Abstract
A real-time PCR assay using Taqman probes was developed to detect and quantify Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum” in feline blood samples. The assay was rapid and sensitive and was successfully used to monitor the in vivo kinetics of cats experimentally infected with each species.
The feline hemoplasmas, Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum,” are hemotropic bacterial species that can induce anemia in cats (4, 6, 7). Conventional PCR assays are known to be more sensitive than cytology for diagnosis of infection (3, 9). The aim of this study was to develop a real-time PCR assay to detect and accurately quantify feline hemoplasma DNA in blood samples from naturally and experimentally infected cats.
EDTA anticoagulated blood samples were obtained both from naturally and experimentally infected cats known to be infected with M. haemofelis and/or “Candidatus M. haemominutum” (n = 30) or from uninfected cats (n = 20), based on cytological examination of blood smears or the results of a previously described specific conventional PCR assay (8). DNA was extracted from 100 μl of blood using the DNeasy Tissue Kit (Qiagen, Crawley, United Kingdom) and was eluted into 200 μl of buffer AE, in accordance with the manufacturer's instructions. Tenfold serial dilutions of genomic DNA template positive for either M. haemofelis or “Candidatus M. haemominutum” were prepared. Plasmids containing cloned 16S ribosomal DNA (rDNA) PCR products from M. haemofelis or “Candidatus M. haemominutum” were also generated. The plasmid copy number was determined by spectophotometry and serial 10-fold dilutions prepared for PCR following linearization with EcoRI. Primers (Hf Forward, 5′-ACGAAAGTCTGATGGAGCAATA-3′; and Hf Reverse, 5′-ACGCCCAATAAATCCGRATAAT-3′) (Life Technologies, Paisley, United Kingdom) known to be specific for feline hemoplasmas (5, 8) were used. Taqman probes specific for either M. haemofelis (hexachloro-fluorescein-TACGAGGGATAATTATGATAGTACTTCGTGA-black hole quencher 1) or “Candidatus M. haemominutum” (6-carboxyfluorescein-AGCTTGATAGGAAATGATTAAGCCTTGA-black hole quencher 1) were designed (purchased from Cruachem Ltd., Glasgow, United Kingdom). PCRs comprised 12.5 μl of 2× Platinum Quantitative PCR Supermix-UDG (Invitrogen Ltd., Glasgow, United Kingdom), a 260 nM concentration of each primer, 300 nM “Candidatus M. haemominutum” probe, 200 nM M. haemofelis probe, 6 mM MgCl2 (final concentration), 1 U of additional Platinum Taq polymerase (2.2 U per 25-μl reaction), and 2 μl of template DNA, made up to a final volume of 25 μl with water. PCR was performed using an iCycler IQ system (Bio-Rad Laboratories Ltd., Hemel Hempstead, London, United Kingdom) with an initial incubation at 50°C for 3 min and then 95°C for 2 min followed by 45 cycles of 95°C for 5 s and 58°C for 30 s. Fluorescence was detected at 530 and 570 nm at each annealing step. DNA samples from noninfected cats and water were subjected to PCR as negative controls. All samples were run in triplicate. Intra-assay precision was measured by performing 10 replicates of each of seven hemoplasma-positive genomic DNA templates in a single PCR run. Interassay precision was measured by performing eight replicates of each of three hemoplasma-positive genomic DNA templates in separate PCR runs. Coefficients of variation were calculated. To enable standardization of hemoplasma DNA to an internal control (2), a real-time PCR assay for 28S rDNA was used. Briefly, forward (5′-CGCTAATAGGGAATGTGAGCTAGG-3′) and reverse (5′-TGTCTGAACCTCCAGTTTCTCTGG-5′) primers were used together with a molecular beacon (Texas red-CGCGCACCCTACTGATGATGTGTTGTTGCCGCGCG-DABCYL, where stem sequences are underlined) to amplify feline 28S rDNA. PCRs consisted of 25-μl reaction mixtures containing 12.5 μl of 2× Platinum Quantitative PCR SuperMix-UDG, a 300 nM concentration of each primer, 170 nM 28S rDNA molecular beacon, 2 μl of template, and water to a final volume of 25 μl, with fluorescence data collected at 620 nm.
To assess the kinetics of M. haemofelis and “Candidatus M. haemominutum” infection in vivo, two healthy female specific-pathogen-free adult cats were inoculated intravenously with blood collected from cats chronically infected with either M. haemofelis or “Candidatus M. haemominutum.” EDTA blood samples (0.2 ml) were collected before inoculation (day 0) and at intervals of 1 to 9 days postinoculation (p.i.) and subjected to DNA extraction. Blood smears were also prepared for cytological examination with Giemsa staining, and hematocrit (HCT) was measured at defined time points (Fig. 1 and 2). DNA samples were analyzed by both hemoplasma and 28S rDNA real-time PCR. The latter was used to standardize hemoplasma PCR threshold cycle values to a specified amount of 28S rDNA. Quantification of hemoplasma DNA was then achieved using the standard curve generated by serial dilutions of plasmid DNA template containing known copy numbers of either M. haemofelis or “Candidatus M. haemominutum.”
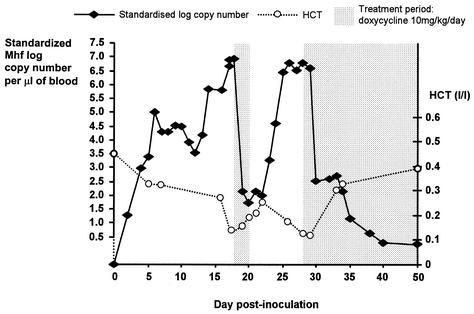
FIG. 1.
Graph plotting standardized log10 of M. haemofelis (Mhf) copy number of DNA template present per microliter of blood versus day postinoculation in a cat experimentally infected with M. haemofelis. Standardization was to 28S rDNA. Hematocrit (HCT) values are also shown.
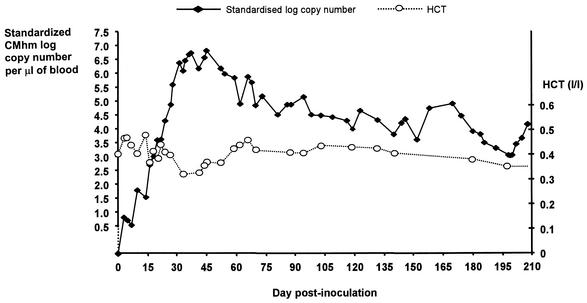
FIG. 2.
Graph plotting standardized log10 of “Candidatus M. haemominutum” (CMhm) copy number of DNA template present per microliter of blood versus day postinoculation in a cat experimentally infected with “Candidatus M. haemominutum.” Standardization was to 28S rDNA. Hematocrit (HCT) values are also shown.
The GenBank accession numbers of the target DNA sequences used for the development of the real-time assays described above were U88564, U95297, AF178677, AF271154, and AF353617.
The assay was able to specifically detect “Candidatus M. haemominutum” and/or M. haemofelis in all genomic DNA samples extracted from cats known to be infected with both or each of the organisms (n = 30). Control samples from uninfected cats (n = 20) and no template water reactions were all negative. In genomic DNA samples, the efficiency of amplification of “Candidatus M. haemominutum” was 99.7% over a 107 range of starting templates with a correlation of 0.992 and that of M. haemofelis was 99.3% over a 106 range of starting templates with a correlation of 0.994. Using plasmid DNA as template, amplification of “Candidatus M. haemominutum” showed 99.7% efficiency over a 108 range of starting templates with a correlation of 0.993, while M. haemofelis amplification was 103.6% efficient over a 109 range of starting templates with a correlation of 0.997. Thus, the efficiencies of hemoplasma DNA amplification in reactions with genomic and plasmid DNA templates are very similar, allowing accurate quantification of unknown DNA samples from the standard curves generated by amplification of plasmid DNA dilutions. The assay was able to detect 3.63 and 1.67 copies of “Candidatus M. haemominutum” and M. haemofelis template, respectively, in triplicate reactions. The intra-assay and interassay coefficients of variation ranged from 1.5 to 2.7% and 1.4 to 2.2%, respectively.
Measurement of the copy number of hemoplasma DNA in experimentally infected cats is shown in Fig. 1 and 2. The hemoplasma copy number increased rapidly following inoculation, reaching a maximum on day 18 p.i. for M. haemofelis and day 45 p.i. for “Candidatus M. haemominutum.” The M. haemofelis-infected cat developed severe hemolytic anemia (with HCT values falling to below the reference range of 0.25 to 0.45 liter/liter) on two occasions, necessitating antibiotic treatment with doxycycline. On both occasions treatment was associated with a fall in the M. haemofelis copy number and rise in HCT. The “Candidatus M. haemominutum”-infected cat showed some fluctuation in HCT values during the 1st month p.i., but values remained within the reference range throughout the study. Definitive cytological evidence of infection was seen only on blood smears collected between days 25 and 29 from the M. haemofelis-infected cat.
The real-time PCR assay described provides a reliable method for diagnosis of infection and quantification of feline hemoplasma DNA in blood samples and offers the advantages of greater speed, increased throughput, and decreased risk of false-positive results over conventional PCR. It is hoped that quantification may help the veterinarian determine the significance of a positive PCR result, which can be problematic with infections such as “Candidatus M. haemominutum” and M. haemofelis, which do not always result in disease in the host. The assay can also be used to monitor response to treatment. The differing pathogenicity of these species previously reported (3, 9) was evident in the present study by the fall in HCT that occurred in association with rises in the M. haemofelis copy number, whereas the HCT values of the “Candidatus M. haemominutum”-infected cat remained within the reference range.
To relate accurately the copy number detected in PCR to the number of organisms present per microliter of blood, the amplification efficiency of the PCR assay and the number of 16S rDNA sequences present in the genome being amplified should be considered. The M. haemofelis copy number of 3.81 × 106 reported on day 29 p.i. equates to either 3.85 × 106 or 1.92 × 106 organisms per μl of blood using an efficiency of amplification of 99.3% and assuming the presence of either one or two copies of 16S rRNA genes in the M. haemofelis genome, as has been reported for other mollicutes (1). On day 29 p.i., cytological examination revealed 2.55 × 106 organisms per μl of blood, suggesting that the copy number provides a good indication of the number of organisms present. The residual discrepancy between these values could be explained by the efficiency of DNA extraction and the fact that cytological enumeration of organisms is unlikely to be very accurate. True correlation between copy number and the number of organisms per microliter of blood requires quantitative culture of organisms. This is not possible for these feline hemoplasma species since they have not yet been successfully cultured.
This real-time PCR assay should provide a valuable molecular tool for the diagnosis and quantification of M. haemofelis and “Candidatus M. haemominutum” DNA in feline blood samples.
Acknowledgments
This work was supported by the Royal College of Veterinary Surgeons Trust Fund West Scholarship for Feline Research.
Simon Cue and Elsa Crawford are thanked for their assistance in cytological examination of blood smears. Michael Lappin is thanked for the kind donation of M. haemofelis-infected blood for use in experimental infection.
REFERENCES
- 1.Amikam, D., G. Glaser, and S. Razin. 1984. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J. Bacteriol. 158:376-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 3.Foley, J. E., S. Harrus, A. Poland, B. Chomel, and N. C. Pedersen. 1998. Molecular, clinical, and pathologic comparison of two distinct strains of Haemobartonella felis in domestic cats. Am. J. Vet. Res. 59:1581-1588. [PubMed] [Google Scholar]
- 4.Foley, J. E., and N. C. Pedersen. 2001. ′Candidatus Mycoplasma haemominutum', a low-virulence epierythrocytic parasite of cats. Int. J. Syst. E vol. Microbiol. 51:815-817. [DOI] [PubMed] [Google Scholar]
- 5.Jensen, W. A., M. R. Lappin, S. Kamkar, and W. J. Reagan. 2001. Use of a polymerase chain reaction assay to detect and differentiate two strains of Haemobartonella felis infection in naturally infected cats. Am. J. Vet. Res. 62:604-608. [DOI] [PubMed] [Google Scholar]
- 6.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2001. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ′Candidatus Mycoplasma haemofelis', ′Candidatus Mycoplasma haemomuris', ′Candidatus Mycoplasma haemosuis' and ′Candidatus Mycoplasma wenyonii'. Int. J. Syst. E vol. Microbiol. 51:891-899. [DOI] [PubMed] [Google Scholar]
- 7.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2002. Revision of haemotrophic Mycoplasma species names. Int. J. Syst. E vol. Microbiol. 52:683.. [DOI] [PubMed] [Google Scholar]
- 8.Tasker, S., S. H. Binns, M. J. Day, T. J. Gruffydd-Jones, D. A. Harbour, C. R. Helps, W. A. Jensen, C. S. Olver, and M. R. Lappin. Use of a polymerase chain reaction assay to assess prevalence and risk factors for Mycoplasma haemofelis and ′Candidatus Mycoplasma haemominutum' in the United Kingdom. Vet. Rec., in press. [DOI] [PubMed]
- 9.Westfall, D. S., W. A. Jensen, W. J. Reagan, S. V. Radecki, and M. R. Lappin. 2001. Inoculation of two genotypes of Hemobartonella felis (California and Ohio variants) to induce infection in cats and the response to treatment with azithromycin. Am. J. Vet. Res. 62:687-691. [DOI] [PubMed] [Google Scholar]