Abstract
One-hundred eight Mycobacterium avium isolates from pigs, humans, birds, and bovines were typed by the IS1245-based restriction fragment length polymorphism (RFLP) method and PCR-restriction enzyme analysis (PRA) of hsp65. Nine clusters of isolates showing more than 80% similarity in their RFLP profiles were detected. The largest cluster (cluster B) included 32 of 79 pig isolates (40.5%), 3 of 25 human isolates (12%), and 1 of 2 bovine isolates, comprising 33% of all isolates. The second largest cluster (cluster A) included 18 pig isolates (22.8%) and 6 human isolates (24%). Six smaller clusters included six pig isolates (clusters C and D), four and two human isolates (clusters E and F, respectively), two pig isolates (cluster I), and two pig isolates plus one bovine isolate and the avian purified protein derivative strain (cluster H). Cluster G represented the “bird-type” profile and included the bird isolate in this series, one pig isolate, plus reference strain R13. PRA revealed four allelic variants. Seventy-seven isolates were identified as M. avium PRA variant I, 24 were identified as M. avium PRA variant II, 6 were identified as M. avium PRA variant III, and 1 was identified as M. avium PRA variant IV. Except for three isolates from cluster B, each of the RFLP clusters was associated with a single PRA pattern. Isolates with unique (nonclustered) RFLP profiles were distributed between PRA variants I and II, and there was one unique isolate of PRA variant IV. These observations are consistent with divergent evolution within M. avium, resulting in the emergence of distinct lineages with particular competence to infect animals and humans.
Disseminated infection due to isolates of the Mycobacterium avium complex (MAC) was rare prior to the human immunodeficiency virus (HIV) pandemic, but disseminated MAC infection emerged as the most prevalent disseminated bacterial infection in patients with advanced AIDS. MAC, which is widespread in the environment (22), includes isolates of the closely related species M. avium and M. intracellulare and probably other less well defined genospecies (7).
Members of MAC can be classified into serotypes on the basis of differences in the C-mycoside glycopeptidolipids. Saito et al. (16), using 16S rRNA probes, assigned serotypes 1 to 6, 8 to 11, and 21 to M. avium and serotypes 7, 12 to 20, and 25 to M. intracellulare. Certain M. avium serotypes (serotypes 1, 4, and 8) have been reported to predominate among isolates from AIDS patients (25). Although these same serotypes have been isolated from the environment, this technique does not have the discriminatory power needed to identify sources of transmission.
Although M. avium has been associated with a variety of diseases in both animals and humans, the specific routes of infection are not well defined. Person-to-person transmission has not been demonstrated. Ingestion of environmental organisms followed by invasion through the gastrointestinal tract has been suggested as the main route of infection in AIDS patients because the organisms are frequently isolated from the stools of these patients (3). There is also an important positive correlation between the presence of MAC in respiratory samples and the subsequent development of disseminated disease (8). Immunocompetent individuals can have asymptomatic respiratory and intestinal colonization but rarely develop disseminated disease (7). No specific virulence factors have been identified to explain why, of the more than 100 species of nontuberculous mycobacteria, M. avium is the most common cause of infection in humans.
Investigation of these questions is complicated by the phenotypic and genotypic heterogeneity of M. avium. On the basis of its host range characteristics and pathogenicity, M. avium was recently subdivided into subspecies: M. avium subsp. avium; M. avium subsp. silvaticum or wood pigeon bacillus; and M. avium subsp. paratuberculosis, which is responsible for Johne's disease in cattle (23). It has been proposed that M. avium isolates from birds represent a separate taxon (14).
A considerable number of genetic tools for identification and typing of MAC isolates have become available in recent years. The most widely used system is a commercial hybridization assay (AccuProbe; Gen-Probe, San Diego, Calif.), in which species-specific labeled DNA probes bind to rRNA sequences. Specific probes are available for the identification of M. avium and M. intracellulare, but some MAC serovars do not hybridize with either species-specific probe and are designated “MAC-other” (19). Amplification of the DT1 and DT6 fragments was considered equally sensitive for species identification of M. avium and M. intracellulare (4). Insertion sequences IS900 (5), IS901 (10), IS1245 (6), and IS1311 (15) can be used as markers to make further subdivisions among M. avium isolates.
PCR-restriction enzyme analysis (PRA) is a method of identifying species of mycobacteria on the basis of analysis of restriction patterns in digests of a PCR-amplified fragment of hsp65 (20). In the initial report of the method, all clinical M. avium isolates examined demonstrated a single PRA type. In our previous studies of isolates obtained from animals and humans (11, 12), we identified three additional allelic variants. In this report we describe a comprehensive genotypic analysis of 108 M. avium isolates from pigs, humans, birds, and bovines by IS1245- and IS1311-based restriction fragment length polymorphism (RFLP) analysis, PRA, and PCR-amplification of DT1 and DT6.
MATERIALS AND METHODS
Isolates.
The study included 108 isolates from various species: 79 pig, 25 human, 2 bovine, and 2 bird isolates. The pig isolates, each of which was from a single infected animal, were cultured from animals slaughtered between November 1997 and July 1998 during an outbreak in an important center of swine meat production in the southern region of Brazil and were obtained from Empresa Brasileira de Pesquisa Agropeudria (Santa Catarina, Brazil).
Of the 25 human isolates, 23 were cultured from individual patients, of whom 7 were HIV positive, 1 was HIV negative, and the remainder were of unknown HIV status. Three of these patients had mixed infections with M. avium plus M. kansasii, M. tuberculosis, and M. lentiflavum, respectively (data not shown). The remaining two human isolates of M. avium were cultured from a single HIV-positive patient with a polyclonal infection (12). All of the clinical isolates were identified at Instituto Adolfo Lutz, São Paulo, Brazil, between July and September 1998, after the implementation of highly active antiretroviral therapy in Brazil.
Two bovine isolates and one bird isolate, cultured in 1996 and 1997, plus avian strain D4, which is used for the production of avian purified protein derivative, were obtained from the collection of the Instituto Biológico, São Paulo, Brazil. Each of the contributing institutions cited represent reference laboratories for the isolation and identification of mycobacteria from animals and humans in Brazil and are engaged in disease control.
Two M. avium reference strains, strain R13 (the “bird-type” strain) and strain IWGMT49 (originally isolated from a pig), were kindly provided by Dick van Soolingen (National Institute of Public Health and the Environment, Bilthoven, The Netherlands) and were included in all gels used for RFLP analysis.
Isolates were kept at −80°C in Middlebrook 7H9 liquid medium (Difco Laboratories, Detroit, Mich.) supplemented with OADC (oleic acid, albumin, dextrose, and catalase; Difco) plus 15% glycerol. Only isolated colonies of M. avium were included in the RFLP analysis.
Identification of mycobacteria.
DNA from all isolates was obtained by three freeze-boil cycles of a loopful of mycobacteria grown on solid medium (Löwenstein-Jensen or Stonebrink) and suspended in 0.4 ml of TE (10 mM Tris, 1 mM EDTA [pH 8.0]) with 1% Triton X-100. Five to 10 μl was used for PCR.
Identification of M. avium was based on the results of PRA (20), amplification of M. avium-specific insertion sequences IS1245 (6) and IS1311 (15), and amplification of MAC-specific fragments DT1 and DT6 (21).
Biochemical identification and probe testing for the detection of rRNA specific for M. avium (AccuProbe; Gen Probe) were performed with selected isolates.
Strain typing.
Strain typing was performed by RFLP analysis, as described by van Soolingen et al. (24). Briefly, isolates were grown in 5 ml of Middlebrook 7H9 liquid medium-OADC. After 7 to 15 days, the culture was transferred to a volume of 50 ml and incubated at 37°C in a shaker for an additional 10 to 15 days. The cells were centrifuged and resuspended in 0.4 ml of TE for DNA extraction, as described previously (24).
DNA was digested with PvuII, and 2 μg of the DNA fragments generated was electrophoretically separated on a 24-cm 0.8% agarose gel until the 872-bp fragment of HaeIII-digested φX174 DNA (external DNA size marker; Invitrogen, Carlsbad, Calif.) reached a distance of 19 cm from the slots of the gel. A mixture of a PvuII-digested supercoiled DNA ladder (Invitrogen) and HaeIII-digested φX174 DNA was used as an internal DNA size marker. DNA was blotted onto nylon membranes (Hybond N-plus; Amersham Biosciences do Brasil Ltda., São Paulo, Brazil) and probed in different PCR experiments with probes obtained by PCR from IS1245 or IS1311. The internal marker was also labeled and used as a probe. The probes were labeled and detected with the ECL direct system (Amersham) or the RPN 3690 kit (Amersham) under stringent conditions. The membranes were exposed to X-ray film (X-OMAT; Kodak, Rochester, N.Y.).
Analysis of RFLP patterns.
RFLP analysis was performed at least twice with each isolate. The best images were selected and analyzed with the GelCompar II (version 2.5) program (Applied Maths, Kortrijk, Belgium). Autoradiograms of the IS1245 fingerprints were superimposed on the autoradiograms of internal markers for normalization. The Dice coefficient of similarity for all pairwise comparisons of patterns was calculated. A dendrogram of pattern relatedness among the strains was constructed by the unweighted pair group method with arithmetic averages clustering method.
RESULTS
The species identification and biochemical characterization of the 108 isolates tested in the present study were reported previously (11, 12, 17, 18) and are summarized in Table 1. Seventy-seven isolates (71.3%) were identified as M. avium PRA allelic variant I, 24 (22.2%) were identified as M. avium PRA allelic variant II, 6 (5.5%) were identified as M. avium PRA allelic variant III, and 1 (0.9%) was identified as M. avium PRA allelic variant IV. Positive amplification with primers from IS1245, IS1311, and DT6 sequences was obtained with DNA from all isolates; and no positive amplification was obtained with primers from DT1 sequence. Biochemical identification and the AccuProbe test with an M. avium-specific probe were performed with selected isolates of each PRA allelic variant. Taken together, the results confirmed the identification of all isolates as M. avium.
TABLE 1.
Isolates included in this studya
| No. of isolates | Host | Source | PRA type | RFLP type | AccuProbe reaction result |
|---|---|---|---|---|---|
| 18 | Pig | Lymph node | I | A | + |
| 29 | Pig | Lymph node | I | B | + |
| 3 | Pig | Lymph node | II | B | |
| 6 | Pig | Lymph node | II | C | + |
| 6 | Pig | Lymph node | III | D | + |
| 1 | Pig | Lymph node | I | G | |
| 2 | Pig | Lymph node | I | H | |
| 2 | Pig | Lymph node | II | I | |
| 5 | Pig | Lymph node | I | Unique | |
| 7 | Pig | Lymph node | II | Unique | |
| 1 | Human, HIV+ | Sputum | I | A | |
| 1 | Human, HIV− | Sputum | I | A | |
| 1 | Human, HIV+ | Bone marrow | I | A | |
| 1 | Human, ND | Bone marrow | I | A | |
| 1 | Human, HIV+ | Blood | I | A | + |
| 1 | Human, ND | Blood | I | A | |
| 1 | Human, HIV+ | Sputum | I | B | |
| 1 | Human, ND | Sputum | I | B | |
| 1 | Human, ND | Blood | I | B | |
| 3 | Human, ND | Sputum | II | E | |
| 1 | Human, ND | Bronchial fluid | II | E | |
| 1 | Human, HIV+ | Gastric fluid | I | F | |
| 1 | Human, HIV+ | Blood | I | F | |
| 1 | Human, HIV+ | Lymph node | I | Unique | |
| 1 | Human, ND | Lymph node | I | Unique | |
| 1 | Human, ND | Sputum | I | Unique | |
| 2 | Human, ND | Sputum | II | Unique | |
| 1 | Human, HIV+ | Bone marrow | IV | Unique | + |
| 1 | Human, ND | Abscess | I | Negative | + |
| 1 | Human, ND | Lymph node | I | Negative | + |
| 1 | Human, ND | Biopsy | I | Negative | + |
| 1 | Human, HIV+ | Blood | I | Negative | + |
| 1 | Strain D4 | I | H | ||
| 1 | Bovine | Lung | I | H | |
| 1 | Bovine | Lymph node | I | B | |
| 1 | Crane | Intestine | I | G |
Abbreviations and symbols: I, II, III, and IV, M. avium PRA variants (11, 12); A to I, groups of isolates showing similarities greater than 80%, as defined by IS1245-RFLP typing (Fig. 1); HIV+, HIV positive; HIV−, HIV negative; ND, nondetermined HIV infection status; +, result of AccuProbe analysis with M. avium-specific probe performed with one isolate in the group.
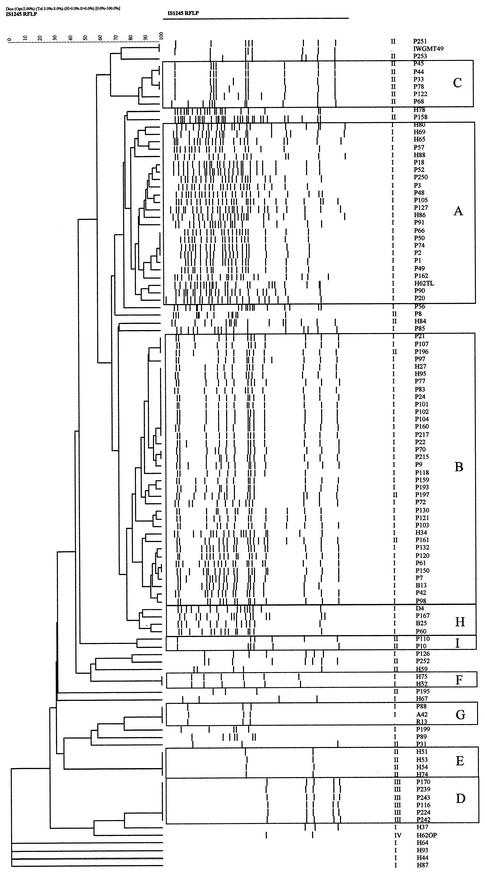
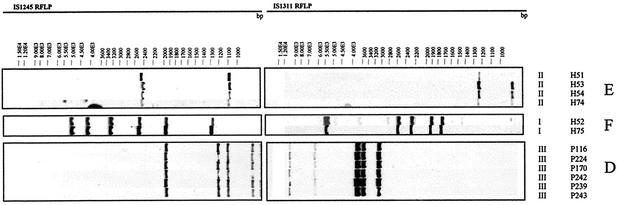
Analysis of the RFLP profiles for all isolates performed with the GelCompar II program provided the dendrogram shown in Fig. 1. Nine clusters of isolates whose RFLP profiles showed more than 80% similarity were detected. The largest cluster (cluster B) included 32 of 79 pig isolates (40.5%), 3 of 25 human isolates (12%), and 1 of 2 bovine isolates, comprising 33% of all isolates. The RFLP pattern of cluster B included a motif of seven bands present in the RFLP pattern of pig isolate IWGMT49, which belongs to serovar 10 (14) (Fig. 2). The PRA patterns for all but three isolates were M. avium PRA variant I. The second largest cluster (cluster A) included 18 of 79 pig isolates (22.8%) and 6 human isolates (24%), all of which were M. avium PRA pattern I. Six smaller clusters included six isolates from pigs (clusters C and D), four and two human isolates (clusters E and F, respectively), two pig isolates (cluster I), and two pig isolates plus one bovine isolate and the avian purified protein derivative strain (cluster H). The clustering of isolates with low IS1245 copy numbers in clusters D, E, and F was confirmed by hybridization with the IS1311-specific probe and PRA (Fig. 3). Cluster G represented the bird-type profile and included, besides reference strain R13, the crane isolate and one pig isolate. Four human isolates lacked IS1245-specific bands in the RFLP experiments, but the result of PCR with IS1245-specific primers was positive and confirmed by sequencing. Isolated colonies of these cultures were retested, with the same results. Lack of hybridization was not related to problems with DNA, as hybridization with the DT6-specific probe was positive (data not shown).
FIG. 1.
Dendrogram of the 108 IS1245-based RFLP profiles of M. avium isolates from pigs, humans, birds, and bovines. The numbers at the top represent percent relatedness. Isolates showing more than 80% similarity are boxed. Letters indicate the cluster to which the isolates belong. The two columns on the right present PRA patterns and isolate names, respectively.
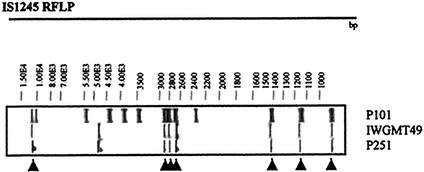
FIG. 2.
IS1245-based RFLP patterns of strain IWGMT49 (serotype 10) and isolate P251, showing identical band patterns, and isolate P101 from cluster B, which shows seven hybridizing bands at the same positions (indicated by arrowheads) as strains IWGMT49 and P251.
FIG. 3.
Isolates with low IS1245 copy numbers belonging to clusters E, F, and D were probed with IS1311. In each cluster, indistinguishable patterns were obtained by RFLP with both probes and by PRA, confirming that the isolates in each cluster represent isolates with closely related genotypes.
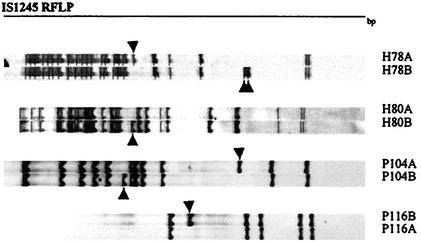
During the replicate RFLP analyses, minor one- or two-band changes were detected among individual colonies of four strains (strains H78, H80, P104, and P44) (Fig. 4).
FIG. 4.
Variations among IS1245-based RFLP patterns of single colonies from isolates H78, H80, P104, and P116. Arrows indicate the positions where changes occurred.
DISCUSSION
Previous studies of M. avium isolates by molecular typing by IS1245-RFLP analysis or pulsed-field gel electrophoresis have emphasized the remarkable diversity of genotypes. In this study, a less stringent cutoff of 80% was chosen to facilitate identification of clusters of isolates that differ at a modest fraction of the insertion sequence loci. In fact, although this study did not systematically investigate the frequency of recent transposition events, we observed one- or two-band changes in the IS1245-based RFLP profiles among subcultures of single colonies of four strains (Fig. 4). Pestel-Caron and Arbeit (13) observed the frequent transposition of IS1245 in vivo among independent isolates collected at a single time, isolates collected from individual patients over time, and isolates collected from multiple patients. Bauer and Andersen (1) confirmed the frequent observation of one- to two-band changes when they analyzed single colonies of M. avium from cultures. Thus, among isolates with ≥80% similarity, at least some of the variations in RFLP profiles could represent relatively recent events.
Although few isolates had indistinguishable IS1245 profiles, the RFLP analysis defined nine clusters that included 87 (80.6%) of the isolates studied. The most prevalent cluster (cluster B) included more than one-third of all isolates and infected pigs, bovines, and humans. Two human isolates and eight pig isolates showed indistinguishable RFLP and PRA patterns. All but three isolates presented M. avium PRA pattern I. Three isolates showing M. avium PRA pattern II could represent convergent evolution of PRA types, which seems implausible for synonymous alleles, or inappropriate assignment of the isolates to cluster B. Sequencing of additional loci might help resolve this issue.
The pattern for the strains in cluster B included the basic seven-band motif found for serotype 10 strains (14). The RFLP profile of the serotype 10 prototype strain was previously found for two of three isolates from pigs from Brazil by Ritacco et al. (14) and in this work (isolate P251). A similar pattern was also previously seen for a dog isolate (6). Patterns like that found for the serotype 10 strains were also found for human and animal isolates from Europe, where this serotype does not seem to be highly prevalent (2, 9, 14). Isolates with this RFLP profile were found in pigs, bovines, and humans and were predominant in our study. The high frequency could be influenced by the inclusion of strains from an epidemic outbreak.
The second most prevalent RFLP profile (the profile for strains in cluster A) was similar to the multiband profiles described in previous studies (2, 9, 14). The majority of human and pig M. avium isolates in those studies belonged in this group, suggesting that strains with genotypes that result in multiband profiles are particularly pathogenic for animals and humans (Fig. 1).
Cluster D represented a set of interesting isolates characterized by a particular PRA pattern (M. avium PRA variant III) and a five-band IS1245-based RFLP pattern; Ritacco et al. (14) did not observe these patterns among the M. avium serovars that they studied. In the present study, all these isolates were obtained from pigs in a restricted geographical region in the south of Brazil, raising the possibility that they represent the local spread of a distinct MAC isolate. However, Smole et al. (19) recently reported on human and environmental isolates that had M. avium PRA pattern III and variable number of bands by IS1245-based RFLP analysis, suggesting that M. avium PRA pattern III isolates have a wider distribution.
In a previous publication (11) we reported that PCR with IS1245-specific primers and lysates of cluster D isolates prepared by the freeze-boil method was unsuccessful. Similar negative amplification of IS1245 was also observed in human isolate H62OP, which exhibited a different PRA pattern (M. avium PRA variant IV) and a nonclustered two-band IS1245-RFLP pattern. The presence of nonspecific PCR inhibitors was ruled out by positive amplifications for other loci (hsp65, DT6, IS1311). In the present studies, we successfully performed PCR with IS1245-specific primers using purified DNA as the template, but we have not determined why our initial attempts were repeatedly negative.
Four isolates that reacted with the AccuProbe reagent specific for M. avium and that were positive for amplification by PCR with IS1245- and IS1311-specific primers were nevertheless repeatedly negative by IS1245-based RFLP analysis. The same membranes with all four isolates were positive when they were subsequently probed by RFLP analysis with a PCR-derived DT6-specific fragment (data not shown), confirming that the restriction digests and transfers were technically successful. Different investigators have described the absence of hybridization with the IS1245-specific probe in RFLP experiments with M. avium isolates, but PCR results have not been reported (2, 14). Komijn et al. (9) stated that the DNA of 25% of human isolates did not hybridize to IS1245, suggesting the existence of subgroups within MAC. At present, we do not have a definitive explanation for the negative hybridization with the IS1245-specific probe, but it will be investigated further.
The bird-type profile was found for the R13 reference strain, an isolate from a bird in a zoo (a crane [Grus virgo Linnaeus 1758]), and a pig isolate. It has been proposed that isolates with this profile represent a distinct subset of M. avium (9, 14). Komijn et al. (9) studied 47 M. avium isolates from birds and showed that they all belonged to a separate taxon within MAC and presented this three-band RFLP pattern. The bird-type RFLP pattern was observed only as an exception among isolates from other hosts. Our results are in accordance with the hypothesis that isolates with this profile represent a distinct subset of M. avium. M. avium isolates from pigs were obtained in a region dedicated to pig and fowl production. Despite an outbreak of M. avium infection in pigs, no concomitant infections were detected in poultry and only one pig isolate presented the bird-type RFLP profile.
In molecular typing studies of M. tuberculosis strains, clustered isolates with the same or highly similar IS6110-based RFLP patterns have typically been interpreted to represent recently transmitted isolates that are epidemiologically related. In contrast, nonclustered isolates have been considered consistent with reactivation from previous infections. In M. avium infections, person-to-person or animal-to-person transmission, as well as the occurrence of latent infections and reactivations, has never been proven. Therefore, the clustering of M. avium RFLP patterns must have different interpretations. Isolates in clusters could represent strains with increased virulence. Otherwise, isolates that are clustered could have better resistance to environmental stress, resulting in greater exposure.
The virulence potential of selected isolates belonging to four major clusters (clusters A, B, C, and D) was analyzed by inoculation of the isolates into hamsters (Mesocricetus auratus). Preliminary results suggest that isolates from clusters A and B (M. avium PRA pattern I) were more virulent than isolates from clusters C and D (M. avium PRA patterns II and III) (data not shown). A recent independent publication reported that isolates with M. avium PRA pattern I had an increased propensity to cause invasive, disseminated infection and that isolates with M. avium PRA patterns II and III were more predominant among environmental isolates (19). These results suggest that the different M. avium genotypes identified by these methods may differ in terms of their virulence determinants that influence the transmission and occurrence of disease.
In conclusion, molecular typing techniques such as IS1245-RFLP analysis and PRA effectively identified M. avium isolates with distinct traits and defined genetic lineages of interest for future pathogenicity studies.
Acknowledgments
Nelson Morés and Eliana Roxo are acknowledged for providing swine, bovine, and bird M. avium isolates; and Suely Yoko Mizuka Ueki is acknowledged for providing human isolates and biochemical identification. Robert Arbeit is acknowledged for fruitful discussions.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
REFERENCES
- 1.Bauer, J., and A. B. Andersen. 1999. Stability of insertion sequence IS1245, a marker for differentiation of Mycobacterium avium strains. J. Clin. Microbiol. 37:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, J., A. B. Andersen, D. Askgaard, S. B. Giese, and B. Larsen. 1999. Typing of clinical Mycobacterium avium complex strains cultured during a 2-year period in Denmark by using IS1245. J. Clin. Microbiol. 37:600-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damsker, B., and E. J. Bottone. 1985. Mycobacterium avium-Mycobacterium intracellulare from the intestinal tracts of patients with the acquired immunodeficiency syndrome: concepts regarding acquisition and pathogenesis. J. Infect. Dis. 151:179-181. [DOI] [PubMed] [Google Scholar]
- 4.Devallois, A., M. Picardeau, K. S. Goh, C. Sola, V. Vincent, and N. Rastogi. 1996. Comparative evaluation of PCR and commercial DNA probes for detection and identification to species level of Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 34:2756-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inderlied, C. B., C. A. Kemper, and L. E. M. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson, M. A., P. C. Hopewell, D. M. Yajko, W. K. Hadley, E. Lazarus, P. K. Mohanty, G. W. Modin, D. W. Feigal, P. S. Cusick, and M. A. Sande. 1991. Natural history of disseminated Mycobacterium avium complex infection in AIDS. J. Infect. Dis. 164:994-998. [DOI] [PubMed] [Google Scholar]
- 9.Komijn, R. E., P. E. W. Haas, M. M. E. Schneider, T. Eger, J. H. M. Nieuwenhuijs, R. J. van den Hoek, D. Bakker, F. G. van Zijd Erveld, and D. van Soolingen. 1999. Prevalence of Mycobacterium avium in slaughter pigs in The Netherlands and comparison of IS1245: restriction fragment length polymorphism patterns of porcine and human isolates. J. Clin. Microbiol. 37:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. McFadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 5:2265-2272. [DOI] [PubMed] [Google Scholar]
- 11.Leão, S. C., M. R. S. Briones, M. P. Sircili, S. C. Balian, N. Mores, and J. S. Ferrreira-Neto. 1999. Identification of two novel Mycobacterium avium allelic variants by PCR-restriction enzyme analysis in pig and human isolates from Brazil. J. Clin. Microbiol. 37:2592-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira, R. S., M. P. Sircili, S. Y. M. Ueki, M. A. Telles, B. Schinabel, M. R. S. Briones, and S. C. Leão. 2000. PCR-restriction enzyme analysis of a bone marrow isolate from a human immunodeficiency virus-positive patient discloses polyclonal infection with two Mycobacterium avium strains. J. Clin. Microbiol. 38:4643-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestel-Caron, M., and R. D. Arbeit. 1998. Characterization of IS1245 for strain typing of Mycobacterium avium. J. Clin. Microbiol. 36:1859-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritacco, V., K. Kremer, T. van der Laan, J. E. M. Pijnenburg, P. E. W. Haas, and D. van Soolingen. 1998. Use of IS901 and IS1245 in RFLP typing of Mycobacterium avium complex: relatedness among serovar reference strains, human and animal isolates. Int. J. Tuberc. Lung. Dis. 2:242-251. [PubMed] [Google Scholar]
- 15.Roiz, M. P., E. Palenque, C. Guerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito, H., H. Tomioka, K. Sato, H. Tasaka, and D. J. Dawson. 1990. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 28:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva, C. F., S. Y. M. Ueki, D. C. P. Geiger, and S. C. Leão. 2000. hsp65 PCR-restriction enzyme analysis (PRA) for identification of mycobacteria in the clinical laboratory. Rev. Inst. Med. Trop. Sao Paulo 43:25-28. [DOI] [PubMed] [Google Scholar]
- 18.Sircili, M. P., E. Roxo, and S. C. Leão. 1999. Discrimination of members of the Mycobacterium avium complex by polymerase chain reaction. Rev. Microbiol. 30:144-148. [Google Scholar]
- 19.Smole, S. C., F. McAleese, J. Ngampasutadol, C. F. von Reyn, and R. Arbeit. 2002. Clinical and epidemiological correlates of genotypes within the Mycobacterium avium complex defined by restriction and sequence analysis of hsp65. J. Clin. Microbiol. 40:3374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thierry, D., V. Vincent, F. Clement, and J. L. Guesdon. 1993. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J. Clin. Microbiol. 31:1048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoen, C. O. 1994. Mycobacterium avium infections in animals. Res. Microbiol. 145:169-172. [DOI] [PubMed] [Google Scholar]
- 23.Thorel, M. F., M. Kirchevsky, and V. Lévy-Frébault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, amended description of Mycobacterium avium, and description of M. avium subsp. avium, M. avium subsp. silvaticum, and M. avium subsp. paratuberculosis. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 24.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leão, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakrus, M. A., and R. C. Good. 1990. Geographic distribution, frequency, and specimen source of Mycobacterium avium complex serotypes isolated from patients with acquired immunodeficiency syndrome. J. Clin. Microbiol. 157:863-867. [DOI] [PMC free article] [PubMed] [Google Scholar]