Abstract
The newly developed Rapid Lumi Eiken/IS60 (RL/IS60) system automatically determines MICs by detecting chemiluminescence produced in the reaction of a chemiluminescent probe and oxygen metabolites from living microorganisms. The present study evaluated this system for accuracy in antimicrobial susceptibility testing. Chemiluminescence intensities after 4 h of cultivation of clinically important strains were plotted against various concentrations of antimicrobial agents, which resulted in curves reflecting the levels of susceptibility. Sixty-percent inhibitory concentrations based on the susceptibility curves agreed with MICs determined by the reference microdilution method. When the MICs of antimicrobial agents for four quality control (QC) strains (Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, and Pseudomonas aeruginosa) were determined by the RL/IS60 system, most (91.1%) of them were within the QC limits proposed by the National Committee for Clinical Laboratory Standards. The system was further assessed for a total of 162 clinical isolates, including E. coli, Citrobacter freundii, Enterobacter cloacae, Klebsiella pneumoniae, Serratia marcescens, Proteus mirabilis, Morganella morganii, P. aeruginosa, Haemophilus influenzae, S. aureus, coagulase-negative staphylococci, Enterococcus faecalis, Enterococcus faecium, and Streptococcus pneumoniae. Overall, there was 90.6% agreement between the RL/IS60 system and the reference microdilution method. Our results suggest that the RL/IS60 system provides rapid and reliable MICs of a variety of antimicrobial agents for clinical isolates as well as QC strains.
The significant increase in the number of drug-resistant microorganisms emphasizes the great need for rapid and accurate methods for determining resistance to antimicrobial agents. Current standard methods, the broth microdilution (12) and disk diffusion tests (13), approved by the National Committee for Clinical Laboratory Standards (NCCLS), require more than 18 h before the final results are known. New methods are desired that could detect the biological activity in a short incubation time and provide MICs equivalent to those determined by the standard tests.
The chemiluminescence assay, which detects photon emissions released from living organisms, is a sensitive method for monitoring viability. Chemiluminescence is produced in the reaction of oxygen metabolites generated during glycolysis with chemiluminescent probes such as luminol and lucigenin (2, 16). We previously showed the application of a luminol-mediated chemiluminescent assay to antimicrobial susceptibility testing for Escherichia coli (17). Chemiluminescence intensity increased during the exponential phase of growth. The reaction was enhanced by a catalyst, menadione. Antimicrobial agents (erythromycin, tetracycline, and oxytetracycline) inhibited chemiluminescence release from E. coli. The effect of antimicrobial agents was detectable within 1.5 h. The results suggested the usefulness of this assay for rapid susceptibility testing.
Recently, we developed a new antimicrobial susceptibility testing system, the Rapid Lumi Eiken/IS60 (RL/IS60). This system consists of a reaction kit (Rapid Lumi Eiken) and robotic equipment (IS60). The Rapid Lumi Eiken uses the testing method of a lucigenin-dependent and menadione-catalyzed chemiluminescence assay. The IS60 automatically regulates sequential events, including incubation of culture plates, addition of lucigenin and NaHCO3-KOH buffer, counting of chemiluminescence, and calculation of MICs. We report here the evaluation of MICs determined by the RL/IS60 assay in comparison with those determined by the standard microdilution method. Quality control (QC) strains and clinical isolates were tested for a variety of antimicrobial agents.
MATERIALS AND METHODS
Bacterial strains.
QC strains tested in this study were E. coli ATCC 25922, Staphylococcus aureus ATCC 29213, Pseudomonas aeruginosa ATCC 27853, and Enterococcus faecalis ATCC 29212. S. aureus ATCC 49619 and EKN4778 as methicillin-resistant S. aureus (MRSA), Streptococcus pneumoniae ATCC 49619 as penicillin-intermediate S. pneumoniae (PISP), E. faecalis ATCC51299 as vancomycin-resistant enterococci (VRE) and as high-level aminoglycoside (gentamicin)-resistant E. faecalis (gentamicin HLAR), and E. coli EKN4119 as extended-spectrum β-lactamase (ESBL)-producing E. coli were used as antimicrobial-resistant strains. S. aureus ATCC 29213, E. faecalis ATCC 29212, S. pneumoniae EKN4713, and E. coli ATCC 25922 were used as corresponding susceptible strains. A total of 162 clinical isolates were obtained from the blood, cerebrospinal fluid, stool, and ascites of patients in the diagnostic laboratory of Tohoku University Hospital. The isolates included 18 E. coli isolates, 3 Citrobacter freundii isolates, 3 Enterobacter cloacae isolates, 19 Klebsiella pneumoniae isolates, 4 Serratia marcescens isolates, 3 Proteus mirabilis isolates, 3 Morganella morganii isolates, 17 P. aeruginosa isolates, 15 Haemophilus influenzae isolates, 20 S. aureus isolates, 10 coagulase-negative staphylococcus isolates, 10 E. faecalis isolates, 9 Enterococcus faecium isolates, and 28 S. pneumoniae isolates. They were grouped into six categories: Enterobacteriaceae, P. aeruginosa, H. influenzae, staphylococci, enterococci, and streptococci. Bacterial strains were stored at −80°C in 10% skim milk. At the time of use, they were removed from storage, streaked on appropriate agar plates, and incubated for 18 h at 35°C.
Antimicrobial agents.
The following 22 antimicrobial agents were tested: ampicillin, penicillin G, piperacillin, oxacillin, cefazolin, cefotiam, ceftazidime, cefpodoxime, cefditoren, cefotaxime, flomoxef, aztreonam, imipenem, amikacin, arbekacin, minocycline, clarithromycin, clindamycin, vancomycin, teicoplanin, levofloxacin, and fosfomycin. Antimicrobial agents were dehydrated in two types of microdilution plates, the Rapid Lumi Eiken plate (Eiken Chemical Co., Tokyo, Japan) for the RL/IS60 system and the Dry Plate Eiken for the broth microdilution method. All antimicrobial agents were tested for four QC strains. Twelve antimicrobial agents were selected for each group of clinical isolates. Oxacillin was tested for screening MRSA. Vancomycin, gentamicin, penicillin G, and cefotaxime and cefotaxime-clavulanic acid were tested for VRE, HLAR, PISP, and ESBL-producing E. coli, respectively.
Antimicrobial susceptibility testing by the Rapid Lumi system. (i) Preparation of inoculum and inoculation.
The Rapid Lumi Eiken system contains a Rapid Lumi Eiken plate, reagent A (broth for Enterobacteriaceae, staphylococci, enterococci, streptococci, and H. influenzae), reagent B (broth for P. aeruginosa), Lumi Supply Eiken (supplement for H. influenzae), lucigenin solution, and NaHCO3-KOH buffer solution (pH 11.0). Several well-separated colonies of Enterobacteriaceae, staphylococci, enterococci, or streptococci were picked from a 5% sheep blood agar plate (Eiken Chemical Co.) and suspended in saline. The bacterial suspension was adjusted to a turbidity equal to that of a 1.0 McFarland standard, and then 0.1 ml of each suspension was diluted with 7.5 ml of reagent A to prepare an inoculum (approximately 2 × 105 CFU/ml). An inoculum of P. aeruginosa was similarly prepared with reagent B. An inoculum of H. influenzae was prepared by suspending colonies from a chocolate-agar plate (Eiken Chemical Co.) in saline and diluting the suspension with the mixture of reagent A and Lumi Supply Eiken just before use. Each well of the Rapid Lumi Eiken plate was filled with a 50-μl inoculum.
(ii) Incubation, reaction, detection, and analysis.
The inoculated Rapid Lumi Eiken plates were placed in an IS60 device (Nagase & Co., Tokyo, Japan) and maintained at 35°C. After aerobic incubation for 4 h, lucigenin solution and NaHCO3-KOH buffer solution were automatically added to each well and total photon emission (chemiluminescence release) was counted. The MIC was read as the concentration of antimicrobial agent at which there was 60% inhibition of chemiluminescence intensity (IC60). The IC90 was used for the combination of E. faecalis and gentamicin.
Reference broth microdilution method.
Reference MICs were determined by the microdilution method currently recommended by the NCCLS (14) using the Dry Plate Eiken. Briefly, each well of a plate was inoculated with a 100-μl inoculum (approximately 5 × 105 CFU/ml), and the plate was incubated for 18 to 24 h at 35°C. The MIC was read as the lowest concentration of antimicrobial agent at which there was no visible growth.
RESULTS
Susceptibility curves.
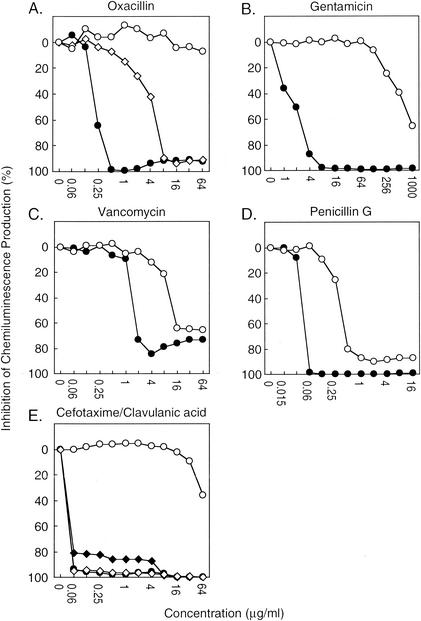
Bacterial strains with various antimicrobial susceptibility were subjected to the chemiluminescent assay using the RL/IS60 system. The plots of relative chemiluminescence intensities at various concentrations of antimicrobial agents produced smooth susceptibility curves of QC and clinical strains (Fig. 1). The susceptibility curves of oxacillin against S. aureus QC strain ATCC 29213, methicillin-resistant S. aureus ATCC 43300, and methicillin-resistant S. aureus EKN4778 were separated from each other, indicating different levels of susceptibility (Fig. 1A). Similarly well-separated curves of susceptible and resistant strains were obtained for gentamicin against E. faecalis (Fig. 1B), vancomycin against E. faecalis (Fig. 1C) and penicillin G against S. pneumoniae (Fig. 1D). Figure 1E shows the effect of clavulanic acid on cefotaxime resistance of ESBL-producing E. coli. When IC60 values of chemiluminescence intensity (IC90 for gentamicin against E. faecalis) were used as MICs, the values were equivalent to the reference MICs determined by the microdilution method (Table 1). As a control experiment, we examined the interactions between antimicrobial agents and the chemiluminescence signal system in reactions containing 5 mM hydrogen peroxide instead of microorganisms. Addition of each antimicrobial agent to the reaction did not show significant effect on the release of chemiluminescence (data not shown).
FIG. 1.
Differential susceptibilities of resistant strains and corresponding susceptible strains to a range of antimicrobial agent doses. The effect of 4 h of antimicrobial treatment on the abilities of various strains to produce chemiluminescence was evaluated by the following equation: percent chemiluminescence production = (chemiluminescent intensity generated by antimicrobial-treated strain/chemiluminescent intensity generated by untreated strain) × 100. The susceptibilities of antimicrobial agents for strains were tested as follows: (A) oxacillin for S. aureus ATCC 29213 (•), S. aureus ATCC 43300 (MRSA) (⋄), and S. aureus EKN4778 (MRSA) (○); (B) gentamicin for E. faecalis ATCC 29212 (♦) and E. faecalis ATCC 51299 (HLAR) (⋄); (C) vancomycin for E. faecalis ATCC 29212 (♦) and E. faecalis ATCC 51299 (VRE) (⋄); (D) penicillin G for S. pneumoniae EKN4713 (♦) and S. pneumoniae ATCC 49619 (PISP) (⋄); (E) cefotaxime for E. coli ATCC 25922 (•) and E. coli EKN4119 (ESBLs) (○); and a combination of cefotaxime and clavulanic acid for E. coli ATCC 25922 (♦) and E. coli EKN4119 (ESBLs) (⋄). Representative data from three independent experiments are shown.
TABLE 1.
MICs determined by the RL/IS60 system and microdilution method
| Antimicrobial agent(s) | Strain | MIC (μg/ml) determined by:
|
|
|---|---|---|---|
| RL/IS60a | Microdilution | ||
| Oxacillin | S. aureus ATCC 29213 | 0.25 | 0.25 |
| S. aureus ATCC 43300 (MRSA) | 4 | 8 | |
| S. aureus EKN4778 (MRSA) | >64 | >64 | |
| Gentamicin | E. faecalis ATCC 29212 | 4b | 8 |
| E. faecalis ATCC 51299 (HLAR) | 1,000b | 1,000 | |
| Vancomycin | E. faecalis ATCC 29212 | 2 | 2 |
| E. faecalis ATCC 51299 (VRE) | 16 | 32 | |
| Penicillin G | S. pneumoniae EKN4716 | 0.06 | 0.06 |
| S. pneumoniae ATCC 49619 (PISP) | 0.5 | 0.5 | |
| Cefotaxime | E. coli ATCC 26922 | <0.06 | <0.06 |
| E. coli EKN4119 (ESBL) | >64 | >64 | |
| Cefotaxime- clavulanic acidc | E. coli ATCC 25922 | <0.06/4 | <0.06/4 |
| E. coli EKN4119 (ESBL) | <0.06/4 | <0.06/4 | |
Determined as drug concentration corresponding to IC60 in antimicrobial susceptibility curves (Fig. 1A and C to E).
Determined as drug concentration corresponding to IC90 in antimicrobial susceptibility curves (Fig. 1B).
Results expressed as MIC of cefotaxime plus clavulanic acid/MIC of clavulanic acid.
MICs determined by the RL/IS60 assay and the QC limits by the NCCLS.
MICs of 22 antimicrobial agents for QC strains were determined by the RL/IS60 system. Most (91.1%) of them agreed with the QC limits proposed by the NCCLS (Table 2). Only four MICs were out of the range, one MIC with 1 dilution lower than the range (aztreonam for P. aeruginosa) and three MICs with 1 dilution higher (clindamycin and levofloxacin for S. aureus and levofloxacin for E. faecalis).
TABLE 2.
MICs determined by the RL/IS60 system and the acceptable QC limitsd
| Antimicrobial agent | MICa (QC limitb) for:
|
|||
|---|---|---|---|---|
| E. coli ATCC 25922 | E. faecalis ATCC 29212 | P. aeruginosa ATCC 27853 | S. aureus ATCC 29213 | |
| Ampicillin | 2 (2-8) | 0.5 (0.5-2) | >8 | 0.5 (0.25-1) |
| Penicillin G | >8 | 4 (1-4) | >8 | 1 (0.25-1) |
| Piperacillin | 2 (1-4) | 2 (1-4) | 1 (1-4) | 1 (1-4) |
| Oxacillin | >16 | 16 (8-32) | >16 | 0.5 (0.12-0.5) |
| Cefazolin | 1 (1-4) | 16 | >16 | 0.25 (0.25-1) |
| Cefotiam | <0.12 | >8 | >8 | 0.5 |
| Ceftazidime | 0.06 (0.06-0.5) | >16 | 1 (1-4) | 8 (4-16) |
| Cefpodoxime | 0.5 (0.25-1) | >16 | >16 | 2 (1-8) |
| Cefditoren | 0.25 | >16 | 16 | 0.25 |
| Cefotaxime | 0.12 (0.03-0.12) | >16 | 8 (8-32) | 2 (1-4) |
| Flomoxef | 0.12 | >16 | >16 | 0.25 |
| Aztreonam | 0.06 (0.06-0.25) | >16 | 1 (2-8) | >16 |
| Imipenem | 0.25 (0.06-0.25) | 2 (0.5-2) | 1 (1-4) | 0.03 (0.015-0.06) |
| Amikacin | 2 (0.5-4) | 64 (64-256) | 2 (1-4) | 4 (1-4) |
| Arbekacin | 1 | >16 | 1 | 1 |
| Minocycline | 0.5 (0.25-1) | 1 (1-4) | 8 | 0.25 (0.06-0.5) |
| Clarithromycin | >16 | 4 | >16 | 0.5 (0.12-0.5) |
| Clindamycin | >4 | 4 (4-16) | >4 | 0.5 (0.06-0.25) |
| Vancomycin | >16 | 2 (1-4) | >16 | 2 (0.25-2) |
| Teicoplanin | >16 | 0.06 (0.06-0.25) | >16 | 1 (0.25-1) |
| Levofloxacin | 0.06 (0.008-0.06) | 4 (0.25-2) | 1 (0.5-4) | 1 (0.06-0.5) |
| Fosfomycinc | 2 | 16 | >16 | 16 |
MICs (in micrograms per milliliter) were determined by chemiluminescent assay with the RL/IS60 system.
Acceptable QC limits of MIC proposed by NCCLS.
The ranges of QC limits against fosfomycin were omitted because they were determined not by broth dilution method but by the agar dilution method.
The data presented are representative of repeated experiments.
MICs for clinical isolates.
The RL/IS60 assay system, its accuracy against QC strains verified, was further assessed for a total of 162 clinical isolates. The isolates were grouped in six species and families, and 12 clinically important antimicrobial agents were selected for each group. Representative results from several repeated tests are shown in Tables 3 to 8. In the group of P. aeruginosa, percentages of isolates for which the MICs were within 1 log2 dilution difference between the RL/IS60 assay and the microdilution assay ranged from 82.4 to 100.0% (Table 3). The overall agreement, the average of percentages for 12 antimicrobial agents, was 95.0%. Results from the other combinations of antimicrobial agents and bacterial categories also showed high levels of agreement. Overall agreement values were as follows: 92.3% for Enterobacteriaceae (Table 4), 92.1% for enterococci (Table 5), 91.4% for staphylococci (Table 6), 87.2% for H. influenzae (Table 7), and 84.5% for S. pneumoniae (Table 8). Low agreement values were found for fosfomycin in Enterobacteriaceae (66.0% [Table 4]), minocycline in enterococci (63.2% [Table 5]), and ceftazidime in H. influenzae (46.7% [Table 7]). Consequently, the total overall agreement for 162 clinical isolates was 90.6%.
TABLE 3.
Distribution of differences in MICs of 12 antimicrobial agents for 17 strains of P. aeruginosa: RL/IS60 method versus microdilution methodc
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree-mentb | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | +1 | +2 | >+2 | ||
| Cefotiam | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 100.0 |
| Piperacillin | 1 | 1 | 10 | 3 | 1 | 1 | 0 | 82.4 |
| Cefazolin | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 100.0 |
| Ceftazidime | 1 | 0 | 7 | 6 | 1 | 0 | 0 | 93.3 |
| Cefpodoxime | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 100.0 |
| Flomoxef | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 100.0 |
| Aztreonam | 0 | 0 | 6 | 3 | 7 | 1 | 0 | 94.1 |
| Imipenem | 0 | 0 | 6 | 6 | 3 | 0 | 2 | 88.2 |
| Amikacin | 1 | 0 | 2 | 9 | 5 | 0 | 0 | 94.1 |
| Minocycline | 0 | 0 | 5 | 11 | 1 | 0 | 0 | 100.0 |
| Levofloxacin | 1 | 1 | 6 | 6 | 3 | 0 | 0 | 88.2 |
| Fosfomycin | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 100.0 |
| Overall | 4 | 2 | 42 | 129 | 21 | 2 | 2 | 95.0 |
Difference in MICs determined by the RL/IS60 system and the microdilution method. Differences are given as log2 dilutions (e.g., −1 indicates a −1-log2 dilution difference).
Percentage of isolates within 1 log2 dilution.
The data presented are representative of repeated experiments.
TABLE 8.
Distribution of differences in MICs of 12 antimicrobial agents for 28 strains of S. pneumoniae: RL/IS60 method versus microdilution method
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree- ment | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | −1 | +2 | >+2 | ||
| Cefditoren | 0 | 0 | 2 | 16 | 7 | 2 | 1 | 89.3 |
| Ampicillin | 1 | 1 | 9 | 17 | 0 | 0 | 0 | 92.9 |
| Penicillin G | 1 | 0 | 3 | 18 | 6 | 0 | 0 | 96.4 |
| Ceftazidime | 1 | 0 | 9 | 15 | 1 | 0 | 2 | 89.3 |
| Flomoxef | 0 | 1 | 11 | 14 | 1 | 0 | 0 | 96.3 |
| Cefpodoxime | 1 | 0 | 4 | 14 | 5 | 1 | 3 | 82.1 |
| Cefotaxime | 0 | 1 | 3 | 17 | 3 | 1 | 3 | 82.1 |
| Imipenem | 0 | 2 | 9 | 14 | 1 | 2 | 0 | 85.7 |
| Minocycline | 0 | 5 | 13 | 7 | 3 | 0 | 0 | 82.1 |
| Clarithromycin | 6 | 2 | 7 | 10 | 3 | 0 | 0 | 71.4 |
| Levofloxacin | 0 | 0 | 0 | 2 | 15 | 7 | 4 | 60.7 |
| Clindamycin | 0 | 0 | 7 | 16 | 1 | 0 | 4 | 85.7 |
| Overall | 10 | 12 | 77 | 160 | 45 | 13 | 17 | 84.5 |
Difference in MICs determined by the RL/IS60 system and the microdilution method. See footnotes to Table 3 for further details.
TABLE 4.
Distribution of differences in MICs of 12 antimicrobial agents for 53 strains of Enterobacteriaceae: RL/IS60 method versus microdilution method
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree- ment | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | +1 | +2 | >+2 | ||
| Cefotiam | 0 | 0 | 8 | 36 | 8 | 0 | 1 | 98.1 |
| Piperacillin | 0 | 3 | 11 | 31 | 8 | 0 | 0 | 94.3 |
| Cefazolin | 0 | 1 | 22 | 40 | 0 | 0 | 0 | 98.4 |
| Ceftazidime | 0 | 1 | 19 | 30 | 3 | 0 | 0 | 98.1 |
| Cefpodoxime | 0 | 0 | 4 | 41 | 6 | 0 | 2 | 96.2 |
| Flomoxef | 0 | 5 | 6 | 40 | 2 | 0 | 0 | 90.6 |
| Aztreonam | 0 | 0 | 18 | 33 | 2 | 0 | 0 | 100.0 |
| Imipenem | 3 | 3 | 22 | 14 | 10 | 0 | 1 | 86.8 |
| Amikacin | 0 | 3 | 25 | 21 | 4 | 0 | 1 | 92.6 |
| Minocycline | 0 | 3 | 23 | 17 | 6 | 3 | 0 | 88.5 |
| Levofloxacin | 0 | 0 | 3 | 26 | 23 | 1 | 0 | 98.1 |
| Fosfomycin | 8 | 9 | 10 | 20 | 5 | 1 | 0 | 66.0 |
| Overall | 11 | 28 | 171 | 349 | 77 | 5 | 5 | 92.3 |
Difference in MICs determined by the RL, IS60 system and the microdilution method. See footnotes to Table 3 for further details.
TABLE 5.
Distribution of differences in MICs of 12 antimicrobial agents for 19 strains of enterococci: RL/IS60 method versus microdilution method
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree- ment | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | +1 | +2 | >+2 | ||
| Cefotiam | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 100.0 |
| Ampicillin | 0 | 0 | 0 | 13 | 6 | 0 | 0 | 100.0 |
| Cefditoren | 0 | 0 | 0 | 17 | 0 | 1 | 1 | 89.5 |
| Flomoxef | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 100.0 |
| Imipenem | 0 | 0 | 0 | 16 | 3 | 0 | 0 | 100.0 |
| Arbekacin | 0 | 1 | 7 | 8 | 3 | 0 | 0 | 94.7 |
| Clarithromycin | 1 | 1 | 4 | 5 | 7 | 1 | 0 | 84.2 |
| Minocycline | 6 | 0 | 5 | 4 | 3 | 1 | 0 | 63.2 |
| Vancomycin | 0 | 0 | 3 | 10 | 6 | 0 | 0 | 100.0 |
| Teicoplanin | 0 | 0 | 7 | 6 | 6 | 0 | 0 | 100.0 |
| Levofloxacin | 0 | 0 | 0 | 9 | 5 | 2 | 3 | 73.7 |
| Oxacillin | 0 | 0 | 0 | 16 | 3 | 0 | 0 | 100.0 |
| Overall | 7 | 2 | 26 | 142 | 39 | 5 | 4 | 92.1 |
Difference in MICs determined by the RL/IS60 system and the microdilution method. See footnotes to Table 3 for further details.
TABLE 6.
Distribution of differences in MICs of 12 antimicrobial agents for 30 strains of staphylococci: RL/IS60 method versus microdilution method
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree- ment | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | −1 | +2 | >+2 | ||
| Cefotiam | 0 | 0 | 5 | 22 | 2 | 1 | 0 | 96.7 |
| Ampicillin | 0 | 2 | 17 | 7 | 3 | 1 | 0 | 90.0 |
| Cefditoren | 0 | 1 | 4 | 24 | 1 | 0 | 0 | 96.7 |
| Flomoxef | 0 | 0 | 7 | 17 | 6 | 0 | 0 | 100.0 |
| Imipenem | 0 | 0 | 2 | 24 | 4 | 0 | 0 | 100.0 |
| Arbekacin | 0 | 0 | 0 | 2 | 21 | 7 | 0 | 76.7 |
| Clarithromycin | 6 | 0 | 1 | 19 | 1 | 3 | 0 | 70.0 |
| Minocycline | 0 | 0 | 3 | 8 | 17 | 2 | 0 | 93.3 |
| Vancomycin | 0 | 0 | 0 | 5 | 25 | 0 | 0 | 100.0 |
| Teicoplanin | 0 | 0 | 6 | 19 | 4 | 1 | 0 | 96.7 |
| Levofloxacin | 0 | 0 | 7 | 9 | 9 | 5 | 0 | 83.3 |
| Oxacillin | 1 | 0 | 1 | 27 | 0 | 0 | 1 | 93.3 |
| Overall | 7 | 3 | 53 | 183 | 93 | 20 | 1 | 91.4 |
Difference in MICs determined by the RL/IS60 system and the microdilution method. See footnotes to Table 3 for further details.
TABLE 7.
Distribution of differences in MICs of 12 antimicrobial agents for 15 strains of H. influenzae: RL/IS60 method versus microdilution method
| Antimicrobial agent | No. of isolates with difference in MICsa
|
% Agree- ment | ||||||
|---|---|---|---|---|---|---|---|---|
| <−2 | −2 | −1 | 0 | +1 | +2 | >+2 | ||
| Cefditoren | 0 | 0 | 0 | 5 | 7 | 3 | 0 | 80.0 |
| Ampicillin | 0 | 0 | 4 | 8 | 3 | 0 | 0 | 100.0 |
| Penicillin G | 0 | 0 | 8 | 5 | 2 | 0 | 0 | 100.0 |
| Ceftazidime | 0 | 0 | 2 | 4 | 1 | 0 | 8 | 46.7 |
| Flomoxef | 0 | 1 | 8 | 4 | 2 | 0 | 0 | 93.3 |
| Cefpodoxime | 0 | 0 | 3 | 6 | 4 | 2 | 0 | 86.7 |
| Cefotaxime | 0 | 0 | 1 | 5 | 5 | 2 | 2 | 73.3 |
| Imipenem | 4 | 0 | 8 | 2 | 1 | 0 | 0 | 73.3 |
| Minocycline | 0 | 0 | 2 | 2 | 11 | 0 | 0 | 100.0 |
| Clarithromycin | 0 | 1 | 7 | 5 | 2 | 0 | 0 | 93.3 |
| Levofloxacin | 0 | 0 | 0 | 12 | 3 | 0 | 0 | 100.0 |
| Clindamycin | 0 | 0 | 0 | 11 | 4 | 0 | 0 | 100.0 |
| Overall | 4 | 2 | 43 | 69 | 41 | 7 | 10 | 87.2 |
Difference in MICs determined by the RL/IS60 system and the microdilution method. See footnotes to Table 3 for further details.
DISCUSSION
The present study verified that the RL/IS60 system is a rapid and accurate antimicrobial susceptibility testing system for a variety of antimicrobial agents against QC strains and clinical isolates. MICs obtained by the RL/IS60 system, obtained after 4 h of incubation, were reproducibly equivalent to those determined by the reference microdilution method.
Previous studies have assessed the rapidity and accuracy of various antimicrobial susceptibility methods. The E-test assay (4, 5, 7) and the frozen testing panel assay (8) optically detect bacterial growth, so that these methods require at least overnight to 24 h of incubation time. A robotic system that periodically reads turbidity determines MICs by analyzing growth kinetics but still requires 7 h of incubation time for accurate testing (9). Chromogenic and fluorogenic methods that detect specific enzyme activity in bacterial cells provide sensitive, accurate, and rapid testing methods (1, 10), but they are not popular in many clinical laboratories because of the difficulty in optimizing reaction conditions. The bioluminescent assay, which detects ATP in bacterial cells, is a highly sensitive method (2a, 11) but has not become an established method because of the instability of ATP during cell lysis and sample preparation.
We demonstrated the accuracy of the RL/IS60 assay by comparing the MICs of 12 antimicrobial agents for four QC strains with the QC limits determined by the reference microdilution method approved by the NCCLS. Although not all the tests were repeated, the data were reproducible with minimum variation. Only a few discrepancies (8.9%) were found. MICs of levofloxacin for E. faecalis and S. aureus were 1 dilution higher than the QC range. Similar discrepancies were observed in the present study for clinical isolates (Tables 5 and 6) and in our studies with other new quinolones, ofloxacin, sparfloxacin, and tosufloxacin (data not shown). It has been reported that new quinolones of concentrations equivalent to 1.0 times the MIC did not extensively reduce viability of S. aureus (6). The observed discrepancies may be caused by a difference in MIC determining procedures between the RL/IS60 method that reads cell viability after 4 h of incubation and the microdilution method that reads turbidity after 18 h of incubation.
The demonstration for the accuracy of our system was further obtained by testing 162 clinical isolates that were grouped into six species and families. Twelve antimicrobial agents were carefully selected for each group. Since the measuring principle of the new RL/IS60 assay system is completely different from that of the microdilution method approved by the NCCLS, we thought that it would be worthwhile to evaluate the new assay system for various combinations. Thus, we ventured to examine combinations such as enterococci and cephalosporins and those of H. influenzae and penicillin, minocycline, and clindamycin. The RL/IS60 system provided MICs equivalent to those determined by the reference microdilution method for most combinations of antimicrobial agents and bacterial species. It is notable that MICs of clinically important antimicrobial agents showed high agreement values; for example, agreement on oxacillin for staphylococci was 93.3%, and that on penicillin G for S. pneumoniae was 96.4%. Some combinations resulted in low agreement due to 2- to 3-dilution-lower or -higher shifts of MICs. The discrepancy of arbekacin for staphylococci (76.7%) may be partly explained by the lower pH (6.9) of the culture medium used in the RL/IS60 system than the pH (7.2) used in the reference method because the activity of arbekacin is weakened at lower pH (15). The low agreement values of ceftazidime (46.7%) and cefotaxime (73.3%) for H. influenzae may be related to the filamentation of bacteria, which was observed by us in a study with standard strains and clinical isolates (data not shown) and by another group with a laboratory strain transformed with chromosome DNA from ampicillin-resistant clinical isolates (3).
Minimum interpretive category errors occurred in the assay of the RL/IS60 system with clinical isolates. The errors included one MRSA instead of MSSA, one penicillin-susceptible S. pneumoniae strain instead of PISP, 3 PISP instead of penicillin-susceptible S. pneumoniae strains, and three ampicillin-susceptible H. influenzae strains instead of intermediately susceptible strains.
Timely and accurate detection of clinical isolates resistant to important antimicrobial agents is the role of microbiology laboratories to provide the first line of defense. Our study demonstrated that the RL/IS60 system meets this purpose. We also showed that the plotting of chemiluminescence intensity data provided clearly separated antimicrobial susceptibility curves, suggesting their usefulness for the screening of resistant strains. In addition, the fully automated system enables us to test large numbers of samples. Assays using the RL/IS60 system may become a choice for efficient antimicrobial susceptibility testing.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Exploratory Research from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Baker, C. N., and F. C. Tenover. 1996. Evaluation of Alamar colorimetric microdilution susceptibility testing method for staphylococci and enterococci. J. Clin. Microbiol. 34:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell, A. K., M. B. Hallett, and I. Weeks. 1985. Chemiluminescence as an analytical tool in cell biology and medicine. Method Biochem. Anal. 31:317-416. [DOI] [PubMed] [Google Scholar]
- 2a.Chapman, A. G., and D. E. Atkinson. 1977. Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv. Microb. Physiol. 15:253-306. [DOI] [PubMed] [Google Scholar]
- 3.Clairoux, N., M. Picard, A. Brochu, N. Rousseau, P. Gourde, D. Beauchamp, T. R. Parr, Jr., M. G. Bergeron, and F. Malouin. 1992. Molecular basis of the non-beta-lactamase-mediated resistance to beta-lactam antibodies in strains of Haemophilus influenzae isolated in Canada. Antimicrob. Agents Chemother. 36:1504-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Bonaventura, G., E. Ricci, N. D. Loggia, G. Catamo, and R. Piccolomini. Evaluation of the E test for antimicrobial susceptibility testing of Pseudomonas aeruginosa isolates from patients with long-term bladder catheterization. J. Clin. Microbiol. 36:824-826. [DOI] [PMC free article] [PubMed]
- 5.Endtz, H. P., N. V. D. Braak, A. V. Belkum, W. H. Goessens, D. Kreft, A. B. Stroebel, and H. A. Verbrugh. 1998. Comparison of eight methods to detect vancomycin resistance in enterococci. J. Clin. Microbiol. 36:592-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto, S., S. Miyazaki, and Y. Ishida. 1992. In vitro and in vivo antibacterial activities of a new quinolone, levofloxacin (DR-3355). Chemotherapy 40:14-26. [Google Scholar]
- 7.Jorgensen, J. H., A. W. Howell, and A. A. Maher. 1991. Quantitative antimicrobial susceptibility testing of Haemophilus influenzae and Streptococcus pneumoniae by using the E-test. J. Clin. Microbiol. 29:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen, J. H., M. L. McElmeel, and S. A. Crawford. 1998. Evaluation of the Dade MicroScan MICroSTREP antimicrobial susceptibility testing panel with selected Streptococcus pneumoniae challenge strains and recent clinical isolates. J. Clin. Microbiol. 36:788-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen, J. H., A. L. Barry, M. M. Traczewski, D. F. Sahm, M. L. McElmeel, and S. A. Crawford. 2000. Rapid automated antimicrobial susceptibility testing of Streptococcus pneumoniae by use of the bioMerieux VITEK 2. J. Clin. Microbiol. 38:2814-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manafi, M., W. Kneifel, and S. Bascomb. 1991. Fluorogenic and chromogenic substrates used in bacteria diagnostics. Microbiol. Rev. 55:335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molin, O., L. Milson, and S. Ansehn. 1983. Rapid detection of bacterial growth in blood cultures: bioluminescent assay of bacterial ATP. J. Clin. Microbiol. 18:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1997. Approved standard M7-A4, M100-S8. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS, Wayne, Pa.
- 13.National Committee for Clinical Laboratory Standards. 1997. Approved standard M2-A6, M100-S8. Performance standards for antimicrobial disk susceptibility tests. NCCLS, Wayne, Pa.
- 14.National Committee for Clinical Laboratory Standards. 2000. Approved standard M7-A5, M100-S10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS, Wayne, Pa.
- 15.Nishino, T., M. Tanaka, Y. Aono, T. Iwai, and M. Otsuki. 1992. In vitro and in vivo antibacterial activity of levofloxacin. Chemotherapy 40:36-50. [Google Scholar]
- 16.Yamashoji, S., T. Ikeda, and K. Yamashoji. 1989. Chemiluminescent assay for determination of viable cell density of yeast, mammalian, and plant cells. Anal. Biochem. 181:149-152. [DOI] [PubMed] [Google Scholar]
- 17.Yamashoji, S., I. Manome, and M. Ikedo. 2001. Menadione-catalyzed O2-production by Escherichia coli: application of the rapid chemiluminescent assay to antimicrobial susceptibility testing. Microbiol. Immunol. 45:333-340. [DOI] [PubMed] [Google Scholar]