Abstract
PCR-based assays were used to evaluate agr locus nucleotide polymorphism for the identification of agr autoinducer receptor specificity groups within a population of Staphylococcus aureus isolates colonizing children and their guardians. All isolates could be assigned to one of three major agr groups that had similar prevalences, regardless of whether isolates were implicated in transmission of S. aureus within families. Among healthy carriers, agr groups I to III appear to be equally fit, which may reflect selection for the coexistence of S. aureus strains in a population.
In Staphylococcus aureus, the accessory gene regulator (agr) globally controls the coordinated production of virulence factors. The agr signaling system consists of a classical two-component signaling system (AgrC as the signal receptor and AgrA as the response regulator) that serves as a quorum-sensing regulon to autoinduce RNA III, the principal effector of the agr response (1). The agr signaling system is driven by an autoinducing peptide pheromone encoded within agrD. S. aureus isolates can be divided into four predominant agr groups on the basis of the specificity of the autoinducing peptide for its membrane sensor (AgrC) (5). Isolates of one agr group are capable of activating the agr response in isolates of the same group, but they usually inhibit it in members of the other groups. Thus, cross-restriction of the agr signaling pathway between isolates constitutes a novel form of bacterial interference (4).
It has been suggested that agr autoinducer receptor specificity groups may influence host ecology by enhancing or inhibiting the ability of an S. aureus isolate to colonize (or compete) in the presence of resident strains (4), including other staphylococci (9). The putative within-host effects of agr polymorphism are also likely to affect the host-to-host transmission and epidemiology of S. aureus clones (11). If agr polymorphism does have these effects, preferential transmission of strains having particular agr groups might occur, reflecting those that succeed in competitive within-host situations. To test this idea, we sought linkage between agr group and clonal S. aureus transmission among family members.
In this study, a rapid PCR-based method was used to identify agr specificity groups among a genotypically characterized collection of colonizing S. aureus isolates obtained from a healthy population of children and their guardians in New York City, New York (13). Study subjects represented a previously described well-patient population visiting a pediatric outpatient clinic (13). The 500 individuals in the study came from 212 households, which varied in size from two to five persons, with one to four children and either one or two guardians. The majority of families (147 individuals) presented as one child and one guardian (convenience sampling was used, i.e., only family members at the clinic were enrolled). S. aureus was isolated (one strain per subject) from 96 of 275 (35%) children and from 64 of 225 (28%) guardians (13). agr specificity groups were identified by PCR amplification of the hypervariable domain of the agr locus using oligonucleotide primers specific for each of the four major specificity groups (obviating the need for restriction fragment length polymorphism analysis commonly employed in previous studies [7, 10, 14]). A forward primer, pan-agr (5′-ATGCACATGGTGCACATGC-3′), corresponding to conserved sequences from the agrB gene, was used in all reactions (Fig. 1A) (primer sequences were obtained from GenBank accession numbers X52543, AF001782, AF001783, and AF288215). Four reverse primers, each specific for amplification of a single agr group based on agrD or agrC gene nucleotide polymorphism, were as follows: agr I, 5′-GTCACAAGTACTATAAGCTGCGAT-3′ (in the agrD gene); agr II, 5′-GTATTACTAATTGAAAAGTGCCATAGC-3′ (in the agrC gene); agr III, 5′-CTGTTGAAAAAGTCAACTAAAAGCTC-3′ (in the agrD gene); and agr IV, 5′-CGATAATGCCGTAATAC CCG-3′ (in the agrC gene).
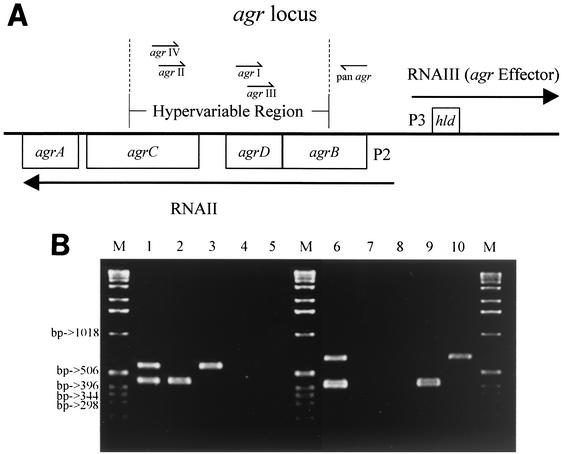
FIG. 1.
PCR assay for the identification of agr specificity groups. (A) Schematic map of the S. aureus agr locus showing the locations of the different primers used for amplification of the hypervariable region. (B) Whole-cell PCR was performed with type strains of each of the four agr specificity groups. To distinguish between the similar-sized products of groups I and III and groups II and IV, two duplex PCR amplifications were performed for each isolate as follows. Lane 1, combined products from PCR of agr type strains I and II using reverse primers agr I and agr II; lanes 2 to 5, PCR products using reverse primers agr I and agr II from agr type strains I to IV (440 bp for agr I and 572 bp for agr II); lane 6, combined products from PCR of agr type strains III and IV using reverse primers agr III and agr IV (406 bp for agr III, and 588 bp for agr IV); lanes 7 to 10, PCR products using reverse primers agr III and agr IV from agr type strains I to IV. Lanes M contain molecular size markers (sizes in base pairs).
agr specificity groups were identified by the expected product sizes (Fig. 1B). PCR was performed by adding 1 μl of a 1:200 dilution of chromosomal template DNA and 24 μl of water to 25 μl of a PCR mixture that includes 2.5 U of AmpliTaq Gold DNA polymerase, 2 mM MgCl2, 350 μM (total) deoxynucleoside triphosphates, and 25 mM KCl in 0.2-ml PCR tubes (Perkin-Elmer Applied Biosystems Division, Norwalk, Conn.). A negative control and control strains from each agr specificity group (PHRI S. aureus collection) were included. Thermal cycling was performed in a GeneAmp 9600 instrument (Perkin-Elmer Applied Biosystems Division); 25 cycles of PCR were done, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 55°C), and extension (1 min at 72°C). PCR products were separated by electrophoresis on 1% agarose gels, which were stained with ethidium bromide (Shelton Scientific, Shelton, Conn.). The lengths of the PCR products were estimated by comparison with the 1-kb DNA ladder molecular size markers (Gibco BRL, Life Technologies, Gaithersburg, Md.).
Genotypes were previously determined for 154 of the S. aureus isolates colonizing children and guardians by macrorestriction analysis using pulsed-field gel electrophoresis (PFGE) analysis and DNA sequence analysis of the protein A gene (spa) hypervariable region (13). One genetic background accounted for more than 11.7% of the colonizations; the average proportion was 1.7% per strain (range, 0.6 to 11.7%), indicating that overall, genotypic diversity was high in the sample.
S. aureus carriage in family members of carriers was similar to carriage in the overall sample: among 115 households (consisting of 276 individuals) where at least one family member was colonized, S. aureus was isolated from only 45 of 154 (29.8%) potential subjects (45 of 154 versus 154 of 500; P = 0.599 by one-tailed t test; statistical analysis performed using SAS version 8 [SAS, Cary, N.C.]). Thus, family members of carriers were not more likely to be colonized themselves.
To estimate the frequency of transmission of S. aureus in families, we compared S. aureus clonal types of host pairs, defined as a colonized child having one colonized guardian. Among the subgroup of 66 isolates from the 33 child-guardian pairs examined, genotypic diversity was high: 29 distinct spa strain types were seen. However, in 22 instances (67% of the cases), a child and guardian within a pair had the same spa type. This high percentage of concurrence of spa types within households when both family members are colonized is greater than would be expected by chance: the kappa statistic equals 0.65 [95% confidence interval of 0.58 to 0.72], where pure-chance concurrence of types would yield a kappa value of 0 and perfect concurrence value of 1.0. Thus, when multiple colonizations do happen, they are likely to occur with S. aureus strains of the same spa type, indicating transmission. Among transmitting isolates, genotypic diversity remained high: 14 types were recovered, one from each of 14 households, and only four spa types occurred in two households. Apparently, the ability to transmit is ubiquitous among S. aureus strains.
Analysis of agrD gene polymorphism showed that all 154 of the S. aureus genotypes could be assigned to one of three major agr specificity groups. Sixty-four isolates (42%) belonged to agr specificity group I, 38 (24%) belonged to group II, and 52 (34%) belonged to group III. Each agr group harbored a diversity of S. aureus genotypes (not shown). The balanced recovery of three of the four agr groups differs from data previously reported (14), perhaps reflecting ecological and geographical structuring (or sampling bias). A fourth agr group, which is common among exfoliatin-producing strains (3), was not identified in our population. No association was observed for agr group with age, gender (for either child or guardian), previous antibiotic usage, outpatient visits, or known underlying condition. However, among children there was an association between agr group I and recent hospitalization (P = 0.02).
The results of analyzing hosts implicated in transmission of S. aureus within families showed that among the 22 child-guardian pairs colonized with strains of the same spa type, the distribution of agr specificity groups was no different from that in the overall sample (P = 0.95) (Table 1). Thus, there was no correlation between agr group and intrafamily staphylococcal transmission.
TABLE 1.
Frequency of agr specificity groups among the S. aureus strains carried
| agr specificity group | No. of agr specificity groups (%)
|
P valuec | ||
|---|---|---|---|---|
| Nontransmitting isolatesa | Transmitting isolatesb | Total | ||
| I | 44 (40) | 20 (45) | 64 (42) | 0.589 |
| II | 28 (25) | 10 (23) | 38 (24) | 0.837 |
| III | 38 (35) | 14 (32) | 52 (34) | 0.851 |
| Total | 110 (100) | 44 (100) | 154 (100) | |
S. aureus isolates with the indicated agr specificity group among households where transmission was not apparent (only one family member was colonized, or family members were colonized by unrelated S. aureus strains).
S. aureus isolates implicated in staphylococcal transmission in families.
P value comparing the number of agr specificity groups in the nontransmitting isolates to the number of agr specificity groups in the transmitting isolates by Fisher's exact (two-tailed) test.
Frequent exposure to the flora of close contacts may lead to mixed-clone S. aureus colonization. Specificity in the agr system could then operate to affect the frequency of agr group recovery even in the absence of transmission of a predominant colonizing strain. The occurrence of different spa types among colonized child-guardian pairs, however, did not predict the agr group (P = 0.87). Stated differently, among child-guardian pairs colonized by different strains, agr-mediated interference did not determine the clonal performance (defined by agr allele frequencies) of predominant colonizing isolates.
Congruence was indicated between genetic markers used for clonal analysis and agr grouping: any two isolates that were identical on the basis of both PFGE and spa typing were invariably of the same agr group. The same result was obtained using spa type alone in all but four occasions, consistent with the slight decrease in specificity observed for spa typing compared to that for PFGE (12).
The rare occurrence of noncongruence between genetic markers and agr contrasts with recent reports that recombination rates are high in S. aureus (2). This observation may be explained by at least three hypotheses. First, primary differentiation of agr group is apparently followed by polymorphism of spa. Second, recombination in the short-term will not obscure relationships among many of the isolates that were implicated in family (i.e., recent) transmission. A third but not mutually exclusive hypothesis is that reassortment events involving agr frequently occur between identical alleles.
In summary, a study of children and their guardians attending an urban pediatric clinic showed evidence that different family members were frequently colonized by a variety of agr types, and the presence of any one agr group in a family did not affect the likelihood of recovering isolates with the same (stimulatory) or different (inhibitory) agr autoinducer. We did not find a predominance of any particular agr group among isolates implicated in transmission of S. aureus in families. Apparently, in a population of healthy carriers, agr groups I to III affect the transmission of S. aureus equally or not at all. Thus, it seems unlikely that one interference group will spread at the expense of another, even among hosts living in close proximity.
The uniform fitness of S. aureus agr groups suggests that they also have comparable competitive ability within the host. Rather than promoting exclusion, it may be that agr-mediated interference functions to create niche (or microniche) separation between staphylococcal strains so that mixed genotype colonizations can occur. Heterogeneity could enhance the fitness of the total population through increased genetic exchange, variability, host defense costs, and transmissibility.
Our conclusions may not apply when considering populations faced with different ecological constraints (such as carriage of hospital-acquired strains or methicillin-resistant strains), or isolates of agr group IV. Despite the unique ability of the group IV pheromone to inhibit the Staphylococcus epidermidis agr response (8), the absence of group IV isolates from this study and other studies (6, 14) suggests that competition does not favor these strains.
The results also showed that the family members of S. aureus carriers are not more likely to be colonized themselves than the overall population. When both members of a child-guardian pair were colonized with S. aureus, however, transmission of staphylococci within the family was indicated, since the colonizing isolates are often (67%) the same strain. Frequency of colonization, but not strain type, is independent of exposure to colonized family members. Thus, among healthy urban carriers, it appears that host-microbe interactions, rather than exposure, determine carrier status.
Acknowledgments
This work was funded in part by the Public Health Research Institute, New York Community Trust, and by a research grant from the Professional Staff Congress, City University of New York (awarded to P.A.).
We thank Karl Drlica, David Dubnau, Larry Koreen, and François Vandenesch for their critical comments on the manuscript.
REFERENCES
- 1.Balaban, N., and R. P. Novick. 1995. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 92:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 5.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore, P. C., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39:2760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullarky, I. K., C. Su, N. Frieze, Y. H. Park, and L. M. Sordillo. 2001. Staphylococcus aureus agr genotypes with enterotoxin production capabilities can resist neutrophil bactericidal activity. Infect. Immun. 69:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto, M., H. Echner, W. Voelter, and F. Gotz. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto, M., R. Sussmuth, C. Vuong, G. Jung, and F. Gotz. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 450:257-262. [DOI] [PubMed] [Google Scholar]
- 10.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 11.Read, A. F., and L. H. Taylor. 2001. The ecology of genetically diverse infections. Science 292:1099-1102. [DOI] [PubMed] [Google Scholar]
- 12.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shopsin, B., B. Mathema, J. Martinez, E. Ha, M. L. Campo, A. Fierman, K. Krasinski, J. Kornblum, P. Alcabes, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Prevalence of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the community. J. Infect. Dis. 182:359-362. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]