynamics, thus providing key insights into the pathogenesis of HIV-1 disease (11, 18, 25).
Several assays are commercially available for the detection and quantitation of HIV-1 RNA in plasma (10). These assays utilize technologies based upon either signal amplification (QUANTIPLEX HIV-1 RNA assay [bDNA], Bayer Nucleic Acid Diagnostics, Norwood, Mass.) or target amplification (AMPLICOR HIV-1 MONITOR Test [Roche Molecular Systems, Inc., Branchburg, N.J.] and NucliSens HIV-1 QT assay [bioMérieux Inc., Durham, N.C.]). Each assay has specific advantages and disadvantages, and therefore, multiple factors should be considered in the selection of the most-appropriate assay for clinical use with specific patient populations. Specific versions of all three assays are currently approved by the Food and Drug Administration (FDA) for monitoring plasma HIV-1 viral load.
The performance characteristics of an assay such as dynamic range, lower LOD and quantitation, accuracy, specificity, reproducibility, and the ability to detect and accurately quantify various HIV-1 subtypes are key factors for selection in clinical and research use. In addition, the concordance of results from one assay to another is critical when comparing data from antiretroviral therapy trials and in cases where the same viral load assay has not been consistently used for patient management.
HIV-1 RNA quantitation with the NucliSens HIV-1 QT assay is based upon an isothermal nucleic acid sequence-based amplification (NASBA) method that is highly suited for the amplification of RNA (13, 24). Total plasma nucleic acids and three internal calibrators of known low, medium, and high RNA concentrations are coextracted according to the method of Boom et al. (2). Single-tube coamplification of wild-type patient HIV-1 RNA (sense RNA) and the internal calibrators is achieved through the coordinated activities of three enzymes (avian myeloblastosis virus reverse transcriptase, RNase H, and T7 RNA polymerase) and two DNA oligonucleotides that are specific for the gag region of the HIV-1 target sequence. The amplification results in the production of large amounts of single stranded RNA that is antisense to the original target RNA. The single-stranded RNA product can then be readily detected using electrochemiluminescence (ECL) after hybridization of wild-type or calibrator-specific ruthenium-labeled oligonucleotide probes. The amount of light generated (ECL units) is proportional to the amount of the product amplificate (in RNA copies per milliliter). Calculation based upon the relative amounts of the four amplificates (wild-type patient sample and three calibrators) determines the original amount of wild-type HIV-1 RNA in the sample. No additional external controls or standards are necessary for the sample quantitation. However, the manufacturer recommends that a high-positive control, a low-positive control, and a negative control be included with the first run of each kit lot and a negative control and a low-positive control be included with each subsequent run of the same kit to verify product performance. The use of an external control can also be used to monitor assay reproducibility from lot to lot, run to run, and technician to technician.
In this multicenter study, we evaluated the performance characteristics of the NucliSens HIV-1 QT assay, which is an enhanced version of the 1st generation NASBA HIV-1 RNA QT assay (bioMérieux, Inc.). The assay parameters analyzed included linear dynamic range, limit of detection (LOD) and limit of quantitation (LOQ), clinical sensitivity, reproducibility, specificity, and subtype detection. In addition, studies were conducted that compared the performance of the NucliSens HIV-1 QT assay with that of the standard and ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, assays.
(This study was presented in part at the 17th Annual Clinical Virology Symposium, Clearwater, Fla., 29 April to 2 May 2001, and presented in part at the 53rd Annual American Association of Blood Bank Meeting, Washington, D.C., 4 to 8 November 2000.)
MATERIALS AND METHODS
Clinical trial sites.
Three clinical trial sites conducted the studies: the Center for Blood Research, the research affiliate of the Sacramento Medical Foundation (Sacramento, Calif.); the North Shore-Long Island Jewish Health System Laboratories (Lake Success, N.Y.)—North Shore University Hospital (Manhasset, N.Y.); and Albany Medical Center Hospital (Albany, N.Y.). All studies were performed under Institutional Review Board approval, and informed consent was obtained prior to the collection of blood samples.
Clinical samples.
Whole-blood samples were collected by venipuncture using VACUTAINER EDTA anticoagulant tubes (Becton Dickinson, Franklin Lakes, N.J.). Tubes were centrifuged for 20 min at 1,200 × g, and 1 ml of cell-free plasma was added to 9.0 ml of NucliSens lysis buffer (bioMérieux Inc.). Tubes were vortexed well and, after lysis at room temperature for 15 min, were stored at −70°C until tested with the NucliSens HIV-1 QT assay and, when indicated, the first-generation NASBA HIV-1 RNA QT assay. The remaining cell-free plasma was removed and immediately frozen in cryovials (Nalgene, Nunc Intl., Roskilde, Denmark) at −70°C until tested with the standard AMPLICOR HIV-1 MONITOR, version 1.0, test (Roche Molecular Systems, Inc.) and/or the ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, test (Roche Molecular Systems, Inc.).
HIV-1 RNA panels.
To characterize the performance of the NucliSens HIV-1 QT assay, various panels were obtained and evaluated.
(i) HIV-1 dilution panel.
A well-characterized HIV-1 subtype B RNA stock (titer = 5.4 × 108 HIV-1 RNA copies/ml) (Rush-Presbyterian-St. Luke's Medical Center, Chicago, Ill.) was diluted in normal human plasma (HIV-1 seronegative) and used to prepare panels for both the linearity study and the LOQ study. The normal human plasma used to prepare the dilutions of the HIV-1 RNA stock was obtained from an HIV-1-seronegative volunteer blood donor and was found to be HIV-1 RNA negative upon testing with both the NASBA assay and the NucliSens assay.
(ii) Reference panel.
A 10-member reference panel consisting of two normal human plasma samples (HIV-1 seronegative) and eight human plasma samples with various concentrations of HIV-1 RNA (range = 101 to 2.5 × 105 copies/ml) was provided by the Center for Biologics Evaluation and Research (CBER) (FDA, Rockville, Md.).
(iii) BBI HIV-1 RNA quantification panel.
The BBI HIV-1 RNA quantification panel QRD702-1.0 (BBI Diagnostics, Boston Biomedica Company, West Bridgewater, Mass.), a six-member panel, consisted of one normal human plasma sample (HIV-1 seronegative) and five human plasma samples with various concentrations of HIV-1 subtype B RNA (range = <4 × 102 to 2 × 105 copies/ml).
(iv) BBI HIV-1 clade performance panel.
The BBI HIV-1 clade performance panel PRD201-1.1 (BBI Diagnostics, Boston Biomedica Company) consisted of a set of eight aliquots of HIV-1 virus from tissue culture representing HIV-1 group M clades A through H. The viral isolates used to prepare the BBI panels were provided by the National Institutes of Health (Bethesda, Md.), the United States Department of Defense (Washington, D.C.), and the European Network for Virological Assessment of New HIV Therapies (Utrecht, The Netherlands). Each viral culture was supplied by BBI diluted in defibrinated human plasma, negative for HIV-1 RNA, to a copy number of approximately 5 × 104 virus particles per ml.
Quantitation of HIV-1 RNA. (i) First-generation NASBA HIV-1 RNA QT assay and NucliSens HIV-1 QT assay.
Nucleic acid isolation, amplification, and detection of plasma HIV-1 RNA were performed with the first-generation NASBA HIV-1 RNA QT assay and NucliSens HIV-1 QT assay according to the manufacturer's instructions provided with each assay kit.
(ii). Standard and ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, tests.
The quantitation of plasma HIV-1 RNA using the standard and/or ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, test was performed according to the manufacturer's instructions. The lower LOQ for the standard AMPLICOR procedure and that of the ultrasensitive procedure, as stated in the package insert, are 400 and 50 HIV-1 RNA copies/ml, respectively.
Linearity studies.
Each member of a 19-member series panel, derived by twofold serial dilutions (range = 2.0 × 101 to 5.4 × 106 HIV-1 RNA copies/ml) from a 1:100 dilution of the HIV-1 RNA stock (Table 1), was tested in duplicate with each of three lots of the NucliSens assay. The order for the NucliSens testing of the panel members was randomized, as was the sequence in which each kit lot was analyzed.
TABLE 1.
Linearity and LOD analysis obtained using dilutions of the VQA stock HIV-1 RNA
| VQA dilution panel | NucliSens HIV-1 QTa
|
NASBA HIV-1 QTb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Panel member | No. of HIV-1 copies/mlc | Log HIV-1 copies/ml | No. of samples tested | No. of samples positive | % Positive results | Mean no. of copies/ml | Mean log copies/ml | No. of samples tested | Mean no. of copies/mld | Mean log copies/ml |
| 1 | 5,400,000 | 6.73 | 6 | 6 | 100 | 6,550,000 | 6.79 | 0 | Not tested | |
| 2 | 2,700,000 | 6.43 | 6 | 6 | 100 | 2,800,000 | 6.44 | 1 | 1,600,000 | 6.20 |
| 3 | 1,350,000 | 6.13 | 6 | 6 | 100 | 1,300,000 | 6.11 | 1 | 650,000 | 5.81 |
| 4 | 675,000 | 5.83 | 6 | 6 | 100 | 595,000 | 5.77 | 1 | 390,000 | 5.59 |
| 5 | 337,000 | 5.53 | 6 | 6 | 100 | 275,000 | 5.44 | 1 | 210,000 | 5.32 |
| 6 | 168,750 | 5.23 | 6 | 6 | 100 | 128,000 | 5.10 | 1 | 96,000 | 4.98 |
| 7 | 84,375 | 4.93 | 6 | 6 | 100 | 64,667 | 4.80 | 1 | 56,000 | 4.75 |
| 8 | 42,187 | 4.63 | 6 | 6 | 100 | 29,833 | 4.47 | 1 | 28,000 | 4.45 |
| 9 | 21,093 | 4.32 | 6 | 6 | 100 | 17,167 | 4.23 | 1 | 14,000 | 4.15 |
| 10 | 10,546 | 4.02 | 6 | 6 | 100 | 8,533 | 3.92 | 1 | 4,900 | 3.69 |
| 11 | 5,273 | 3.72 | 6 | 6 | 100 | 4,683 | 3.65 | 1 | 2,200 | 3.34 |
| 12 | 2,636 | 3.42 | 6 | 6 | 100 | 2,400 | 3.38 | 1 | 1,200 | 3.08 |
| 13 | 1,318 | 3.12 | 6 | 6 | 100 | 1,033 | 3.00 | 1 | 540 | 2.73 |
| 14 | 659 | 2.82 | 6 | 6 | 100 | 433 | 2.57 | 1 | 820 | 2.91 |
| 15 | 329 | 2.52 | 6 | 6 | 100 | 455 | 2.51 | 1 | <LDLe | |
| 16 | 164 | 2.21 | 6 | 5 | 83 | 97 | 1.91 | 1 | <LDL | |
| 17 | 82 | 1.91 | 6 | 3 | 50 | 42 | 1.61 | 1 | <LDL | |
| 18 | 41 | 1.61 | 6 | 4 | 67 | 90 | 1.93 | 1 | <LDL | |
| 19 | 20 | 1.30 | 6 | 1 | 17 | 31 | 1.49 | 1 | <LDL | |
NucliSens HIV-1 QT results for each panel member represent the mean of duplicate testing with each of three kit lots.
NASBA HIV-1 RNA QT results were obtained by testing each panel member one time.
Nominal values.
Observed values.
<LDL, less than the detectable limit (400 copies/ml).
LOD and LOQ studies.
Five of the HIV-1 stock panel specimens (dilutions 14 through 18) representing a range of HIV-1 RNA concentrations (6.59 × 102 to 4.1 × 101 copies/ml) were selected based on the linearity study results, and each was tested 66 additional times using three lots of the NucliSens assay. The order for the NucliSens testing of the panel members was randomized within a given run, as was the sequence in which each lot was analyzed. Each panel member was also tested once with the NASBA assay.
Terminal dilution of clinical specimens.
Clinical specimens from HIV-1-infected individuals with various HIV-1 RNA concentrations (range = 2.0 × 103 to 4.7 × 106 copies/ml) were diluted with normal human plasma, and each dilution was tested with each of three lots of the NucliSens assay and once with the NASBA assay. The order for the NucliSens testing of the diluted specimens was randomized within a given run, as was the sequence in which each lot was analyzed.
Determination of clinical sensitivity.
The clinical sensitivity of the NucliSens assay was assessed by testing 106 specimens collected from patients at North Shore University Hospital. Samples from these patients contained low copy numbers of HIV-1 RNA (<1,000 copies/ml) or no detectable HIV-1 RNA copy numbers (<400 copies/ml) when tested with the NASBA assay. Each specimen was tested with a 1.0-ml plasma volume using the NASBA assay and the NucliSens assay.
Specificity studies.
The specificity of the NucliSens assay was assessed by evaluating 502 specimens from a volunteer whole-blood donor population. The NucliSens results were compared to those obtained with an FDA-licensed enzyme immunoassay (EIA) for detection of HIV-1 and HIV-2 antibodies (HIV-1/2 EIA; Abbott Laboratories, Abbott Park, Ill.).
Reproducibility and precision studies.
A reproducibility panel consisting of four specimens containing different levels of input HIV-1 (range, 106 to 103 copies/ml) was evaluated in multiple runs with three different NucliSens kit lots. The panel specimens were prepared by dilution in normal HIV-1-seronegative plasma of the HIV-1 RNA stock (titer = 5.4 × 108 copies/ml). The order of panel member testing for each lot was randomized, as was the sequence in which each lot was analyzed. The study was designed to estimate interassay variability due to sites, lots, technicians, and days. To estimate intra-assay precision, each specimen was run in duplicate in each run with the NucliSens assay. The total number of tests for each specimen was 72 (three sites times three lots times one technician times four days times two replicates per run).
Comparison with the AMPLICOR HIV-1 MONITOR, version 1.0, tests.
Specimens from the HIV-1 panels (described above) and from clinical subjects were used in this study. Each specimen was tested once with each assay. Specimens with <400 HIV-1 RNA copies/ml with the standard AMPLICOR procedure were reanalyzed with the ultrasensitive AMPLICOR procedure as specimen volume permitted.
Detection and quantification of HIV-1 clades A through H.
Each member of the nine-member panel (clades A through H and diluent negative control) was tested once with the NucliSens assay and once with the standard AMPLICOR procedure.
Statistical analysis. (i) Linear range.
The linear range was determined with the y variable equal to the logarithm of the actual number of observed copies and the x variable equal to the logarithm of the expected number of input copies based on the original value assigned to the HIV-1 stock of 5.4 × 108 RNA copies/ml. Test results at each x value were assumed to be independent and to follow a Gaussian distribution.
Different data sets were formed from the results obtained with the three lots of NucliSens HIV-1 QT. Each data set represented a different range of the x variable for the analysis. For each data set, a test of linearity of the results in the range was performed with linear regression analysis to partition the residual variation into two components: one representing error due to lack of fit and the other representing pure error. The ratio of expected mean squares of these sources of error follows an F distribution. For each range, an F value estimating this ratio was computed, and the probability (P) of obtaining an F value greater than the observed value under a null hypothesis of no lack of fit to a linear model was calculated. A P of less than 0.15 was taken to indicate evidence of nonlinearity.
(ii) LOD and LOQ.
The LOD is defined as the HIV-1 RNA input level at which at least 95% of the tests produce a quantifiable result. The LOQ is defined as the HIV-1 RNA input level at which at least 95% of the tests produce quantifiable results with good precision (coefficient of variation [CV] = <35%). The percentage of positive results for each nominal HIV-1 RNA concentration of the panel was calculated, and the LOD was determined by logistic regression with PROC LOGISTIC (SAS/STAT version 6.12).
(iii) Precision and reproducibility.
Four sources of variation were identified: between lot, between laboratory, between run, and within run. Due to the study design, between-laboratory and between-operator variability coincide. The variance components model used was the following: yi,j,k,m = μ + νi + λj + δi,j,k + ɛi,j,k,m, where yi,j,k,m is the logarithm of observed number of HIV-1 RNA copies obtained at laboratory i, lot j, day (run) k, and replicate m; μ is the overall mean; νi is the random effect from laboratory (operator) i; λj is the random effect from lot j; δi,j,k is the random effect from day k in laboratory i with lot j; and ɛi,j,k,m is the random effect from replicate m of laboratory i, lot j, and day k.
Variance components were quantified using restricted maximum-likelihood techniques using the statistical software package SAS (PROC VARCOMP). The total variability was assessed as the sum of the individual variance components. To illustrate the between-lot and between-site variability, graphs were constructed showing the mean results of each lot over the sites and the mean results of each site over the lots as a function of the observed overall mean. In addition, a graph was constructed showing the total variability as a function of the input.
(iv) Specificity studies.
To determine the specificity of the assay, the observed fraction of false-positive (or true-negative) results were calculated with exact two-sided 95% confidence intervals based on the binomial probability distribution.
(v) Performance comparison with the standard and ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, assays.
HIV-1 copy numbers reported by the NucliSens assay and the AMPLICOR assays were transformed to base 10 logarithms and analyzed to estimate the Pearson product moment coefficient. The confidence intervals for the correlation were determined using Fischer's method.
RESULTS
NucliSens HIV-1 QT assay linearity.
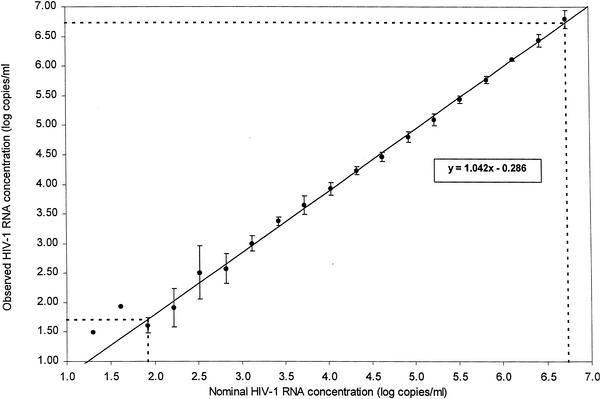
Testing of 19 specimens derived by twofold serial dilutions of the HIV-1 RNA stock (Table 1) indicated a direct proportional relationship between the dilutions tested and the number of HIV-1 RNA copies reported by the NucliSens assay (Fig. 1). The number of HIV-1 RNA copies consistently decreased as a function of increasing dilution of the stock. The linear range of the assay, as determined using the combined data from three individual kit lots and linear regression analysis of the log observed HIV-1 RNA copies regressed on the log expected HIV-1 RNA copies (Fig. 1), extended from panel member 1 to panel member 17. Based on the regression equation, the mean assay result was 5,390,000 HIV-1 RNA copies/ml for specimen 1, and 51 HIV-1 RNA copies/ml for specimen 17. While the performance response of the assay was similar for each of the three NucliSens kit lots evaluated, some variation was observed for values outside the linear range of the assay.
FIG. 1.
Assay linearity determined by the relationship between nominal and observed HIV-1 RNA concentrations. Values for log nominal HIV-1 RNA copies per milliliter are plotted against the means of the observed concentrations (log10 HIV-1 RNA copies per milliliter). Error bars represent 1 SD. Regression analysis is shown by the solid line (y = 1.042x − 0.286). Dotted lines indicate linear dynamic range of the assay.
NucliSens HIV-1 QT LOD and LOQ.
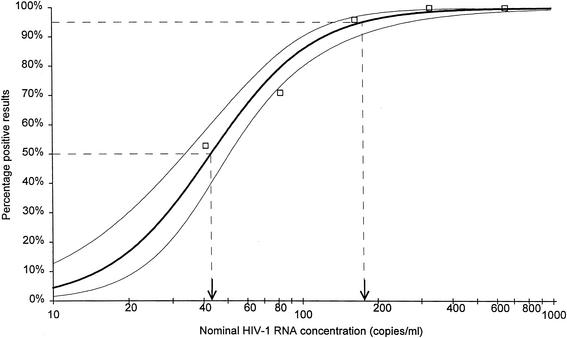
The LOD is defined as the lowest HIV-1 RNA input level at which at least 95% of the tests produce a result indicative of reactivity of the input sample for HIV-1. Logistic regression analysis of the results presented in Table 1 indicated a LOD of 176 HIV-1 RNA copies/ml, with a detection rate of 95% (Fig. 2). The lowest observed concentration that was reported by the NucliSens assay was 25 HIV-1 RNA copies/ml (nominal value of 41 HIV-1 RNA copies/ml). Further studies with panels of specimens representing a low range of HIV-1 RNA concentrations indicated that the detection of HIV-1 RNA was a function of concentration, with the highest percentage of detection (100%) at the highest concentrations tested (329 and 659 HIV-1 RNA copies/ml) and the lowest detection rate (53%) at the lowest concentration tested (41 HIV-1 RNA copies/ml) (Table 2). The LOQ is defined as the concentration with at least a 95% detection rate with good precision (CV = <35%). Observed CVs for log copies for panel members 15 (nominal input = 329 HIV-1 RNA copies/ml) and 16 (nominal input = 164 HIV-1 RNA copies/ml) were 10 and 16%, respectively. The observed standard deviations (SD) for log copies for these panel members are 0.28 and 0.36, respectively. Based on these results, the LOQ is equal to the LOD, i.e., 176 HIV-1 RNA copies/ml.
FIG. 2.
LOD and LOQ. Logistic regression analysis of results is represented by the bold line. The two fine lines represent the upper and lower confidence intervals. Arrows indicate 50 and 95% detection limits, which are 41 and 176 HIV-1 RNA copies/ml, respectively.
TABLE 2.
NucliSens HIV-1 QT assay LOD measurements
| Nominal concna (HIV-1 RNA copies/ml) | Log HIV-1 RNA copies/ml | NucliSens HIV-1 QT result
|
||
|---|---|---|---|---|
| No. of samples with <LDLb | No. of samples with >25 copies/ml | % Positive | ||
| 41 | 1.61 | 34 | 38 | 53 |
| 82 | 1.91 | 21 | 51 | 71 |
| 164 | 2.21 | 3 | 69 | 96 |
| 329 | 2.52 | 0 | 72 | 100 |
| 659 | 2.82 | 0 | 72 | 100 |
Nominal concentration was derived from serial dilutions of a highly concentrated HIV-1 RNA stock (n = 72 per dilution) (see Materials and Methods).
<LDL, result below the cutoff limit (25 copies/ml).
Clinical sensitivity.
One hundred six clinical specimens collected from persons known to be HIV-1 positive were evaluated with both the NucliSens and the NASBA assays. The lowest reportable concentration of the NASBA assay is 400 copies/ml with a 1-ml specimen input volume compared to 25 copies/ml for the NucliSens assay. A significant difference in the number of specimens reported with HIV-1 RNA was observed when the results obtained with both assays were compared (data not shown). The NucliSens assay detected HIV-1 RNA in 82 of 106 specimens (77%), whereas the NASBA assay detected HIV-1 RNA in approximately three times fewer specimens (26 of 106 [25%]). Both assays detected HIV-1 RNA in 25 specimens, and neither detected HIV-1 RNA in 23 specimens. Based on the previously described results for the terminal dilution study (Table 1) and low-copy-number replication study (Table 2), the RNA copy number in those 23 specimens with a NucliSens result less than the detectable limit was either <25 HIV-1 RNA copies/ml or within the range of detection (25 to 176 copies/ml) that demonstrated a detection rate of <95%. Fifty-seven clinical specimens had an HIV-1 RNA copy number reported with the NucliSens assay only, and one specimen had an HIV-1 RNA copy number (1,300 copies/ml) reported with the NASBA assay only. For these 57 specimens, the following distribution was noted: 13 samples contained 25 to 99 HIV-1 RNA copies/ml, 20 samples contained 100 to 499 HIV-1 RNA copies/ml, 11 contained 500 to 999 HIV-1 RNA copies/ml, and 13 samples contained >1,000 HIV-1 RNA copies/ml. The mean copy number was 628, with a median of 330 and an SD of 716. The range of HIV-1 RNA copy numbers was 27 to 3,300 copies/ml.
To further evaluate the relative clinical sensitivity of the NucliSens assay, six plasma specimens from HIV-1-infected individuals were diluted to terminal endpoint with normal human plasma. In each case, the response in copy number was a function of the dilution tested, with a decrease of HIV-1 RNA copy number observed with increasing dilution (Table 3). The detection of HIV-1 RNA was equivalent for each of the three NucliSens kit lots evaluated, although some fluctuation in response was observed in the dilution immediately preceding the extinction dilution for three of the six specimens. The sensitivity of NucliSens assay was improved significantly over that of NASBA assay in that detection of HIV-1 RNA was obtained at 1 lower dilution for NucliSens with each of the six clinical specimens. In addition, testing of the HIV-1 RNA stock panel with the NASBA assay demonstrated that the last sample with detectable RNA was sample 14 (659 copies/ml) (Table 1). In contrast, the NucliSens assay demonstrated superior sensitivity in detecting HIV-1 RNA at low concentrations, as shown by the results obtained for specimens 15 through 19 (expected values = 329 to 25 copies/ml).
TABLE 3.
HIV-1 RNA Copy numbers reported for clinical specimens diluted to terminal endpointa
| Specimen | Dilution tested | NucliSens HIV-1 QT (copies/ml)
|
NASBA HIV-1 RNA QT (copies/ml) | ||
|---|---|---|---|---|---|
| Lot 1 | Lot 2 | Lot 3 | |||
| 9040321 | Undiluted | 470,000 (5.67) | 340,000 (5.53) | 470,000 (5.67) | 360,000 (5.56) |
| 1:300 | 2,500 (3.40) | 1,200 (3.08) | 1,700 (3.23) | 2,000 (3.30) | |
| 1:3,000 | 81 (1.91) | 150 (2.18) | 41 (1.61) | <LDLb | |
| 1:30,000 | <Lower limitc | <Lower limit | <Lower limit | <LDL | |
| 9040423 | Undiluted | 200,000 (5.30) | 92,000 (4.96) | 170,000 (5.23) | 240,000 (5.38) |
| 1:100 | 2,100 (3.32) | 1,200 (3.08) | 1,600 (3.20) | 3,900 (3.59) | |
| 1:1,000 | 270 (2.43) | 81 (1.91) | 470 (2.67) | <LDL | |
| 1:10,000 | <Lower limit | <Lower limit | <Lower limit | <LDL | |
| 9040318 | Undiluted | 73,000 (4.86) | 38,000 (4.58) | 82,000 (4.91) | 84,000 (4.92) |
| 1:100 | 910 (2.96) | 580 (2.76) | 360 (2.56) | 1,300 (3.11) | |
| 1:1,000 | 42 (1.62) | <Lower limit | 120 (2.08) | <LDL | |
| 1:10,000 | <Lower limit | <Lower limit | <Lower limit | <LDL | |
| 9040338 | Undiluted | 13,000 (4.12) | 6,300 (3.80) | 6,800 (3.83) | 16,000 (4.20) |
| 1:10 | 950 (2.98) | 790 (2.90) | 1,200 (3.08) | 1,100 (3.04) | |
| 1:100 | 35 (1.54) | <Lower limit | <Lower limit | <LDL | |
| 1:1,000 | <Lower limit | <Lower limit | <Lower limit | <LDL | |
| 9040500 | Undiluted | 4,400 (3.64) | 2,800 (3.45) | 7,800 (3.89) | 6,600 (3.82) |
| 1:5 | 950 (2.98) | 760 (2.88) | 1,300 (3.12) | 1,100 (3.04) | |
| 1:50 | 200 (2.30) | 28 (1.45) | 360 (2.56) | <LDL | |
| 1:500 | <Lower limit | <Lower limit | <Lower limit | <LDL | |
| 9040428 | Undiluted | 4,000 (3.60) | 2,000 (3.30) | 3,700 (3.57) | 3,700 (3.57) |
| 1:5 | 740 (2.87) | 190 (2.28) | 500 (2.70) | 1,200 (3.08) | |
| 1:20 | 280 (2.45) | 130 (2.11) | <Lower limit | <LDL | |
| 1:200 | <Lower limit | <Lower limit | <Lower limit | <LDL | |
Values in parentheses are log10 HIV-1 copies/ml.
<LDL, result below the detectable limit for NASBA HIV-1 QT (400 copies/ml).
Below lower limit for NucliSens HIV-1 QT (25 copies/ml).
Assay specificity.
Five hundred two (502) plasma specimens from low-risk whole-blood donors were tested with the NucliSens assay and the FDA-licensed Abbott HIV-1/2 antibody EIA. Neither HIV-1 RNA- nor HIV-1-specific antibodies were detected in the samples. The specificity of the NucliSens assay relative to the licensed antibody test was 100% (95% confidence limits of 99.27 to 100%). The distribution of the NucliSens assay wild-type ECL signal detection values for these specimens indicated that the likelihood of reporting a false positive is minimal. Of the wild type ECL values obtained for these specimens, 72% were <20 ECL units.
Reproducibility and precision studies.
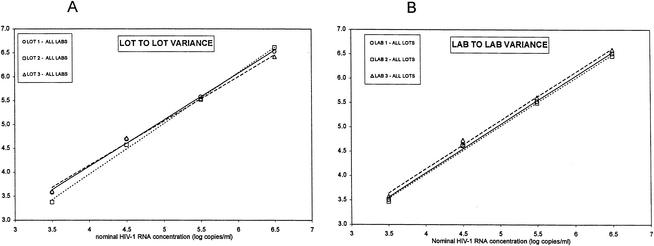
Overall between-run and within-run precision were determined in a reproducibility study in which the effects of multiple lots and multiple technicians/sites were evaluated with a series of four specimens containing HIV-1 RNA ranging from 106 to 103 copies/ml prepared from the HIV-1 RNA stock (Table 4). The NucliSens assay lot-to-lot variance (Fig. 3, panel A), interlaboratory variance (Fig. 3B), between-run variance (range = 0.033 to 0.081 log10 copies/ml), and within-run variance (range = 0.062 to 0.100 log10 copies/ml) were low for all specimens in the medium- to high-copy-number range, with a combined SD of 0.13 log10 copies/ml (range, 0.024 to 0.18 log10 copies/ml) from the observed mean values (Table 4). The total combined precision rate at the low-end concentrations (1.98 to 2.93 log10 copies/ml), expressed as the SD from the mean observed values, was 0.32 log10 copies/ml (Table 4). The total SDs of the observed HIV-1 RNA concentrations (log10 copies per milliliter) versus the mean observed HIV-1 RNA concentrations (log10 copies per milliliter) for all eight samples was 0.225 log10 copies/ml. In terms of CVs, when test results were measured on a logarithmic scale, estimated total precision ranged from 2 to 17%, interassay variability ranged from 1 to 4%, and intra-assay variability ranged from 1 to 17%. The lot-to-lot precision was 0.5 to 3% and the operator-to-operator precision ranged from 1 to 3%.
TABLE 4.
NucliSens HIV-1 QT assay precision estimates
| HIV-1 RNA stock dilutiona | Nominal mean value (log10 copies/ml) | No. of specimens that tested positive | Observedb mean value (log10 copies/ml) | Overall SDc (log10 copies/ml) |
|---|---|---|---|---|
| 2 | 6.43 | 72 | 6.52 | 0.14 |
| 5 | 5.53 | 72 | 5.54 | 0.09 |
| 8 | 4.63 | 72 | 4.66 | 0.12 |
| 12 | 3.42 | 72 | 3.52 | 0.18 |
| 14 | 2.82 | 72 | 2.93 | 0.29 |
| 15 | 2.52 | 72 | 2.66 | 0.28 |
| 16 | 2.21 | 69 | 2.26 | 0.36 |
| 17 | 1.91 | 51 | 1.98 | 0.34 |
Serial dilutions were made from a well-characterized HIV-1 RNA stock (titer = 5.4 × 108 HIV-1 RNA copies/ml) as described in Materials and Methods.
Observed mean value obtained for each dilution derived from replicate tests per sample.
Overall value for all 552 replicate tests, 0.225.
FIG. 3.
Linear regression analysis on the four mean observed concentrations generated by all three laboratories using the three specific lot numbers (A) and by all three lots at the three specific laboratories (B). Mean observed HIV-1 RNA concentrations (log10 copies per milliliter) (on the y axis) are depicted against nominal HIV-1 RNA concentrations.
Performance comparison with the standard AMPLICOR HIV-1 MONITOR, version 1.0, assay. (i) Clinical samples.
The clinical sensitivity of the NucliSens assay was compared to the standard AMPLICOR assay by testing 76 clinical specimens with HIV-1 RNA copy numbers ranging from 1.43 log10 copies/ml to 5.75 log10 copies/ml (27 to 560,000 copies/ml) as determined by the NucliSens assay (Table 5). The standard AMPLICOR assay reported an HIV-1 RNA copy number for 63 of the 76 samples (83%). For one specimen in which HIV-1 RNA was detected with both assays, only the NucliSens assay reported a valid copy number of 5.75 log10 copies/ml (560,000 HIV-1 RNA copies/ml) while the standard AMPLICOR assay result was >5.88 log10 copies/ml (750,000 HIV-1 RNA copies/ml), which exceeded the linear range of the assay. The average difference in the reported HIV-1 RNA copy numbers for both assays was 0.210 log.
TABLE 5.
Sensitivity of NucliSens HIV-1 QT and AMPLICOR HIV-1 MONITOR assays
| Group | No. tested | No. of specimens reported with copy no. (% positive)
|
|
|---|---|---|---|
| NucliSens HIV-1 QT | AMPLICOR HIV-1 MONITOR | ||
| HIV-1 RNA stock dilution | 14 | 14 (100) | 9 (64) |
| Clinical specimen | 76 | 76 (100) | 63 (83) |
| BBI and CBER panela | 13 | 12 (92) | 8 (62) |
| Total | 103 | 102 (99) | 80 (78) |
Number tested only includes those samples which contained HIV-1 RNA (8 of 10 CBER samples and four of five BBI samples).
(ii) HIV-1 panel dilutions.
Fourteen specimens (specimens 3 through 16) used for the linearity-LOQ studies (Table 1) were tested with the standard AMPLICOR assay. In contrast to the results obtained with NucliSens assay which produced a copy number for each specimen tested, the standard AMPLICOR assay detected HIV-1 RNA in 78.6% (11 of 14) of the specimens but reported a copy number for only 64% (9 of 14) (Table 5). Of the five specimens for which HIV-1 RNA copy numbers were not reported with the standard AMPLICOR assay, two were >750,000 HIV-1 RNA copies/ml and three were <400 HIV-1 RNA copies/ml. For the nine specimens for which HIV-1 RNA copy numbers were detected with both assays, the HIV-1 RNA copies were very similar for both assays, with a 0.086 mean log difference and an SD of 0.26 log.
BBI quantification panel.
Results of testing the BBI quantification panel (BBI-QRD702) with the standard AMPLICOR assay and the NucliSens assay are listed for all six specimens in Table 5. The NucliSens assay gave reportable results for five of the six samples. Panel member 1 was the diluent only (normal human plasma) without any HIV-1 RNA, and neither assay detected HIV-1 RNA with this panel member. Panel member 2 contained a low concentration of HIV-1 RNA and was reported positive by the NucliSens assay (71 copies/ml) but not by the standard AMPLICOR assay. The remaining four specimens contained HIV-1 RNA (range, 103 to 2 × 105) and were correctly reported with both the standard AMPLICOR assay and the NucliSens assay.
CBER reference panel.
Testing with the CBER reference panel indicated that the NucliSens assay was more sensitive in detecting HIV-1 RNA than was the AMPLICOR assay (Table 5). With this panel (copy number ranging from 0 to 25,000 copies/ml), neither assay reported HIV-1 RNA for the two specimens negative for HIV-1 RNA and the one specimen that contained 10 HIV-1 RNA copies/ml. Three additional specimens, with HIV-1 RNA ranging from 50 to 500 copies/ml, were detected with the NucliSens assay only. Both assays detected the remaining four members of the panel with expected HIV-1 RNA copy numbers ranging from 2,500 to 250,000 copies/ml. The average difference in the reported HIV-1 RNA copy numbers for the two assays was 0.1761 log.
Performance comparison with the ultrasensitive AMPLICOR HIV-1 MONITOR, version 1.0, assay.
Further analysis was performed with 17 specimens that were present at <400 HIV-1 RNA copies/ml in the standard AMPLICOR assay by evaluation with the ultrasensitive AMPLICOR procedure. Of the 17 specimens evaluated with the ultrasensitive procedure, an HIV-1 RNA copy number was obtained with 15 (88%), while the NucliSens assay detected 16 of these specimens (94%). An HIV-1 RNA copy number was not obtained with either test for one specimen which contained only 10 HIV-1 RNA copies/ml. The average difference in the reported HIV-1 RNA copy numbers for the two assays was 0.2241 log.
The results from the NucliSens assay and both AMPLICOR assays were found to be highly correlated when the mean observed values of the NucliSens assay and either AMPLICOR assay were plotted against the difference of the two observed values (data not shown). An estimate of the Pearson product moment correlation, on a log copy basis, was 0.96 (P < 0.0001). A 95% confidence interval for the correlation is 0.94 to 0.98. The overall mean difference between all assays, i.e., standard or ultrasensitive AMPLICOR versus NucliSens on every reported sample was 0.0097 log10 copies/ml, with an SD of 0.26 log10 copies/ml, indicating that the variation in results from the two assays on a specific specimen is not that different from the individual interassay variation. However, the NucliSens assay reported more specimens with a copy number than did the standard AMPLICOR assay, indicating greater sensitivity (Table 5), but the results were equivalent when tested with the ultrasensitive AMPLICOR assay.
Detection and quantification of group M clades A to H HIV-1 RNA.
The NucliSens assay was compared to the standard AMPLICOR assay for the detection and quantitation of HIV-1 RNA derived from a panel of samples containing HIV-1 virus (approximately 5 × 104 virus particles/ml) from group M clades A, B, C, D, E, F, G, and H. The NucliSens assay was able to detect and quantitate HIV-1 RNA derived from all eight clades. Overall, the HIV-1 RNA copy numbers obtained with the NucliSens assay were highly comparable with the expected values determined by BBI, with an overall mean difference of 0.094 log10 copies/ml. The HIV-1 RNA values for the clade A sample (3.38 log10 copies/ml) and clade C sample (3.97 log10 copies/ml) were significantly less (>0.5 log10 copies/ml difference) than the expected values (approximately 4.69 log10 copies/ml). The AMPLICOR assay detected seven of the eight panel members, with a result less than the LOD (<400 copies/ml) for the panel member containing HIV-1 clade A virus. The HIV-1 RNA values for the clade E sample (4.09 log10 copies/ml), clade G sample (3.92 log10 copies/ml), and clade H sample (3.18 log10 copies/ml) were significantly less than the expected values (approximately 4.48 to 4.69 log10 copies/ml). The AMPLICOR assay had a higher overall observed difference (0.655 log) from the expected HIV-1 RNA values determined by the manufacturer than did the NucliSens assay (0.0936 log). Both assays gave undetectable results for the panel member containing plasma diluent only.
DISCUSSION
The NucliSens HIV-1 QT assay is a modified version of the NASBA HIV-1 RNA QT assay. The modifications to the original assay included lower internal calibrator values (for the NucliSens assay, 104.8, 104.2, and 103.6 versus, for the NASBA assay, 106.0, 105.0, and 104.0), enhanced stability of the enzyme formulation through lyophilization, a new software algorithm for the calculation of results, and second excitation during ECL detection for over range signals. A secondary ECL signal is used for calculations when the primary signal for the highest input calibrator (104.8) and/or the patient sample exceeds the instrument's upper LOD. Since the values of the primary and secondary signals are linear over a broad range, redetection of the amplified sample is not required. The goals of the modifications were to improve accuracy across the dynamic range, including the high input range of 107, 1 log increased sensitivity (from 103 to 102) and better positive-negative discrimination. The purpose of this multicenter study was to establish the performance characteristics of the NucliSens assay and evaluate the assay in comparison to another approved FDA assay, the AMPLICOR HIV-1 MONITOR, version 1.0, test.
The linear range of the NucliSens assay was determined to be 51 to 5,390,000 HIV-1 RNA copies/ml, a dynamic range that is approximately 10-fold greater than both the standard AMPLICOR, version 1.0, assay and the QUANTIPLEX bDNA v3.0 assay, and approximately 100-fold greater than the ultrasensitive AMPLICOR, version 1.0, assay. Importantly, the data clearly demonstrated that the assay has excellent precision and reproducibility over the entire dynamic range. The ability to quantitate HIV-1 RNA over a 5-log span using a single test format is a significant advantage of the NucliSens assay, particularly when testing a variety of patient populations with a wide range of viral loads. This is important when test runs include samples from newly diagnosed untreated patients, patients with histories of multiple drug failures and pediatric patients, who generally have a higher viral load than HIV-1 infected adults (21). This feature also decreases the number of times incorrect tests are ordered (standard versus ultrasensitive), eliminates the need for reflex repeat testing, improves turnaround time to results, and is associated with significant savings in both technical time and reagent costs. This is particularly important when the number of reimbursable viral load tests per patient per year are limited by either insurance carriers or state or federal health agencies. Finally, the wide dynamic range enables clinicians to accurately and precisely assess the scope of the viral load decline and evaluate virological response to therapy in accordance with published therapeutic guidelines by the International AIDS Society (5) and the U.S. Department of Health and Human Services (23).
The LOD of the assay with positivity rates of 50 and 95% were determined to be 41 and 176 HIV-1 RNA copies/ml, respectively. The lowest concentration that was reported by the assay was 25 HIV-1 RNA copies/ml, which is the lower LOD as defined by the instrument software. With both clinical specimens and panels containing a broad concentration range of HIV-1 RNA, the NucliSens assay demonstrated superior clinical sensitivity over a larger dynamic range for quantitation of HIV-1 RNA in direct comparison with the standard AMPLICOR assay. Interestingly, this study clearly demonstrated that both the NucliSens and the ultrasensitive AMPLICOR assays have equivalent clinical sensitivity despite their different claims for lower LOD (ultrasensitive AMPLICOR assay, 50 HIV-1 RNA copies/ml; NucliSens assay, 176 HIV-1 RNA copies/ml). Note that the detection rate for the ultrasensitive AMPLICOR procedure at 50 copies/ml has never been reported. Studies from one of our laboratories (K. A. Stellrecht et al., unpublished data) demonstrated a detection rate of approximately 50% for samples with fewer than 80 copies/ml with the ultrasensitive AMPLICOR procedure.
Although the present NucliSens assay was validated for a 1-ml specimen input volume, both the nucleic acid extraction format and detection software are designed to easily permit the use of a wide range of plasma input volumes (10 μl to 2 ml). A 0.9-ml lysis buffer tube is used for small volume sample extractions (10 μl to 200 μl), and the standard 9.0-ml lysis buffer tube is used for volumes ranging from >200 μl up to 2 ml. Independent of the specimen input volume, the final extraction volume remains constant at 50 μl. The detection software allows the user to input the plasma volume and all results are then extrapolated to a value reported as HIV-1 RNA copies/ml. The ability to use variable volumes with highly correlative results was demonstrated in a study by Witt et al. (26). Results obtained with both stock HIV-1 RNA dilution panels and clinical specimens indicated that the reported HIV-1 RNA copy number were directly proportional to the specimen input volumes across the entire dynamic range of the assay. The absolute number of reported detectable results increased proportionally with increased specimen input volume, the direct result of concentrating more RNA target per amplification reaction. Studies from one of our laboratories (C. C. Ginocchio et al., unpublished data) have demonstrated that increasing the plasma input volume from 1 to 2 ml increased the sensitivity of the assay to 50 HIV-1 RNA copies/ml, with a detection rate of 90%. The ability to detect HIV-1 RNA levels down to 50 copies/ml is highly advantageous and is in accordance with current viral load testing guidelines published by the International AIDS Society (5) and the U.S. Department of Health and Human Services (23). Conversely, the ability to decrease the input volume permits flexibility when testing clinical or research samples containing less plasma than the standard required volume or very high HIV-1 RNA titers, as often seen in specimens from HIV-1-seropositive pediatric patients (7, 21). The NucliSens assay is currently the only commercial assay that offers this important feature. The AMPLICOR assays requires a fixed volume (200 μl for the standard assay and 500 μl for the ultrasensitive protocol), and the QUANTIPLEX bDNA assay requires 1 ml.
The results of the NucliSens and the AMPLICOR assays were highly correlative (P = <0.0001). The variation between results obtained by both assays was within the range of single-assay variation (<0.3 log10 copies/ml) and well below what would be considered a significant change in viral load (±0.5 log10 copies/ml) (3). This close correlation is important for several reasons including the ability to switch from one assay to another without the need to refigure the baseline for all patients and for the comparison of viral load data (obtained using different assays) from therapeutic clinical trial studies designed to assess response to antiretroviral therapy. The NucliSens assay was able to detect HIV-1 RNA derived from a commercially prepared panel representative of group M clades A through H and accurately quantitated clades B, D, E, F, and G. The standard AMPLICOR assay did not detect the HIV-1 RNA from the clade A sample and appeared to underquantitate the HIV-1 RNA levels in samples from clades E, G, and H, noting that this was found in one observation. The inability to either detect or appropriately quantitate HIV-1 RNA from certain non-B clade samples has been described previously for both the NucliSens assay and the AMPLICOR 1.0 assay (1, 6, 12, 16, 17, 22). In comparison to the NucliSens and AMPLICOR assays, the QUANTIPLEX assay has shown the most consistent results in detecting non-B subtypes (17).
Currently, HIV-1 viral load assays are not FDA approved for the diagnosis of HIV disease. Despite this fact, in clinical practice the assays have been used “off label” to identify persons with acute HIV-1 disease prior to seroconversion or for the resolution of HIV-1 status in individuals with indeterminate Western blot analyses. Therefore, it is extremely important for an assay to be highly specific and limit the number of false-positive samples due to either high background signal or cross-contamination of negative samples with HIV-1 RNA from positive samples. Low-level false-positive results have been reported for persons who were not HIV-1 infected when tested with the QUANTIPLEX 2.0 and 3.0 assays (4, 9, 19; F. M. Hecht, B. D. Rawal, J. O. Kahn, M. Swanson, M. A. Chesney, J. A. Levy, and M. P. Busch, abstr. 178, 6th Conf. Retrovir. Opportunistic Infect. 1999) and the ultrasensitive AMPLICOR assay (4, 19). A multicenter study which evaluated the performance characteristics of the QUANTIPLEX 3.0 found 6.2% of the HIV-1 negative replicate samples had ≥50 HIV-1 RNA copies/ml reported (9). Roland et al. (M. E. Roland, J. N. Martin, D. Chernoff, B. McGovern, J. Bamberger, M. M. Katz, T. J. Coates, and J. O. Kahn, abstr. 179, 6th Conf. Retrovir. Opportunistic Infect. 1999) identified a false positive rate of 4.4% for the QUANTIPLEX 2.0 assay and 9.5% for the 3.0 version of the assay, when testing patients recently exposed to HIV-1, who participated in the San Francisco postexposure prevention program. In addition, two studies identified false positive results obtained from infants who were not HIV-1 infected (7, 8). In our studies, the NucliSens assay was very specific as no false-positive results were obtained with any of the 502 samples derived from HIV-1-seronegative persons. The high specificity of the NucliSens assay for true positive HIV-1 RNA containing specimens may be attributed to both the new software algorithms that clearly delineate between positive and negative ECL cut off values and the solid-phase nucleic acid extraction method which greatly reduces the potential for sample cross-contamination. However, despite the high specificity of the assay, clinicians must be aware that the assay is not FDA approved for this application and must be cautioned about the interpretation of viral load results, particularly low HIV-1 RNA levels, for the diagnosis of HIV-1 infection.
In summary, the NucliSens HIV-1 QT assay has excellent reproducibility and good sensitivity. The assay gives comparable quantitative results when compared to both the standard and ultrasensitive AMPLICOR assays. In addition, the clinical sensitivity of the assay is comparable to that of the ultrasensitive AMPLICOR assay. The broad linear range and flexible specimen input volume permit a single test format for a wide variety of patient populations with variable viral loads. The assay detected and quantitated representative samples from group M clades A to H. Finally, the assay is highly specific, with an isolation format and excellent positive-negative signal discrimination that greatly reduce the chances of sample cross-contamination and false-positive results.
Acknowledgments
bioMérieux, the manufacturer of the NucliSens HIV-1 QT assay, funded this study. C. C. Ginocchio was partly funded for this study by the Jane and Dayton Brown and Dayton Brown, Jr., Molecular Diagnostics Laboratory.
We are indebted to the following individuals for their technical assistance and expertise: at the North Shore-Long Island Jewish Health System Laboratories, Fan Zhang, and at the Albany Medical Center, Ryan Bennet. We thank James Bremer, Rush Presbyterian-St Luke's Medical Center, for kindly supplying the HIV-1 RNA stock. For assistance with statistical analysis we thank Karen McDonald and Andy Stead (bioMérieux Inc., Durham, North Carolina) and Jos Weusten (bioMérieux, Boxtel, The Netherlands). We thank Frank Simons for critical review of the manuscript.
REFERENCES
- 1.Alaeus, A., K. Lidman, A. Sonnerborg, and J. Albert. 1997. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS 11:859-865. [DOI] [PubMed] [Google Scholar]
- 2.Boom, R., C. J. Sol, M. M. Salisman, C. J. Lansen, P. M. E. Bertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brambilla, D., P. S. Reichelderfer, J. W. Bremer, D. E. Shapiro, R. C. Hershow, D. A. Katzenstein, S. M. Hammer, B. Jackson, A. C. Collier, R. S. Sperling, M. G. Fowler, R. W. Coombs, et al. 1999. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. AIDS 13:2269-2279. [DOI] [PubMed] [Google Scholar]
- 4.Brown, A. E., B. Jackson, S. A. Fuller, J. Sheffield, M. A. Cannon, and J. R. Lane. 1997. Viral RNA in the resolution of human immunodeficiency virus type 1 diagnostic serology. Transfusion 37:926-929. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, C. C., D. A. Cooper, M. A. Fischl, J. M. Gatell, B. G. Gazzard, S. M. Hammer, M. S. Hirsch, D. M. Jacobsen, D. A. Katzenstein, J. S. Montaner, D. D. Richman, M. S. Saag, M. Schechter, R. T. Schooley, M. A. Thompson, S. Vella, P. G. Yeni, and P. A. Volberding. 2000. Antiretroviral therapy in adults. Updated recommendations of the International AIDS Society-USA Panel. JAMA 283:381-390. [DOI] [PubMed] [Google Scholar]
- 6.Chew, C. B., B. L. Herring, F. Zheng, C. Browne, N. K. Saksena, A. L. Cunningham, and D. E. Dwyer. 1999. Comparison of three commercial assays for the quantification of HIV-1 RNA in plasma from individuals infected with different HIV-1 subtypes. J. Clin. Virol. 14:87-94. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, C. K., T. T. Charbonneau, K. Song, D. Patterson, T. Sullivan, T. Cummins, and B. Poiesz. 1999. Comparison of human immunodeficiency virus 1 DNA polymerase chain reaction and qualitative and quantitative RNA polymerase chain reaction in human immunodeficiency virus 1-exposed infants. Pediatr. Infect. Dis. 18:30-35. [DOI] [PubMed] [Google Scholar]
- 8.Delamare, C., M. Burgard, M. J. Mayaux, S. Blanche, A. Doussin, S. Ivanoff, M. L. Chaix, C. Khan, C. Rouzioux, et al. 1997. HIV-1 RNA detection in plasma for the diagnosis of infection in neonates. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:121-125. [DOI] [PubMed] [Google Scholar]
- 9.Erice, A., D. Brambilla, J. Bremer, J. Brooks Jackson, R. Kokka, B. Yen-Lieberman, and R. W. Coombs. 2000. Performance characteristics of the QUANTIPLEX HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginocchio, C. C. 2001. HIV-1 viral load testing. Lab. Med. 32:142-152. [Google Scholar]
- 11.Ho, D. D., A. U. Neuman, A. S. Perelson, W. Chen, J. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 12.Holguin, A., C. de Mendoza, and V. Soriano. 1999. Comparison of three different commercial methods for measuring plasma viremia in patients infected with non-B HIV-1 subtypes. Eur. J. Clin. Microbiol. Infect. Dis. 18:256-259. [DOI] [PubMed] [Google Scholar]
- 13.Kievits, T., B. van Gemen, D. van Strijp, R. Schukkink, M. Dircks, H. Adriaanse, L. Malek, R. Sooknanan, and P. Lens. 1991. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods 35:273-286. [DOI] [PubMed] [Google Scholar]
- 14.Marschner, I. C., A. C. Collier, R. W. Coombs, R. T. D'Aquila, V. DeGruttola, M. A. Fischl, S. M. Hammer, M. D. Hughes, V. A. Johnson, D. A. Katzenstein, D. D. Richman, L. M. Smeaton, S. A. Spector, and M. S. Saag. 1998. Use of changes in plasma levels of human immunodeficiency virus type 1 RNA to assess the clinical benefit of antiretroviral therapy. J. Infect. Dis. 177:40-47. [DOI] [PubMed] [Google Scholar]
- 15.Mellors, J. W., A. M. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 16.Pantaleo, G., and J. Schupbach. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1 Swiss HIV Cohort Study. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 17.Parekh, B., S. Phillips, T. C. Granade, J. Baggs, D. J. Hu, and R. Respess. 1999. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res. Hum. Retrovir. 20:133-142. [DOI] [PubMed] [Google Scholar]
- 18.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 19.Rich, J. D., N. A. Merriman, E. Mylonakis, T. C. Greenough, T. Flanifan, B. J. Mady, and C. C. J. Carpenter. 1999. Misdiagnosis of HIV infection by HIV-1 plasma viral load testing: a case series. Ann. Intern. Med. 130:37-39. [DOI] [PubMed] [Google Scholar]
- 20.Saag, M. S., M. Holodniy, D. R. Kuritzkes, W. A. O'Brien, R. Coombs, M. E. Poscher, D. M. Jacobsen, G. M. Shaw, D. D. Richman, and P. A. Volberding. 1996. HIV viral load markers in clinical practice. Nat. Med. 348:625-629. [DOI] [PubMed] [Google Scholar]
- 21.Shearer, W. T., T. C. Quinn, P. LaRussa, J. F. Lew, L. Mofenson, S. Almy, K. Rich, E. Handelsman, C. Diaz, M. Pagano, V. Smeriglio, L. A. Kalish, and the Women and Infants Transmission Study Group. 1997. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N. Engl. J. Med. 336:1337-1342. [DOI] [PubMed] [Google Scholar]
- 22.Triques, K., J. Coste, J. L. Perret, C. Segarra, E. Mpoudi, J. Reynes, E. Delaporte, A. Butcher, K. Dreyer, S. Herman, J. Spadoro, and M. Peeters. 1999. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J. Clin. Microbiol. 37:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.U.S. Department of Health and Human Services Panel on Clinical Practice for Treatment of HIV Infection. 2002. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. U.S. Department of Health and Human Services, Washington, D.C.
- 24.van Gemen, B., T. Kievits, R. E. Schukkin, D. van Strijp, L. T. Malek, R. Sooknanan, H. G. Huisman, and P. Lens. 1993. Quantification of HIV-1 RNA in plasma using NASBA during primary infection. J. Virol. Methods 43:177-188. [DOI] [PubMed] [Google Scholar]
- 25.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emmini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 26.Witt, D. J., M. Kemper, A. Stead, C. C. Ginocchio, and A. M. Caliendo. 2000. Relationship of incremental specimen volumes and enhanced detection of human immunodeficiency virus type 1 RNA with nucleic acid amplification technology. J. Clin. Microbiol. 38:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]