Abstract
Neisseria meningitidis is the causative agent of meningococcal sepsis and meningitis. Neisseria polysaccharea is a nonpathogenic species. N. polysaccharea is able to use sucrose to produce amylopectin, a starch-like polysaccharide, which distinguishes it biochemically from the pathogenic species N. meningitidis. The data presented here indicate that this may be an insufficient criterion to distinguish between these two species. The nonencapsulated Neisseria strain 93246 expressed a phenotype of amylopectin production similar to that of N. polysaccharea. However, strain 93246 reacted with N. meningitidis serotype 4 and serosubtype P1.14 monoclonal antibodies and showed the N. meningitidis L1(8) lipo-oligosaccharide immunotype. Further analyses were performed on four genetic loci in strain 93246, and the results were compared with 7 N. meningitidis strains, 13 N. polysaccharea strains, and 2 N. gonorrhoeae strains. Three genetic loci, opcA, siaD, and lgt-1 in strain 93246, were the same as in N. meningitidis. Particularly, the siaD gene encoding polysialyltransferase responsible for biosynthesis of N. meningitidis group B capsule was detected in strain 93246. This siaD gene was inactivated by a frameshift mutation at the poly(C) tract, which makes strain 93246 identical to other nonencapsulated N. meningitidis strains. As expected, the ams gene encoding amylosucrase, responsible for production of amylopectin from sucrose, was detected in strain 93246 and all 13 N. polysaccharea strains but not in N. meningitidis and N. gonorrhoeae strains. These data suggest that strain 93246 is nonencapsulated N. meningitidis but has the ability to produce extracellular amylopectin from sucrose. The gene for amylopectin production in strain 93246 was likely imported from N. polysaccharea by horizontal genetic exchange. Therefore, we conclude that genetic analysis is required to complement the traditional phenotypic classification for the nonencapsulated Neisseria strains.
Neisseria meningitidis is a pathogenic species of the genus Neisseria causing meningococcal septicaemia and meningitis (15). N. meningitidis expresses different capsular polysaccharides that determine the meningococcal serogroups (17, 18). Neisseria polysaccharea is a nonpathogenic species that has been isolated from the throats of healthy children (19, 23). The most prominent biochemical feature of N. polysaccharea is the production of α-D-glucan from sucrose, distinguishing it phenotypically from N. meningitidis (5, 23, 24). The gene coding the extracellular amylosucrase that uses sucrose to produce this amylopectin has been cloned and sequenced (7, 9).
Initially we examined 14 strains of N. polysaccharea from bacterial culture collections of the National Microbiology Laboratory of Health Canada for the opcA gene coding an outer membrane protein (20, 33). The results revealed that two N. polysaccharea strains, 85322 and 89357, contained a novel opcA orthologous gene (N. polysaccharea opcA) that differed from the opcA genes reported for N. meningitidis (20) and Neisseria gonorrhoeae (33). Interestingly, we found that an N. polysaccharea strain, 93246, contained an N. meningitidis opcA gene. PCR-restriction fragment length polymorphism analysis of 20 single colonies of this culture yielded the same results for opcA, suggesting that the culture of strain 93246 was not likely to be a mixture of N. polysaccharea and N. meningitidis. This suggested that strain 93246 might be N. meningitidis rather than N. polysaccharea. Therefore, the phenotypic and genetic characteristics of strain 93246 have been further investigated by several methods. Strain 93246 showed phenotypic characteristics similar to those of N. polysaccharea, and its genome also contained the gene encoding amylosucrase. Nevertheless, the results from three genetic loci, opcA, siaD, and lgt-1, showed that these genes in strain 93246 were the same as those in N. meningitidis. This suggests that strain 93246 has the basic genome framework of N. meningitidis and acquired an amylosucrase gene from N. polysaccharea.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA isolation.
The bacterial strains used in this study are shown in Table 3. Five N. meningitidis strains, M986, 126E, M158, S4383, and 6304, were obtained from the culture collections of C. E. Frasch, Center for Biologics Evaluation and Research (CBER), U.S. Food and Drug Administration (FDA), Bethesda, Md. Other strains were from authors' collections. Among them, strain 93246 was isolated from the vagina of a 27-year-old female in 1993. The isolate was submitted to the National Microbiology Laboratory of Health Canada for differentiation between N. meningitidis and N. polysaccharea. This isolate was classified as N. polysaccharea because it behaved typically for N. polysaccharea, including production of amylopectin from sucrose as well as the ability to grow on Thayer-Martin selective medium containing the antimicrobial agents vancomycin, colistin, and amphotericin B. The growth and biochemical characteristics of strain 93246 were confirmed by the standard method. The bacterial strains were grown on brain heart infusion agar plates at 37°C in 5% CO2 for 16 h. The chromosomal DNA was isolated using a modified phenol-chloroform extraction method (20, 26).
TABLE 3.
The genetic characteristics of strain 93246
| Strain | opcAa | siaD/orf2b | amS |
lgt-1
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| lgtA | lgtB | lgtC | lgtD | lgtE | lgtZ | lgtH | ||||
| N. meningitidis | ||||||||||
| Z2491 | + (m) | + (A) | − | + | + | − | − | − | − | + |
| MC58 | + (m) | + (B) | − | + | + | − | − | + | − | − |
| M986 | − | + (B) | − | + | + | − | − | − | − | + |
| 126E | + (m) | + (C) | − | − | − | + | + | + | + | − |
| M158 | − | + (C) | − | + | + | − | − | + | − | − |
| S4383 | + (m) | + (W135) | − | + | + | − | − | − | − | + |
| 6304Y | − | + (Y) | − | + | + | − | − | + | − | − |
| 93246 | + (m) | + (B) | + | − | − | + | + | − | + | + |
| N. polysaccharea | ||||||||||
| 85321 | − | − | + | + | + | − | − | − | − | + |
| 85322 | + (p) | − | + | + | + | − | − | − | − | + |
| 85323 | − | − | + | + | + | − | − | − | − | + |
| 87042 | − | − | + | + | + | − | − | − | − | + |
| 87043 | − | − | + | + | + | − | − | − | − | + |
| 87188 | − | − | + | + | + | + | + | − | − | + |
| 87190 | − | − | + | + | + | + | + | − | − | + |
| 89353 | − | − | + | + | + | − | − | − | − | + |
| 89354 | − | − | + | + | + | − | − | − | − | + |
| 89355 | − | − | + | + | + | − | − | − | − | + |
| 89357 | + (p) | − | + | + | + | − | − | − | − | + |
| 90400 | − | − | + | + | + | − | − | − | − | + |
| 91275 | − | − | + | + | + | − | − | − | − | + |
| N. gonorrhoeae | ||||||||||
| FA1090 | + (g) | − | − | + | + | + | + | + | − | − |
| F62 | + (g) | − | − | + | + | + | + | + | − | − |
Phenotypic analysis. (i) Amylopectin production from sucrose.
Bacterial strains were grown on tryptic soy broth (Difco) agar containing 1% sucrose at 37°C for 48 h and tested for the production of amylopectin with Lugol's iodine solution using the Centers for Disease Control and Prevention's method (http://www.cdc.gov/ncidod/dastlr/gcdir/NeIdent/Polysacc.html).
(ii) Capsular production.
Bacterial strains were grown on antiserum agar containing 6% N. meningitidis serogroup B serum against capsular polysaccharide (horse 46 globulin; CBER, FDA) at 37°C for 24 h, and the presence of capsular polysaccharide was detected by immunoprecipitation (8). Serogroup B antiserum was used because the siaD gene detected in strain 93246 was specific for the group B capsule (see below) (2, 28).
(iii) Immunotype of lipooligosaccharide (LOS).
LOS samples were analyzed by using the sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining method (30). Immunoblotting was performed using the antibodies specific to N. meningitidis LOS (31, 32).
(iv) Serotyping and subtyping.
N. meningitidis serotyping and subtyping were performed using monoclonal antibodies and whole-cell enzyme-linked immunosorbent assay (1).
Genetic analysis. (i) PCR.
Twelve primers for PCR are shown in Table 1. The primers for the opcA locus were designed from an alignment of four opcA sequences from N. meningitidis strains Z2491 (GenBank accession number AJ242841) and MC58 (accession number AE002456) and N. gonorrhoeae strains FA1090 (accession number AJ242839) and MS11 (accession number AJ242840) (21, 29, 33). The primers at the siaD region were designed from the reported sequence of N. meningitidis strain B1940 (M95053) (11). Five primer pairs for detecting siaD and orf-2 loci in N. meningitidis were the same as those described by Taha (serogroups A, W135, and Y) (28) and Arreaza et al. (serogroups B and C) (2) except for the reverse primer for group B siaD (P64) (Table 1). For the purpose of this study, the gene encoding amylosucrase is designated ams. The primers at the ams region were designed from the reported ams sequence of N. polysaccharea strain ATCC 43768 (accession number AJ011781) (9). The primers for detecting seven lgt genes at the lgt-1 locus were the same as in our previous report (35, 36). Seven N. meningitidis reference strains (Z2491, MC58, M986, 126E, M158, S4383, and 6304Y) and two N. gonorrhoeae reference strains (FA1090 and F62) served as controls in the PCR analysis.
TABLE 1.
Primers for PCR amplification of the opcA, siaD, and ams loci
| Primer | Sequence (5′ - 3′) | Specificitya | Strandb | Positionc | Accession no. | PCR sized |
|---|---|---|---|---|---|---|
| P103 | TTCGTTACCTCCGGCATCCG | opcA (E) | F | 1356-1375 | AJ242839 | 2,744 (P103/P104) |
| P61 | ACCATCAAATGAATATCCAT | opcA (E) | R | 4080-4099 | AJ242839 | |
| P51 | GCCGCCATCTCCGGTACTGC | opcA (I) | F | 2976-2995 | AJ242839 | 662 (P51/P52) |
| P52 | TGACGGTGTTTGTAGAACGG | opcA (I) | R | 3618-3637 | AJ242839 | |
| P124 | GTCGCTGCTTGCAATATTCG | siaD, (E) | F | 2991-3010 | M95053 | 1,770 (P124/P123) |
| P125 | TACAGCAGCTCTGTTGTCGA | siaD, (E) | R | 4741-4760 | M95053 | |
| P63 | CTCTCACCCTCAACCCAATGTC | siaD (I) | F | 4141-4160 | M95053 | 453 (P63/P64) |
| P64 | TCGGCGGAATAGTAATAATGTT | siaD (I) | R | 4572-4593 | M95053 | |
| P114 | CGCCGGTCGGAAACTTCAGA | ams (E) | F | 101-120 | AJ011781 | 1,985 (P114/P115) |
| P115 | CCGTCTGAAACGGTTCAGAC | ams, (E) | R | 2066-2085 | AJ011781 | |
| P116 | CAAGTCGGCGGCGTGTGCTA | ams (I) | F | 451-470 | AJ011781 | 1,300 (P116/P117) |
| P117 | CTGCGGTCGACGGATCGTTG | ams (I) | R | 1731-1750 | AJ011781 |
E, external primer at the flanking region; I, internal primer at the coding region.
F, forward; R, reverse.
Position on the sequence from the following strains: AJ242839 from strain FA1090 (33); AJ011781, strain ATCC 43768 (25); M95053, strain B1940 (11).
Numbers indicate the expected size of the PCR product (base pairs). External or internal primer pairs are indicated in parentheses.
The PCR mixtures contained 1 μl of 10 mM deoxynucleoside triphosphates, 10 pmol of each primer, 0.1 μg of chromosomal DNA, 5 μl of 10× PCR buffer, and 1.5 U of Taq DNA polymerase (Perkin-Elmer), and sterile redistilled H2O in a final volume of 50 μl. PCR amplification was performed using the following protocol: denaturation at 94°C for 2 min, 30 cycles of amplification at 94°C for 30 s, 56°C for 30 s and 72°C for 2 min, and a final extension at 72°C for 4 min. The PCR products were analyzed by electrophoresis on a 1% agarose gel and stained with ethidium bromide.
(ii) RT-PCR.
Bacteria were grown in a 10-ml liquid brain heart infusion culture that was shaken at 160 rpm for 6 h. Bacteria cells were collected by centrifugation at 3,000 × g for 20 min, and total bacterial RNA was isolated from the pellet using TRIzol Reagent (Life Technologies) using the manufacturer's protocol. The total RNA sample was treated using DNase I (Life Technologies) to remove the genomic DNA, and reverse transcription-PCR (RT-PCR) was performed using the Titan One tube RT-PCR system (Roche Molecular Biochemicals) as described by the manufacturer. Two internal primers, P51 and P52, were used for RT-PCR analysis of opcA expression. The genomic DNA from strain 93246 was used as a positive control, and RNA from 93246 without RT was used as a negative control. The template of reverse transcription from RNA was used for examination of opcA expression.
(iii) DNA sequencing.
The whole regions of opcA, siaD, and ams were amplified using the flanking primers and internal primers (complete list available on request to the corresponding author). DNA sequences were determined from both strands of three independent PCR products for each strain as described previously (36). DNA sequences were analyzed with the Genetics Computer Group package (GCG10.2-Unix; University of Wisconsin) (10) and Molecular Evolutionary Genetics Analysis software (MEGA2.1; Arizona State University) (16).
Nucleotide sequence accession numbers.
The nucleotide sequences of the opcA, siaD, and ams regions from strain 93246 were determined and have been submitted to GenBank under accession numbers AY099332 to AY099334. The sequence of ams from strain 85322 was determined as well (GenBank accession no. AY099335).
RESULTS
Phenotypic features of strain 93246.
The phenotypic features of strain 93246 were tested and compared with those of N. meningitidis strain M986 and N. polysaccharea strain 89357 (Table 2). Strain 93246 showed the same characteristics as N. polysaccharea strain 89357, including production of amylopectin from sucrose, negative growth in Catlin meningococcal defined medium, and negative reaction with serogroup B horse serum. Strain 93246 reacted with N. meningitidis serotype 4 and serosubtype P1.14 monoclonal antibodies. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed that strain 93246 has two LOS bands. The estimated size of the major band is approximately 3.8 kDa, and that of the minor band is approximately 3.6 kDa. The major LOS band reacted with N. meningitidis L1 antibody and the minor band reacted with N. meningitidis L8 antibody in immunoblotting analysis, indicating that strain 93246 expresses a N. meningitidis LOS of L1(8) pattern.
TABLE 2.
Phenotypic characteristics of strain 93246
| Strain | Amylopectin from sucrosea | Growth in Catlin medium | Precipitation of B capsule | Serotype, serosubtypeb | LOS immunotypec |
|---|---|---|---|---|---|
| N. meningitidis | |||||
| M986 | − | + | + | 2a, P1.2,5 | L3,7 |
| 93246 | + | − | − | 4, P1.14 | L1, (8) |
| N. polysaccharea | |||||
| 89357 | + | − | − | − | UD |
12 N. polysaccharea strains showed the same phenotype as strain 89357; 2 N. meningitidis control strains, A1 (serogroup A) and 126E (group C) and N. cinerea control strain 81176 showed the same phenotype as strain M986 (B).
Serotyping and serosubtyping using monoclonal antibodies against N. meningitidis PorB and PorA proteins.
Strain 93246 showed major L1 and minor L8 bands as LOS prototype strain 126E. UD, undetermined.
PCR amplification of selected genes.
Eleven genes at four loci, whose specificity for Neisseria has been well characterized in a natural diverse population, were chosen for this study. These genes in strain 93246 were evaluated by PCR, and the results were compared with those for 13 N. polysaccharea strains, 8 N. meningitidis strains, and 2 N. gonorrhoeae strains whose genes were partially or fully described previously (12, 14, 21, 29, 33, 36, 37) (Table 3). The PCR products of opcA, siaD, ams, lgtZ, lgtC, lgtD, and lgtH genes were detected in strain 93246. The results of this gene pattern suggest that strain 93246 is a group B N. meningitidis strain.
Among the strains from the different species, a striking diversity in distribution of genes was observed for siaD/orfA, ams, lgtZ, and lgtE. A PCR product of siaD/orfA was amplified in all 7 N. meningitidis control strains, but not in the 13 N. polysaccharea strains or the 2 N. gonorrhoeae control strains, using a set of primer pairs previously reported (2, 28). The results from N. meningitidis control strains were consistent with their serogroup classification except that a group C siaD product was detected from strain M158, originally reported as the prototype serogroup D strain (6). The ams gene was detected in all 13 N. polysaccharea strains but not in the 7 N. meningitidis strains and 2 N. gonorrhoeae strains. The lgtZ gene is rare and was only found in N. meningitidis. The lgtE gene was not amplified in 13 N. polysaccharea strains.
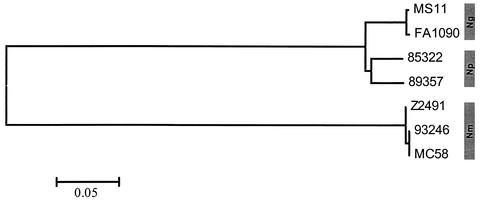
A 3,088-bp region of opcA from strain 93246 showed a genetic organization similar to that of N. meningitidis strain MC58, with a single copy of the IS1106 element upstream of opcA (29). Strain 93246 had an opcA coding region identical to that of MC58, but they differed at the promoter region. Strain 93246 has a C9 poly(C) tract, which suggests that opcA is not expressed (26). RT-PCR analysis showed no transcript of opcA in strain 93246, consistent with the poly(C) sequence prediction. The 2,122-bp upstream region in 93246 had 96.9% homology to the corresponding region in MC58. A phylogenetic tree was constructed from the nucleotide sequences of the opcA coding region for strain 93246 and six representative strains, including N. polysaccharea 85322 and 89357, N. meningitidis Z2491 and MC58, and N. gonorrhoeae FA1090 and MS11 (Fig. 1). The opcA from strain 93246 is identical to MC58 within the N. meningitidis opcA branch.
FIG. 1.
Phylogenetic relationships of the opcA genes. The diagram shows an unrooted tree constructed using neighboring-joining methods (MEGA2.1 program) from the nucleotide sequences of the coding region of the opcA gene.
A 1,715-bp sequence, containing 1,489 bp of the siaD coding region and a 226-bp flanking region from strain 93246, was identical to the corresponding region of siaD in N. meningitidis serogroup B strain B1940 (accession no. M95053) (11), except that strain 93246 had one more cytidine nucleotide at the poly(C) tract (G8) in the coding region. This G8 tract would cause a frameshift mutation and therefore inactivate the siaD gene in strain 93246.
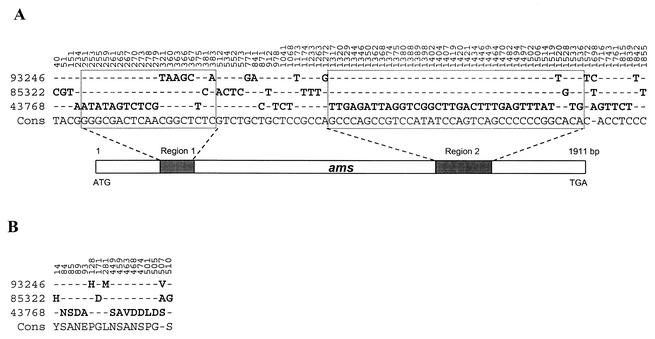
A 1,967-bp sequence, containing 1,911 bp of the ams coding region and a 56-bp flanking region from strain 93246, had 96.4% homology to the corresponding region previously reported in N. polysaccharea strain ATCC 43768 (accession no. AJ011781) (9). To understand the natural variation of ams gene in N. polysaccharea species, the 1,967-bp ams region from N. polysaccharea strain 85322 was sequenced as well (accession no. AY099335). The ams sequence from strain 89322 has 96.2 and 98.5% homology to the sequences from strain ATCC 43768 and strain 93246, respectively. Among the three sequences, 83 sites are polymorphic (4.3%) in the 1,911-bp coding region (Fig. 2A), resulting in 17 substitution sites in 636 amino acids of the Ams protein (2.7%) (Fig. 2B). Fifty-six nucleotide polymorphisms (67.5%) were located at two regions of 133 and 220 bp at positions 251 to 383 and 1317 to 1536 from the ATG start codon. These two fragments accounted for only 18.5% of the coding region, suggesting that they might be two hypervariable regions in ams or hot spots that serve for intragenic recombination. Seventeen amino acid substitutions were observed among three Ams protein variants (Fig. 2B), but they were not at the five critical positions for the enzymatic activity (25).
FIG. 2.
Details of polymorphic sites in the aligned ams genes (A) and the deduced proteins (B) from N. meningitidis strain 93246, N. polysaccharea strains 85322 (accession no. AY099335), and ATCC 43768 (accession no. AJ011781) (25). Numbers above indicate the position. Strain names are on the left. Cons indicates the consensus sequence, and a dash (-) indicates the nucleotide and amino acid identical to Cons. Each polymorphic site is in bold. Two highly polymorphic regions of 133 and 220 bp (Region 1 and Region 2) are indicated with a shadowed box within the ams gene.
DISCUSSION
In 1983, Riou et al. described 13 isolates of a new taxon of Neisseria sp. provisionally named Neisseria polysaccharea (23). Characteristics that differentiated N. polysaccharea from meningococcal isolates were D-glucan production when cultured in the presence of 5% sucrose, gamma-glutamylaminopeptidase activity, and a requirement for cysteine or cystine to be able to grow in the Catlin defined medium (5, 23, 24). Among these characteristics, the amylopectin production from starch-free medium containing sucrose is considered the most critical feature for differentiating between N. meningitidis and N. polysaccharea. However, the data from strain 93246 indicate that results from this kind of phenotypic test may not accurately speciate strains.
The orthologous opcA genes have been described for three Neisseria species, including N. meningitidis, N. gonorrhoeae, and N. polysaccharea (20, 33, 37). Most N. gonorrhoeae strains have an opcA gene, and about 60% N. meningitidis strains have opcA (27, 33). However, the opcA gene in N. polysaccharea species is rare, and interspecies diversity of opcA distinguishes opcA from different Neisseria species (37). The opcA gene was flanked by the IS1106 element at the upstream region in N. meningitidis but with orfX and orfY in N. gonorrhoeae and N. polysaccharea (33, 34, 37). Strain 93246 contains not only the N. meningitidis opcA gene but also an IS1106 element at the upstream region. Therefore, both the coding region and the genetic organization at the opcA locus identify strain 93246 as belonging to N. meningitidis.
The classification of N. meningitidis serogroups is based on the structural and immunological differences in the capsular polysaccharide. Traditionally, N. meningitidis serogrouping has been done by immunologic methods using polyclonal and monoclonal antibodies. However, the serogroups of some nonencapsulated and nongroupable N. meningitidis strains can be determined by PCR using a set of primers for the siaD gene and orf-2 (2, 28). Strain 93246 contains a siaD gene of serogroup B N. meningitidis, whereas all 13 N. polysaccharea strains tested do not contain siaD or orf-2, which strongly suggests that strain 93246 is N. meningitidis. Hammerschmidt et al. (13) reported that capsule phase variation in N. meningitidis serogroup B is regulated by a slipped-strand mispairing mechanism in the siaD gene. For all nonencapsulated strains analyzed, they found an insertion or deletion of cytidine at the poly(C) stretch within siaD, resulting in a frameshift and loss of capsule formation. The sequence analysis in this study revealed that the siaD gene in strain 93246 has also been inactivated by a frameshift mutation at the poly(C) tract, make it genetically identical to other nonencapsulated N. meningitidis.
The genetic organization of the lgt-1 locus, responsible for biosynthesis of the α chain of lipooligosaccharide, has been investigated in number of strains representing 14 Neisseria species (36). The genetic composition and arrangement at the lgt-1 locus in strain 93246 (lgtZCDH) is similar to those for N. meningitidis L1 prototype strain 126E (lgtZCDE) but differ from those for all 13 N. polysaccharea strains (lgtABH and lgtABCDH). Strain 93246 differs from 126E in that it has an lgtH gene at the 3′ end of the lgt-1 locus whereas strain 126E has a lgtE gene. However, the lgtE and lgtH genes probably have the same function (35). Use of immunoblotting in this study further showed that strains 93246 and 126E have the same major L1 LOS phenotypes. This suggested that both the LOS genotype and phenotype of strain 93246 are classified as N. meningitidis species. The expressed LOS pattern of strain 93246 is the same as that for N. meningitidis L1 prototype strain 126E.
In addition, our study confirms that strain 93246 does produce amylopectin from sucrose and also shows other phenotypic characteristics of N. polysaccharea. We also detected the ams gene encoding amylosucrase in strain 93246 but not in the other N. meningitidis and N. gonorrhoeae strains tested. Nonetheless, the GC content of the ams region (56.7%) in strain 93246 was slightly higher than the average GC contents of two meningococcal genomes (51.8 and 51.5%) (21, 29). The ams gene in strain 93246 was probably imported from N. polysaccharea species through horizontal genetic exchange, as the data from serotyping, serosubtyping, and three other genetic loci, opcA, siaD, and lgt-1, suggest that strain 93246 is N. meningitidis rather than N. polysaccharea. Strain 93246 was isolated from a vaginal specimen from a 27-year-old female in Canada in 1993. Isolation of N. meningitidis from the urogenital tract was considered unusual, but an increasing number of such cases were reported in the past years (3, 4, 22). This study added further information that N. meningitidis strain 93246 isolated from the vagina was able to produce amylopectin from sucrose.
In summary, the opcA, siaD, and lgt-1 loci are scattered on the meningococcal genome, with large distances of approximately 321 to 996 kb, respectively, separating these genes (21, 29), and therefore the basic framework of strain 93246 reassembles the N. meningitidis genome rather than N. polysaccharea. The amylosucrase gene was probably imported into strain 93246, which altered the strain's phenotypic characteristic. Strain 93246 represents an interesting example of the potential discrepancy between genotype and phenotype in bacterial classification. Thus, we conclude that the genetic analysis using multiple genetic loci is required to complement the traditional phenotypic classification for complete identification of atypical bacterial strains.
Acknowledgments
P. Zhu was supported by an NIH Fogarty Postdoctoral Fellowship (369VFFD018551).
We thank Carl E. Frasch for providing the bacterial strains of N. meningitidis, Michael J. Klutch for DNA sequencing, and Freyja Lynn for helpful comments.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1987. Whole-cell ELISA for typing Neisseria meningitidis with monoclonal antibodies. FEMS Microbiol. Lett. 48:367-371. [PubMed] [Google Scholar]
- 2.Arreaza, L., B. Alcala, C. Salcedo, and J. A. Vazquez. 2001. Interruption of siaD in a meningococcal carrier isolate mediated by an insertion sequence. Clin. Diagn. Lab. Immunol. 8:465-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell, C., H. Young, and S. S. Bain. 1978. Isolation of Neisseria meningitidis and Neisseria catarrhalis from the genitourinary tract and anal canal. Br. J. Vener. Dis. 54:41-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohler-Sommeregger, K., C. Poitschek, G. Furnsinn, S. Schuller-Petrovic, and A. Hirschl. 1987. Isolation of Neisseria meningitidis from the urethra and cervix. Wien. Klin. Wochenschr. 1:25-27. [PubMed] [Google Scholar]
- 5.Boquette, M. T., C. Marcos, and J. A. Saez-Nieto. 1986. Characterization of Neisseria polysaccharea sp. nov. (Riou, 1983) in previously identified noncapsular strains of Neisseria meningitidis. J. Clin. Microbiol. 23:973-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branham, S. E. 1958. Reference strains for the serologic groups of meningococcus. Int. Bull. Bacteriol. Nomencl. Taxon. 8:1.
- 7.Buttcher, V., T. Welsh, L. Willmitzer, and J. Kossmann. 1997. Cloning and characterization of the gene for amylosucrase from Neisseria polysaccharea: production of a linear α-1,4-glucan. J. Bacteriol. 179:3324-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, D. E., C. E. Frasch, J. B. Robbins, and H. A. Feldman. 1978. Serogroup identification of Neisseria meningitidis: comparison of an antiserum agar method with bacterial slide agglutination. J. Clin. Microbiol. 7:410-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Montalk, G. P., M. Remaud-Simeon, R. M. Willemot, V. Planchot, and P. Monsan. 1999. Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 181:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frosch, M., U. Edwards, K. Bousset, B. Krausse, and C. Weisgerber. 1991. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol. Microbiol. 5:1251-1263. [DOI] [PubMed] [Google Scholar]
- 12.Gotschlich, E. C. 1994. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J. Exp. Med. 180:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 14.Jennings, M. P., Y. N. Srikhanta, E. R. Moxon, M. Kramer, J. T. Poolman, B. Kuipers, and P. van der Ley. 1999. The genetic basis of the phase variation repertoire of lipopolysaccharide immunotypes in Neisseria meningitidis. Microbiology 145:3013-3021. [DOI] [PubMed] [Google Scholar]
- 15.Knapp. J. S. 1988. Historical perspectives and identification of Neisseria and related species. Clin. Microbiol. Rev. 1:415-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 17.Liu, T. Y., E. C. Gotschlich, E. K. Jonssen, and J. R. Wysocki. 1971. Studies on the meningococcal polysaccharides. I. Composition and chemical properties of the group A polysaccharide. J. Biol. Chem. 246:2849-2858. [PubMed] [Google Scholar]
- 18.Liu, T. Y., E. C. Gotschlich, F. T. Dunne, and E. K. Jonssen. 1971. Studies on the meningococcal polysaccharides. II. Composition and chemical properties of the group B and group C polysaccharide. J. Biol. Chem. 246:4703-4712. [PubMed] [Google Scholar]
- 19.Okada, G., and E. J. Hehre. 1974. New studies on amylosucrase, a bacterial α-d-glucosylase that directly converts sucrose to a glycogen-like α-glucan. J. Biol. Chem. 249:126-135. [PubMed] [Google Scholar]
- 20.Olyhoek, A. J., J. Sarkari, M. Bopp, G. Morelli, and M. Achtman. 1991. Cloning and expression in Escherichia coli of opc, the gene for an unusual class 5 outer membrane protein from Neisseria meningitidis (meningococci/surface antigen). Microb. Pathog. 11:249-257. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 22.Reiss-Levy, E., and J. Stephenson. 1976. Vaginal isolation Neisseria meningitidis in association with meningococcaemia. Aust. N. Z. J. Med. 6:487-489. [DOI] [PubMed] [Google Scholar]
- 23.Riou, J. Y., M. Guibourdenche, and M. Y. Popoff. 1983. A new taxon in the Genus Neisseria. Ann. Microbiol. (Paris) 134:257-267. [DOI] [PubMed] [Google Scholar]
- 24.Riou, J. Y., M. Guibourdenche, M. B. Perry, L. L. MacLean, and D. W. Griffith. 1986. Structure of the exocellular D-glucan produced by Neisseria polysaccharea. Can. J. Microbiol. 32:909-911. [DOI] [PubMed] [Google Scholar]
- 25.Sarcabal, P., M. Remaud-Simeon, R. Willemot, G. Potocki de Montalk, B. Svensson, and P. Monsan. 2000. Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474:33-37. [DOI] [PubMed] [Google Scholar]
- 26.Sarkari, J., N. Pandit, E. R. Moxon, and M. Achtman. 1994. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol. Microbiol. 13:207-217. [DOI] [PubMed] [Google Scholar]
- 27.Seiler, A., R. Reinhardt, J. Sarkari, D. A. Caugant, and M. Achtman. 1996. Allelic polymorphism and site-specific recombination in the opc locus of Neisseria meningitidis. Mol. Microbiol. 19:841-856. [DOI] [PubMed] [Google Scholar]
- 28.Taha, M. K. 2000. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J. Clin. Microbiol. 38:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 278:1809-1815. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, C. M., and C. I. Civin. 1991. Eight lipooligosaccharides of Neisseria meningitidis react with a monoclonal antibody which binds Lacto-N-neotetraose (Galβ1-4GlcNAcβ1-3Galβ1-Glc). Infect. Immun. 59:3604-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai, C. M., L. F. Mocca, and C. E. Frasch. 1987. Immunotype epitopes of Neisseria meningitidis lipooligosaccharide types 1 through 8. Infect. Immun. 55:1652-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, P., G. Morelli, and M. Achtman. 1999. The opcA and ΨopcB regions in Neisseria: genes, pseudogenes, deletions, insertion elements and DNA islands. Mol. Microbiol. 33:635-650. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, P., A. van der Ende, D. Falush, N. Brieske, G. Morelli, B. Linz, T. Popovic, I. G. Schuurman, R. A. Adegbola, K. Zurth, S. Gagneux, A. E. Platonov, J. Y. Riou, D. A. Caugant, P. Nicolas, and M. Achtman. 2001. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl. Acad. Sci. USA 98:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, P., M. J. Klutch, and C. M. Tsai. 2001. Genetic analysis of conservation and variation of lipooligosaccharide expression in two L8-immunotype strains of Neisseria meningitidis. FEMS Microbiol. Lett. 203:173-177. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, P., M. J. Klutch, M. J. Bash, R. Tsang, L. K. Ng, and C. M. Tsai. 2002. Genetic diversity of three lgt loci for biosynthesis of lipooligosaccharide (LOS) in Neisseria species. Microbiology 148:1833-1844. [DOI] [PubMed] [Google Scholar]
- 37.Zhu, P., M. J. Klutch, J. P. Derrick, S. M. Prince, R. S. W. Tsang, and C. M. Tsai. Identification of opcA gene in Neisseria polysaccharea: interspecies diversity of Opc protein family. Gene, in press. [DOI] [PubMed]