Abstract
Rapid identification of microbial pathogens reduces infection-related morbidity and mortality of hospitalized patients. Raman spectra and Fourier transform infrared (IR) spectra constitute highly specific spectroscopic fingerprints of microorganisms by which they can be identified. Little biomass is required, so that spectra of microcolonies can be obtained. A prospective clinical study was carried out in which the causative pathogens of bloodstream infections in hospitalized patients were identified. Reference libraries of Raman and IR spectra of bacterial and yeast pathogens highly prevalent in bloodstream infections were created. They were used to develop identification models based on linear discriminant analysis and artificial neural networks. These models were tested by carrying out vibrational spectroscopic identification in parallel with routine diagnostic phenotypic identification. Whereas routine identification has a typical turnaround time of 1 to 2 days, Raman and IR spectra of microcolonies were collected 6 to 8 h after microbial growth was detected by an automated blood culture system. One hundred fifteen samples were analyzed by Raman spectroscopy, of which 109 contained bacteria and 6 contained yeasts. One hundred twenty-one samples were analyzed by IR spectroscopy. Of these, 114 yielded bacteria and 7 were positive for yeasts. High identification accuracy was achieved in both the Raman (92.2%, 106 of 115) and IR (98.3%, 119 of 121) studies. Vibrational spectroscopic techniques enable simple, rapid, and accurate microbial identification. These advantages can be easily transferred to other applications in diagnostic microbiology, e.g., to accelerate identification of fastidious microorganisms.
The time required for the identification of pathogens is an important determinant of infection-related mortality rates of hospitalized patients. Rapid identification techniques significantly reduce mortality and costs associated with infectious diseases (3). Most commercially available identification systems in routine use in hospitals are based on the physiological and nutritional characteristics of microorganisms. These systems require a pure microbial culture and a large inoculum (13). Consequently, a turnaround time of 24 h (e.g., for Staphylococcus aureus) to up to 5 days (for Candida species) between receipt of patient material and presentation of identification results to the clinician is common. Therefore, empirical treatment with broad-spectrum antibiotics is often started while awaiting further identification of the pathogens. It has been reported that, as a result of this, 10 to 30% of patients suffering from bloodstream infections in intensive care units (ICUs) do not initially receive the correct antimicrobial therapy (9, 23). Mortality rates in this group have been reported to be 30 to 60% higher than in the group that promptly receives appropriate therapy (6, 9). Apart from the risk that the empirical treatment may not be effective at all, this practice may lead to adverse toxic side effects (21) and is known to aggravate problems with resistance to antimicrobial agents (7). Early identification enables the clinician to precisely target a pathogen with the most effective antimicrobial agent.
Novel genotypic approaches to the rapid identification of clinically relevant microorganisms are finding their way into the field of clinical diagnostic microbiology (17, 19). For example, amplification of specific gene sequences by PCR assay enables very sensitive methods to be developed (17). Furthermore, microbes can be detected and identified in complex matrices by using fluorescence in situ hybridization targeting the conserved 16S rRNA (1).
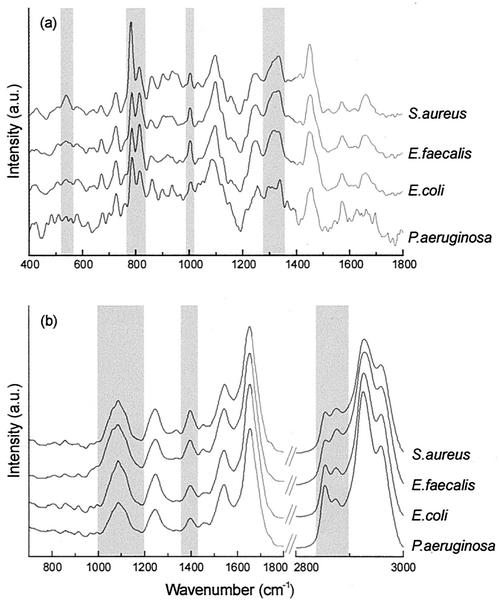
A radically different approach to the development of identification methods is based on spectroscopic techniques (4, 14, 15). These techniques are characterized by a minimum of sample handling: no extractions, amplifications, labeling, or staining steps of any kind are required. We have developed Raman and Fourier transform (FT)-infrared (IR) spectroscopic techniques for the rapid and accurate identification of clinically relevant microorganisms. Vibrational spectra reflect the overall molecular composition of a sample. Since different organisms differ in overall molecular composition, their Raman and FT-IR spectra will also be different (Fig. 1). The spectra can serve as spectroscopic fingerprints that enable highly accurate identification of microorganisms (14). Enterococcus species, such as Enterococcus hirae, E. durans, and the vancomycin-resistant species E. casseliflavus and E. gallinarum, were correctly identified by both FT-IR and Raman spectroscopy, while most of the routinely used identification systems perform poorly (8). Moreover, only a very small inoculum is required to obtain spectra. Within 6 to 8 h after starting a culture on a standard solid culture medium, most commonly encountered pathogens develop microcolonies that are 10 to 100 μm in diameter from which reproducible Raman and FT-IR spectra can be obtained. By Raman spectroscopy, spectra can be collected directly from microcolonies on solid culture medium. Confocal signal detection (18) is used to adapt the measurement volume to the thin microcolonies, thereby minimizing signal contributions from the culture medium. A specially designed subtraction algorithm corrects for remaining signal contributions from the culture medium (11). FT-IR spectra can be collected from imprints of microcolonies on an IR-transparent substrate (14). Microcolonies are transferred from the solid culture medium to the substrate with a special stamping device (15a), and then the imprint is allowed to dry.
FIG. 1.
Typical Raman (a) and FT-IR (b) spectra of four microorganisms included in the reference database used for the identification of pathogens isolated from blood. The spectra have been displaced vertically on the intensity axis. Some marked spectral differences between the species have been highlighted. a.u., arbitrary units.
In order to optimize the spectroscopic contrast between different microorganisms, other sources of variation in molecular composition must be minimized. This requires standardization of culturing conditions, in particular, the culture medium, incubation temperature, and culturing time. Culturing time is important because microbial colonies growing on a solid culture plate become increasingly heterogeneous with respect to molecular composition. Such heterogeneity is absent in small microcolonies obtained after 6 to 8 h of culturing (2).
Standardization of culturing conditions and instrument parameters enables the creation of reference libraries of microorganism spectra. Such libraries are the foundation of microorganism identification algorithms based, for example, on linear discriminant analysis (LDA) (10) or on artificial neural networks (ANNs) (22). These enable the identification of an unknown microorganism on the basis of its Raman or FT-IR spectrum. Here we present results of the first prospective clinical study in which the causative pathogens of blood infections were identified by vibrational spectroscopic methods.
MATERIALS AND METHODS
Sample preparation.
For Raman spectroscopy, the reference strains were seeded at 103 CFU/ml in 10-ml blood samples from healthy volunteers. Aerobic culture vials (bacteria) or mycosis culture vials (yeasts) of the automated BACTEC 9240 blood culture system (Becton Dickinson, Cockeysville, Md.) were inoculated with these samples. When the culture vials were flagged as positive by the system, 100 μl of the liquid culture medium was plated on Mueller-Hinton medium (Merck, Darmstadt, Germany), which supports the growth of a wide range of bacteria, or Sabouraud medium plus 2% glucose (Merck) for yeasts. These cultures were incubated for 6 h at 37 or 30°C, respectively, prior to Raman spectroscopy of microcolonies. Patient blood samples that were positive according to the BACTEC system were treated in the manner described above.
For IR measurements of reference strains, three calibrated platinum loops (1 mm in diameter) of biomass from an overnight culture were suspended in 10 ml of prewarmed Luria broth (pancreatic digest of casein, NaCl, yeast extract; Merck). The suspensions were diluted 100-fold for yeasts and 1,000-fold for bacteria, and an aliquot of 100 μl of this dilution was spread onto prewarmed CASO agar plates (Merck) (bacteria) or Sabouraud agar plates plus 2% glucose (Merck) (yeasts). Positive patient samples from the BacT/Alert system (Organon Teknika, Eppelheim, Germany) were diluted 100-fold in Luria broth in order to remove the charcoal particles from the culture medium. Two aliquots of 100 μl were than spread on CASO medium and Sabouraud medium plus 2% glucose, respectively. After incubation for 6 to 8 h at 37°C, the microcolonies were transferred from the agar plate onto a ZnSe substrate with a specially designed stamping device (Ngo Thi et al., Biomed. Spectrosc.: Vib. Spectrosc. Other Novel Tech., Proc. Soc. Prof. Ind. Eng.). After drying in air for 15 min, the microbial spots deposited onto the IR-transparent plate were measured.
Phenotypic identification.
Microbial identification in routine clinical diagnostic laboratories was performed by phenotypic identification with the API and Vitek systems (both from bioMérieux, Marcy-l'Etoile, France).
Confocal Raman microspectroscopy.
Raman spectroscopic measurements were performed as previously described (10, 11). Briefly, the solid culture medium with microcolonies was placed directly under the microscope of a System 1000 Raman microspectrometer (Renishaw plc, Wotton-under-Edge, United Kingdom). An 80× near-IR objective (MIR Plan 80×/0.75; Olympus) was used to focus 100 to 150 mW of laser light (830 nm) on the sample and collect scattered light from the sample. Five microcolonies per plate were selected. Within each microcolony, spectra were obtained from 10 randomly chosen locations with a signal collection time of 30 s per measurement. For each sample measured, the 50 spectra thus obtained were averaged.
FT-IR microspectroscopy.
FT-IR spectra were recorded on an FT-IR microscope (IR Scope II interfaced with an IFS 28/B spectrometer; Bruker Optics, Karlsruhe, Germany), equipped with a motorized x-y stage, a 15× Cassegrain objective, and a broadband mercury cadmium telluride detector (12). All spectra were acquired over 256 scans, and 10 microcolonies per imprint were measured, resulting in a total measurement time of 18 min per sample.
Analysis of data.
Analysis of data was performed as described before (8, 10, 11; Opus I. R. Handbook, Bruker Optics). Reduction of data was performed by principal-component analysis. The maximum number of n − 1 principal components was calculated (n is the number of spectra in the analysis), typically accounting for 99 to 100% of the variation in the set of data. The reduced data served as the input for a hierarchical cluster analysis (HCA), an LDA, and an ANN analysis. HCA is a means of objectively analyzing groups in a set of data on the basis of spectral similarities. LDA and ANN analysis are both techniques by which to classify unknown samples into predetermined groups. On the basis of HCA groupings, LDA and ANN analysis models were constructed for species identification (see below).
Reference databases of microorganism spectra and development of microorganism identification models.
Separate databases of reference Raman and FT-IR spectra were created representing approximately 85% of the microbial species most commonly encountered in blood infections of patients treated in the ICUs of the University Hospital Rotterdam and the Rudolf Virchow Hospital (Berlin, Germany) (see Table 1). The strains included in the reference databases were either well-characterized clinical isolates or were obtained from culture collections.
TABLE 1.
Results of leave-one-out evaluation of the Raman and infrared prediction models on the basis of strains in the reference databases
| Organism(s) | Raman spectroscopy
|
IR spectroscopy
|
||
|---|---|---|---|---|
| No. (%) identified correctly | Misidentification (no. of strains) | No. (%) identified correctly | Misidentification (no. of strains) | |
| S. aureus | 9 (100) | 13 (100) | ||
| CNSa | 25 (96.2) | S. aureus (1) | 17 (94.4) | S. aureus (1) |
| E. coli | 16 (97.3) | E. cloacae (1) | 12 (100) | |
| E. cloacae | 9 (81.8) | E. aerogenes (2) | 6 (100) | |
| E. aerogenes | 7 (77.8) | E. cloacae (2) | 6 (100) | |
| P. aeruginosa | 9 (100) | 5 (100) | ||
| E. faecalis groupa | 8 (100) | 9 (100) | ||
| E. faecium groupa | 8 (100) | 9 (100) | ||
| Streptococcus spp.a | 8 (88.9) | E. faecalis (1) | 9 (82) | E. faecalis group (1) |
| Candida albicans | 6 (85.7) | C. kefyr (1) | 7 (87.5) | C. dubliniensis (1) |
| Candida dubliniensis | 5 (100) | |||
| Candida glabrata | 5 (100) | 5 (100) | ||
| Candida kefyr | 7 (100) | 4 (100) | ||
| Candida krusei | 7 (100) | 4 (100) | ||
| Candida tropicalis | 8 (100) | 5 (83.3) | C. albicans (1) | |
In the different species groups, the following species were included (number of strains in Raman database/number in IR database): CNS, S. epidermidis (10/6), S. schleiferi (3/2), S. saprophyticus (3/2), S. haemolyticus (3/2), S. capitis (3/2), S. lugdunensis (2/0), S. warneri (2/2), and S. hominis (0/2); Streptococcus spp., S. agalactiae (1/0), S. oralis (2/3), S. salivarus (2/2), S. pneumoniae (1/2), S. pyogenes (3/3), and Streptococcus variant Gr. A (0/1); Enterococcus faecium group, E. faecium (6/5), E. hirae (1/2), and E. durans (1/2); Enterococcus faecalis group (identified as such for IR analysis only), E. faecalis (6), E. casseliflavus (1), and E. gallinarum (2).
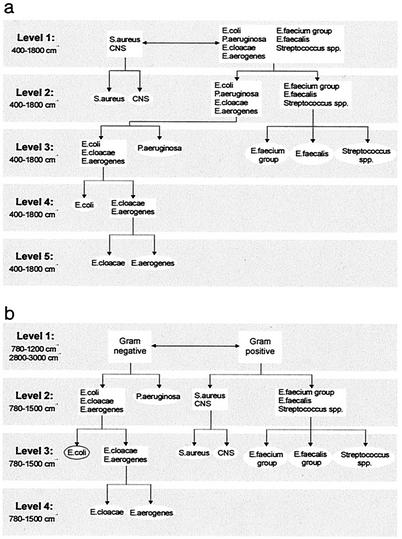
For Raman identification of bacteria, we used an approach similar to that described previously for the identification of Candida species (10). Briefly, HCA of the spectra in the reference library was used as a nonsubjective method by which to determine the major groupings in the set of data. These groupings were used as the starting point for the development of an identification tree based on LDA (Fig. 2a). An LDA model was developed for each division in the tree. Identification of a microorganism occurs by entering its spectrum into this identification tree at level 1. Depending on the outcome, it proceeds to one of the models at level 2, etc. Likewise, a multilayered ANN analysis (Fig. 2b), consisting of one top-level net and several subsequent subnets, was developed for identification based on IR spectra.
Analysis of clinical samples.
Over a 4-month period, all consecutive positive blood cultures from patients in the ICUs and a random selection of positive blood cultures from patients in other wards of the University Hospital Rotterdam were used to test the Raman spectroscopic identification method. Similarly, all positive blood cultures from the ICUs of the Rudolf Virchow Hospital were collected over a 3-month period to test the FT-IR microspectroscopic identification approach.
After the period of incubation on solid culture medium, cell morphology was inspected by direct microscopy to distinguish bacteria from yeasts. On the basis of this distinction, vibrational spectra were collected from isolates on the culture medium that best supported the growth of that organism, e.g., Mueller-Hinton or CASO medium for bacteria and Sabouraud medium plus 2% glucose for yeasts. Raman and IR spectra thus obtained from patient samples were entered into the respective identification trees for species identification as described above (Fig. 2).
FIG. 2.
Sequential identification schemes used for the identification of bacteria. (a) Sequential LDA model used for identification of microorganisms on the basis of their Raman spectra. (b) Schematic diagram of the hierarchical network used to identify bacteria on the basis of their IR spectra. See the text for details.
When a strain was identified by the routine methods as a member of a genus not included in the reference spectral databases, it was excluded from the comparison between routine identification methods and vibrational spectroscopic identification.
RESULTS
Reference databases of microorganism spectra and development of microorganism identification models.
The database of Raman spectra contained the spectra of 106 individual bacterial strains and 34 yeast strains (Table 1). In the IR database, 121 spectra were included (89 from bacterial strains and 32 from yeast strains). Marked spectral differences can be observed in the Raman and IR spectra of different microbial species. Some of the spectral regions in which the species differ are shaded in Fig. 1. Although the spectra of different microorganisms could be analyzed in terms of their biochemical differences, the analysis of large sets of data would become very complicated. Interpreting the spectra as fingerprints and using pattern recognition methods allows the development of automated identification models.
Separate identification models were developed on the basis of the Raman and IR databases. A first validation of the LDA and ANN analysis models was performed by a leave-one-out method (20) in which the spectra of all but one of the reference strains were used to generate the respective models. The strain that was left out was identified on the basis of its spectrum in order to test the prediction models. This procedure was repeated for each strain. The results in Table 1 show that, at the genus level, the models resulted in nearly perfect identification with both databases. In the Raman database, the exceptions were one Streptococcus strain that was identified as E. faecalis and one Escherichia coli strain that was identified as E. cloacae. Misidentifications also occurred in the IR database for two Streptococcus strains that were identified as E. faecalis. At the species level, the models performed nearly as well, with one coagulase-negative Staphylococcus (CNS) strain being misclassified as S. aureus in both the Raman and IR databases. Only the separation of E. aerogenes and E. cloacae at level 5 of the Raman identification proved more problematic, with 80.0% correct identification in the leave-one-out evaluation. For the yeast strains included in the databases, high identification accuracy was achieved as well (Table 1).
Analysis of clinical samples.
We collected 135 blood cultures from 92 patients at the University Hospital Rotterdam for analysis by the Raman method. Bacteria were isolated from 129 blood cultures and 6 were positive for yeast. Similarly, 138 blood cultures were examined from 121 patients of the Rudolf Virchow Hospital; of these, 131 contained bacteria and 7 contained yeast. These samples were analyzed by FT-IR spectroscopy. For both the Raman and FT-IR tests, 17 strains were excluded from the comparison because the phenotypic identification yielded a species not included in the database. In addition, three samples containing mixed cultures with very similar cell morphologies, such as E. coli and E. aerogenes, were excluded from the comparison between Raman spectroscopy and routine identification, as it was not obvious which species was measured in the Raman experiments. However, in all cases, the Raman identification corresponded to one of the components in the mixed culture.
The results of the comparison between the vibrational spectroscopies and the phenotypic identifications are presented in Table 2. Raman spectroscopy correctly identified 92.2% (106 of 115) of the microorganisms included in the comparison, and 98.3% (119 of 121) were accurately identified by IR spectroscopy.
TABLE 2.
Comparison of phenotypic and vibrational spectroscopic identifications of patient samples included in the prospective studya
| Organism(s) | Raman spectroscopy
|
IR spectroscopy
|
||
|---|---|---|---|---|
| No. (%) identified correctly | Misidentification(s) (no. of strains) | No. (%) identified correctly | Misidentification (no. of strains) | |
| S. aureus | 19 (100) | 27 (96.4) | S. aureus (1) | |
| CNS | 37 (97.4) | Streptococcus sp. (1) | 52 (98.1) | CNS (1) |
| E. coli | 22 (91.7) | E. aerogenes (2) | 12 (100) | |
| E. cloacae | 2 (40) | E. aerogenes (3) | 2 (100) | |
| E. aerogenes | 3 (100) | |||
| P. aeruginosa | 5 (100) | 1 (100) | ||
| E. faecalis group | 3 (100) | 6 (100) | ||
| E. faecium group | 1 (100) | 5 (100) | ||
| Streptococcus spp. | 8 (72.7) | E. faecalis (2), E. aerogens (1) | 7 (100) | |
| Candida albicans | 6 (100) | 5 (100) | ||
| Candida glabrata | 1 (100) | |||
| Candida tropicalis | 1 (100) | |||
For Raman spectroscopy, 115 samples were included, and for IR spectroscopy, 121 samples were included.
DISCUSSION
The work presented here was a feasibility study. However, the results of this first prospective study illustrate the enormous potential of vibrational spectroscopic methods for rapid and accurate microbial identification. The fundamental strength of this approach is emphasized by the fact that this study was performed at two hospitals employing different spectroscopic methods, different sample-handling protocols, different methods of signal analysis, and different prediction models but yielding equally good results. Correct identification of 92.2 and 98.3% of the samples (by the Raman and IR methods, respectively) was achieved for spectra that were available 6 to 8 h after signaling by an automated blood culture system.
For 106 of the remaining 115 samples, Raman spectroscopic identification corresponded to the phenotypic identification of the routine diagnostic test. Four of the nine misidentifications can be explained. The two E. coli isolates identified as E. aerogenes were collected from one patient only 5 h apart, making it very likely that the strains were identical. A similar situation occurred for two of the three E. cloacae isolates identified by the Raman method as E. aerogenes. Two of the three Streptococcus species that were misidentified (Streptococcus mitis and S. anginosus) were not included in the reference database. IR spectroscopy correctly identified 119 of 121 samples. Nearly perfect identification of the main contributors to the bloodstream infections (staphylococci and E. coli) was obtained by both methods. The perfect identification of the samples with Candida species 6 (Raman) to 8 (IR) h after a positive signal was obtained from the automated blood culture systems is particularly encouraging, as routine phenotypic identification required an additional 48 h.
Although these results are very encouraging, much larger databases of vibrational spectra of a wider range of microbial genera and species, as well as a larger number of isolates per species, must be established. It will then become possible to recognize whether a new isolate belongs to a species represented in the database or not (i.e., is an unknown). The relatively low number of isolates per species in the current reference database does not enable reliable outlier detection. Extension of the reference spectral databases to include a wider range of microorganisms (genera, species, and strains) will also enable development of other targeted medical microbiological applications. Intraabdominal infections with Candida species, for example, are associated with high mortality rates (5). Rapid identification is important because some species are intrinsically resistant to antifungal agents of the azole group, which are usually the agents of first choice in treating this kind of infections. We have previously shown that highly accurate rapid identification (97%) of Candida species by vibrational spectroscopic methods is possible (10). A clinical pilot study of prospective Candida species identification in intraabdominal infections by Raman spectroscopy is under way. Another potential application for spectroscopic techniques that makes use of the fact that very little biomass is needed is the identification of fastidious microorganisms, e.g., mycobacteria. Routinely used phenotypic identification methods can take between 2 and 8 weeks to be completed for these slow-growing bacteria (16). However, rapid identification of Mycobacterium species is becoming increasingly important because of their increased incidence over the last decade. Rapid discrimination between Mycobacterium tuberculosis and M. avium, currently the topic of an FT-IR spectroscopic study, is of prime importance for effectively guiding the choice of antibiotic therapy, as the two life-threatening infections in immunocompromised patients require different types of management and therapy.
The signal collection times used in this study (25 min for Raman spectroscopy and 18 min for FT-IR spectroscopy) limit the sample throughput. However, with further optimization of the instrumentation, a reduction in signal collection time to only a few minutes is feasible. This will also facilitate the analysis of mixed cultures by performing the identification on more microcolonies selected on the basis of cell and colony morphology. Apart from the clinical significance, a practical reason for our choice to target blood infections first was that they are nearly always due to a single pathogen. Development of dedicated culture media, which will enhance the rate of microcolony development, is expected to further shorten the necessary cultivation time.
Apart from enabling rapid identification, vibrational spectroscopic techniques require virtually no sample handling or consumables, which is in sharp contrast to other rapid identification techniques that are under development, such as molecular genetic approaches. This implies that vibrational spectroscopic techniques are well suited to automation, can be used by nonexperts, and are relatively inexpensive to use. Moreover, vibrational spectroscopy also offers possibilities for the development of rapid drug susceptibility testing (12). We conclude that Raman and FT-IR spectroscopies provide a novel answer to the need for rapid microbial identification in a clinical diagnostic setting.
Acknowledgments
The first two authors contributed equally to this study.
REFERENCES
- 1.Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. [DOI] [PubMed] [Google Scholar]
- 2.Choo-Smith, L.-P., K. Maquelin, T. van Vreeswijk, H. A. Bruining, G. J. Puppels, N. A. Thi, C. Kirschner, D. Naumann, D. Ami, A. M. Villa, F. Orsini, S. M. Doglia, H. Lamfarraj, G. D. Sockalingum, M. Manfait, P. Allouch, and H. P. Endtz. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doern, G. V., R. Vautour, M. Gaudet, and B. Levy. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodacre, R., and D. B. Kell. 1996. Pyrolysis mass spectrometry and its applications in biotechnology. Curr. Opin. Biotechnol. 7:20-28. [DOI] [PubMed] [Google Scholar]
- 5.Hoerauf, A., S. Hammer, B. Muller-Myhsok, and H. Rupprecht. 1998. Intra-abdominal Candida infection during acute necrotizing pancreatitis has a high prevalence and is associated with increased mortality. Crit. Care Med. 26:2010-2015. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahim, E. H., G. Sherman, S. Ward, V. J. Fraser, and M. H. Kollef. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146-155. [DOI] [PubMed] [Google Scholar]
- 7.Jones, R. N. 2001. Resistance patterns among nosocomial pathogens: trends over the past few years. Chest 119:397S-404S. [DOI] [PubMed] [Google Scholar]
- 8.Kirschner, C., K. Maquelin, P. Pina, N. A. Ngo Thi, L.-P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Mainfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kollef, M. H. 2000. Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin. Infect. Dis. 31(Suppl. 4):S131-S138. [DOI] [PubMed] [Google Scholar]
- 10.Maquelin, K., L.-P. Choo-Smith, H. P. Endtz, H. A. Bruining, and G. J. Puppels. 2002. Rapid identification of Candida species by confocal Raman microspectroscopy. J. Clin. Microbiol. 40:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maquelin, K., L.-P. Choo-Smith, T. van Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining, and G. J. Puppels. 2000. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal. Chem. 72:12-19. [DOI] [PubMed] [Google Scholar]
- 12.Maquelin, K., C. Kirschner, L.-P. Choo-Smith, N. A. Ngo-Thi, D. Naumann, and G. J. Puppels. 2002. Vibrational spectroscopic studies of microorganisms, p. 3308-3334. In P. R. Griffiths (ed.), Handbook of vibrational spectroscopy, 1st ed., vol. 5. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 13.Miller, J. M., and C. M. O'Hara. 1999. Manual and automated systems for microbial identification, p. 193-201. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 14.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 15.Nelson, W. H., R. Manoharan, and J. F. Sperry. 1992. UV resonance Raman studies of bacteria. Appl. Spectrosc. Rev. 27(1):67-124. [Google Scholar]
- 15a.Ngo Thi, N. A., C. Kirschner, and D. Naumann. 2000. FT-IR microspectrometry: a new tool for characterizing microorganisms, p. 36-44. In A. Mahadevan-Jansen and G. J. Puppels (ed.), Biomedical spectroscopy: vibrational spectroscopy and other novel techniques 26-27 January 2000, San Jose, California. SPIE-International Society for Optical Engineering, Bellingham, Wash.
- 16.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller, M. A. 2001. Molecular approaches to diagnosing and managing infectious diseases: practicality and costs. Emerg. Infect. Dis 7:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puppels, G. J., W. Colier, J. H. F. Olminkhof, C. Otto, F. F. M. de Mul, and J. Greve. 1991. Description and performance of a highly sensitive confocal Raman spectrometer. J. Raman Spectrosc. 22:217-225. [Google Scholar]
- 19.Sakallah, S. A. 2000. Molecular diagnostics of infectious diseases: state of the technology. Biotechnol. Annu. Rev. 6:141-161. [DOI] [PubMed] [Google Scholar]
- 20.Stone, M. 1974. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. B 36:111-147. [Google Scholar]
- 21.Thompson, R. L., and A. J. Wright. 1998. General principles of antimicrobial therapy. Mayo Clin. Proc. 73:995-1006. [DOI] [PubMed] [Google Scholar]
- 22.Udelhoven, T., D. Naumann, and J. Schmitt. 2000. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 54:1471-1479. [Google Scholar]
- 23.Wheeler, A. P., and G. R. Bernard. 1999. Treating patients with severe sepsis. N. Engl. J. Med. 340:207-214. [DOI] [PubMed] [Google Scholar]