Abstract
In the major pathway for protein disulfide-bond formation in the endoplasmic reticulum (ER), oxidizing equivalents flow from the conserved ER-membrane protein Ero1p to secretory proteins via protein disulfide isomerase (PDI). Herein, a mutational analysis of the yeast ERO1 gene identifies two pairs of conserved cysteines likely to form redox-active disulfide bonds in Ero1p. Cys100, Cys105, Cys352, and Cys355 of Ero1p are important for oxidative protein folding and for cell viability, whereas Cys90, Cys208, and Cys349 are dispensable for these functions. Substitution of Cys100 with alanine impedes the capture of Ero1p-Pdi1p mixed-disulfide complexes from yeast, and also blocks oxidation of Pdi1p in vivo. Cys352 and Cys355 are required to maintain the fully oxidized redox state of Ero1p, and also play an auxiliary role in thiol–disulfide exchange with Pdi1p. These results suggest a model for the function of Ero1p wherein Cys100 and Cys105 form a redox-active disulfide bond that engages directly in thiol–disulfide exchange with ER oxidoreductases. The Cys352–Cys355 disulfide could then serve to reoxidize the Cys100–Cys105 cysteine pair, possibly through an intramolecular thiol–disulfide exchange reaction.
INTRODUCTION
The formation of native protein disulfide bonds is a critical step in the folding of many secretory proteins. Protein disulfide bond formation in the endoplasmic reticulum (ER) of eukaryotic cells requires two essential proteins, Ero1p (ER oxidation 1) and protein disulfide isomerase (PDI). Ero1p is a novel and conserved glycoprotein that is associated with the ER membrane and that introduces the oxidizing equivalents necessary for protein disulfide-bond formation into the ER lumen (Frand and Kaiser, 1998; Pollard et al., 1998).
PDI serves as the principal catalyst of thiol–disulfide exchange in the lumen of the ER. The oxidoreductase activity of PDI depends on two pairs of cysteines, each of which is found in the motif Cys-Xa-Xb-Cys, a hallmark of the redox-active cysteines in oxidoreductases of the thioredoxin superfamily (Martin, 1995; Chivers et al., 1997). The PDI1 gene of yeast is essential for cell viability, and Pdi1p has been implicated in the catalysis of both disulfide bond formation and isomerization in vivo (Scherens et al., 1991; LaMantia and Lennarz, 1993; Laboissière et al., 1995; Holst et al., 1997; Frand and Kaiser, 1999).
Genetic analysis in Saccharomyces cerevisiae has recently defined the core pathway for protein disulfide-bond formation in the ER, whereby oxidizing equivalents flow from Ero1p to secretory proteins via Pdi1p (Frand et al., 2000). Mixed-disulfide complexes between Ero1p and Pdi1p have been captured in yeast, and these complexes are likely to represent intermediates in the direct oxidation of Pdi1p by Ero1p in vivo (Frand and Kaiser, 1999). Consistent with this model, mutational inactivation of ERO1 leads to complete reduction of the active-site cysteines of Pdi1p, whereas these cysteines are found predominantly in the disulfide (oxidized) form in wild-type cells. Moreover, cells depleted of Pdi1p display a defect in protein oxidation, indicating that Pdi1p performs a critical function as an oxidase in vivo (Frand and Kaiser, 1999).
The oxidative activity of Ero1p is further coupled to the production of oxidized glutathione in the ER, but the oxidation of glutathione does not correspond to an intermediate step necessary for protein disulfide-bond formation (Frand and Kaiser, 1998; Cuozzo and Kaiser, 1999). Rather, glutathione competes with protein thiols for oxidizing equivalents derived from Ero1p (Bader et al., 1999b; Cuozzo and Kaiser, 1999).
The discovery that Ero1p engages directly in thiol–disulfide exchange with Pdi1p predicted that at least one redox-active disulfide bond would be required for the function of Ero1p. We therefore sought to identify the redox-active cysteines in Ero1p through a mutational analysis of the seven cysteine residues that are absolutely conserved in the eukaryotic homologues of Ero1. This analysis identified two pairs of conserved cysteines essential for the oxidative activity of Ero1p.
MATERIALS AND METHODS
S. cerevisiae strains were grown and genetically manipulated by using standard techniques (Kaiser et al., 1994). YPD is rich medium with 2% glucose. SMM is minimal medium supplemented with amino acids and 2% glucose. SMM Raf/Gal is minimal medium with 2% raffinose and 2% galactose. A 1 OD600 U corresponds to 2 × 107 cells. Table 1 lists the genotypes of strains used in this study.
Table 1.
Yeast
| Strain | Genotype | Source |
|---|---|---|
| CKY8 | MATα ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY10 | MATa ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY263 | MATa GAL2 ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY559 | MATα ero1-1 ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY561 | MATa ire1Δ∷URA3 ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY598 | MATa GAL2 ero1-1 ura3-52 leu2-3,112 | Kaiser lab collection |
| CKY599 | MATa/α ero1-Δ1-500∷LEU2/ERO1 leu2-3,112/leu2-3,112 ura3-52/ura3-52 | This study |
| CKY600 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF82] | This study |
| CKY601 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF98] | This study |
| CKY602 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF131] | This study |
| CKY603 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF99] | This study |
| CKY604 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF120] | This study |
| CKY605 | MATα ire1Δ∷URA3 ero1-1 ura3-52 leu2-3,112 | This study |
| CKY686 | MATα ero1-Δ1-500∷LEU2 ura3-52 leu2-3,112[pAF131] | This study |
Plasmids and Strain Constructions
The alleles specifying alanine substitution mutants of Ero1p were each generated by site-directed mutagenesis (Kunkel et al., 1991) on pAF82 [CEN URA3 ERO1-myc] (Frand and Kaiser, 1998). The plasmids pAF98, pAF131, pAF122, pAF99, pAF120, pAF96, and pAF95 correspond, respectively, to ero1-A90-myc, ero1-A100-myc, ero1-A105-myc, ero1-A208-myc, ero1-A349-myc, ero1-A352-myc, and ero1-A355-myc. The genomic inserts from these plasmids were cloned into pRS305–2μ [2μ, LEU2] (Sikorski and Hieter, 1989) by homologous recombination in vivo, generating, respectively, pAF124, pAF125, pAF126, pAF127, pAF128, pAF129, and pAF130. The ero1-A90-myc, ero1-A100-myc, ero1-A208-myc, and ero1-A349-myc alleles were inserted into pRS315 [CEN LEU2] (Sikorski and Hieter, 1989) by homologous recombination in CKY605 to generate pAF153, pAF154, pAF155, and pAF156. Plasmids pAF89 [2μ LEU2 ERO1-myc] and pAF85 [CEN LEU2 ERO1-myc] were described previously (Frand and Kaiser, 1998), as were pAF132 [CEN URA3 PGAL1-pdi1–1966] and pAF123 [CEN URA3 PGAL1-mpd2-CQHA] (Holst et al., 1997; Frand and Kaiser, 1999).
To generate a chromosomal deletion of ERO1, 1.5 kilobases (kb) of ERO1-coding sequence as well as 0.8 kb of upstream sequence (devoid of other open reading frames) was removed from pAF82 by digestion with BamHI and HindIII, and replaced with a 2.0-kb BamHI-HindIII fragment from pJJ252 containing the LEU2 marker gene (Jones and Prakash, 1990). The ero1-Δ1-500::LEU2 allele was then isolated as an SphI-KpnI restriction fragment and introduced into a ura3-52, leu2-3,112 diploid generated by mating CKY8 and CKY10. Sporulation of a Leu+ transformant, CKY599, produced no more than two viable Leu− spores per tetrad. When CKY599 was transformed with pAF82 [CEN URA3 ERO1-myc] and sporulated, viable Leu+ Ura+ segregants were recovered. A representative segregant, CKY600, was shown to depend on the episomal ERO1 allele for viability by the failure to grow on medium containing 5-fluoroorotic acid (5-FOA; Toronto Research Laboratories, Downsview, Ontario, Canada). CKY601, CKY603, and CKY604 were generated in a similar manner by sporulation of CKY599 transformed with pAF98, pAF99, or pAF120, respectively. CKY602 and CKY686 are isogenic spore clones isolated after sporulation of CKY599 transformed with pAF131. To construct CKY605, the ire1Δ::URA3 fragment from pCS109A (Cox et al., 1993) was introduced at the IRE1 locus of CKY559 by one-step gene replacement. Transformants of CKY605 were cultured exclusively at 24°C because the strain is inviable at 30°C.
Complementation of Phenotypes Associated with Loss of ERO1 Function
To test for complementation of the temperature-sensitive growth defect of ero1-1 cells, CKY559 was transformed with pAF122, pAF96, or pAF95, and CKY605 was transformed with pAF85, pAF153, pAF154, pAF155, or pAF156. These strains were grown selectively to exponential phase, and then plated on YPD at a density of 1 OD600 U/ml and incubated at 38°C overnight. To test for rescue of the inviability associated with a chromosomal deletion of ERO1, CKY599 (ero1-Δ1-500::LEU2/ERO1 leu2-3,112/leu2-3,112 ura3-52/ura3-52) was transformed with pAF82, pAF98, pAF131, pAF122, pAF99, pAF120, pAF96, or pAF95 and then sporulated. The recovery of Leu+ Ura+ spore clones that were unable to grow on plates containing 5-FOA indicated that the episomal allele of ERO1 could restore growth to ero1-Δ1-500::LEU2 spores. An episomal allele of ERO1 was considered to lack rescuing activity when dissection of at least 18 asci produced tetrads with no more than two viable Leu− spore clones (even though Ura+ segregants could be identified in several tetrads, verifying inheritance of the plasmid). To test the ability of ero1-A100-myc to rescue the inviability of ero1-Δ1-500::LEU2 spores in an ire1-Δ strain background, CKY602 (ero1-Δ1-500::LEU2 [pAF131]) was crossed to CKY561, and the resulting diploid sporulated. Tetratype and nonparental ditype tetrads inheriting pAF131 were identified by the segregation of Leu− Ura+ clones viable on medium containing 5-FOA. Phenotypes of the viable spore clones in these tetrads allowed assignment of the genotype ero1-Δ1-500::LEU2 ire1Δ::URA3 to inviable spores. Kinetic analysis of the maturation of carboxypeptidase Y (CPY), and assays for the sensitivity of yeast strains to dithiothreitol (DTT) were performed as described (Frand and Kaiser, 1998).
Trapping Mixed Disulfides between Ero1p-myc and CGHS–CGHS Pdi1p or CQHA Mpd2p
The capture of mixed disulfides between Ero1p-myc and either CGHS–CGHS Pdi1p or CQHA Mpd2p was performed essentially as described (Frand and Kaiser, 1999). Primary anti-myc immunoprecipitates were prepared from the extract of 10 OD600 U of cells that had been radiolabeled with [35S]methionine and cysteine and then suspended in 10% (wt/vol) trichloroacetic acid (TCA). Ten percent of each sample was saved for analysis by nonreducing SDS-PAGE, whereas the remainder was boiled in 100 mM DTT to reduce disulfide bonds. From the reduced portion of the sample, 15% was reimmunoprecipitated with excess anti-myc antibody and 85% reimmunoprecipitated with excess anti-Pdi1p antibody (kindly provided by Tom Stevens, Eugene, OR). As a control for the expression of PGAL1-pdi1-1966 in all experimental samples, supernatants from the primary anti-myc immunoprecipitation were immunoprecipitated with anti-Pdi1p antibody under reducing conditions. All immunoprecipitations were performed as described (Frand and Kaiser, 1998). Strains. derived from CKY598 (ero1-1 GAL2) were grown to exponential phase at 24°C and then shifted to 38°C for 12 min prior to radiolabeling. After harvesting by centrifugation, these cells were suspended in 100 μl of growth medium and returned to 38°C for 5 min prior to the addition of 10 μl of 100% (wt/vol) TCA.
Assays for Oxidation State of Ero1p and Pdi1p
The oxidation states of Ero1p-myc and Pdi1p were assessed essentially as described (Frand and Kaiser, 1999). For the analysis of Pdi1p, cells were grown to early exponential phase in YPD and then resuspended in SMM at a concentration of 3 OD600 U/ml. After incubation at 30°C for 40 min, cells were collected by centrifugation and suspended in 100 μl of 10% (wt/vol) TCA. This suspension was divided prior to the collection of cellular proteins by centrifugation at 4°C. Samples were washed with 1 ml of acetone and rapidly suspended in 50 μl of buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, 6 M urea, 1 mM phenylmethylsulfonyl fluoride, bromophenol blue) with or without 20 mM 4′-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS; Molecular Probes, Eugene, OR). Proteins were solubilized by manual pipetting and the samples adjusted to near neutral pH by the gradual addition of 1 M Tris-HCl, pH 7.5. Samples were incubated on ice for 15 min, at 37°C for 20 min, and boiled for 2 min. Insoluble material was removed by centrifugation prior to resolution of the samples by nonreducing SDS-PAGE. Pdi1p was detected by Western blotting as described (Frand and Kaiser, 1999).
RESULTS
Conserved Cysteines Essential for Oxidative Function of Ero1p
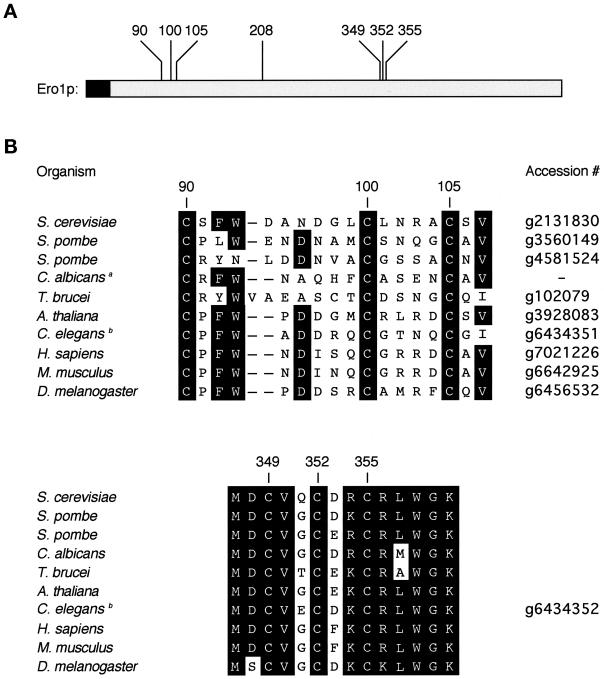
Seven of the 14 cysteines present in yeast Ero1p are absolutely conserved among six full-length sequence homologues of Ero1. These conserved cysteine residues are at positions 90, 100, 105, 208, 349, 352, and 355 of yeast Ero1p (Figure 1A). The three C-terminal cysteines occupy a highly conserved region of the protein, in which the human and yeast homologues of Ero1 are 65% identical at the amino acid level (Figure 1B; Frand and Kaiser, 1998; Pollard et al., 1998).
Figure 1.
Conserved cysteines in Ero1. (A) Seven cysteines are absolutely conserved among the eukaryotic sequence homologues of Ero1. These residues are at positions 90, 100, 105, 208, 349, 352, and 355 of the primary amino acid sequence of yeast Ero1p. The N-terminal hydrophobic sequence of Ero1p (shaded) is likely to serve as a membrane anchor or signal sequence. (B) Predicted amino acid sequences of the homologues of Ero1. The regions shown correspond to residues 90–107 and 347–357 of yeast Ero1p. aThe Candida albicans sequence is derived from an unfinished genome (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). bIn the Caenorhabditis elegans genome, two adjacent sequences are predicted to specify proteins homologous to the N- and C-terminal portions of yeast Ero1p. It remains to be determined whether these sequences correspond to one or two genes.
To assess the importance of each conserved cysteine residue for the function of Ero1p, seven new alleles of ERO1 were generated, replacing each conserved cysteine residue in Ero1p with alanine. To facilitate detection of these proteins, the desired mutations were introduced into ERO1-myc, a fusion gene specifying an epitope-tagged, functional version of Ero1p (Frand and Kaiser, 1998). The new Ero1p mutants are referred to as CAX Ero1p-myc, where X designates the position of the alanine substitution in the primary sequence of Ero1p. The corresponding alleles of ERO1 are likewise referred to as ero1-AX-myc. All seven of the cysteine-to-alanine substitution mutants of Ero1p-myc were expressed at approximately equal levels as wild-type Ero1p-myc in pulse-labeling and steady-state experiments (Figures 3 and 5).
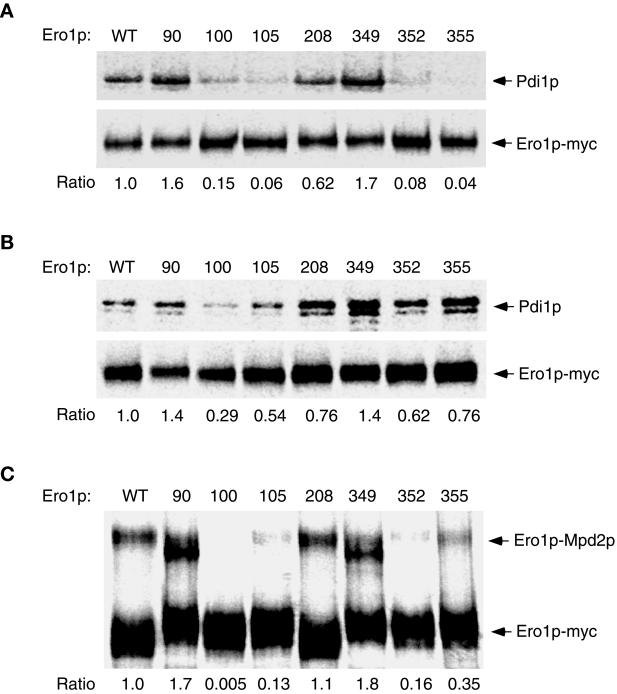
Figure 3.
The capture of mixed-disulfide complexes between each alanine substitution mutant of Ero1p-myc and CGHS–CGHS Pdi1p or CQHA Mpdp2. Cells overproducing a single alanine-substitution mutant of Ero1p-myc in addition to CGHS–CGHS Pdi1p were labeled with [35S]methionine and cysteine and then suspended in 10% TCA to block further thiol–disulfide exchange in vivo. Ero1p-Pdi1p mixed disulfides were isolated by modifying free thiols with NEM prior to immunoprecipitation with anti-myc antibody under nonreducing but denaturing conditions. The primary immunoprecipitates were reduced with 100 mM DTT prior to reimmunoprecipitation with either anti-myc (1× loading) or anti-Pdi1p (7.5× loading) antibody. Samples were resolved by SDS-PAGE and analyzed with a 445si phosphorimager. The efficiency of mixed-disulfide capture is expressed as the ratio of the band intensity of reimmunoprecipitated Pdi1p to that of reimmunoprecipitated Ero1p-myc (per OD600 U of extract), normalized to the value obtained with wild-type Ero1p-myc. (A) Strains derived from CKY598 (ero1-1 GAL2) were grown in SMM Raf/Gal and labeled at restrictive temperature (38°C). (B) Otherwise isogenic strains derived from CKY263 (GAL2) were labeled at 30°C. The strains shown were transformed with pAF132 [PGAL1-pdi1-1966] in addition to the following: WT, pAF89 [2μ ERO1-myc]; 90, pAF124 [2μ ero1-A90-myc]; 100, pAF125 [2μ ero1-A100-myc]; 105, pAF126 [2μ ero1-A105-myc]; 208, pAF127 [2μ ero1-A208-myc]; 349, pAF128 [2μ ero1-A349-myc]; 352, pAF129 [2μ ero1-A352-myc]; or 355, pAF130 [2μ ero1-A355-myc]. (C) Mixed disulfides between CQHA Mpd2p and each alanine substitution mutant of Ero1p-myc were isolated as described above from wild-type cells (CKY263) transformed with pAF123 [PGAL1-mpd2p-CQHA] instead of pAF132, and the complexes resolved directly by nonreducing SDS-PAGE. The efficiency of mixed-disulfide capture is expressed as the ratio of the band intensity of the Mpd2p-Ero1p-myc complexes to free Ero1p-myc, normalized to the value obtained with wild-type Ero1p-myc.
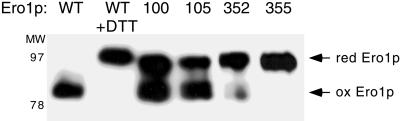
Figure 5.
Cys352 and Cys355 are required for oxidation of Ero1p in vivo. The CA352, CA355, CA100, and CA105 mutants of Ero1p-myc were overproduced in wild-type cells (CKY263), and the in vivo oxidation state of these proteins assessed by the modification of free thiols with AMS after the TCA-precipitation of cellular proteins. Samples were resolved by nonreducing SDS-PAGE and Ero1p-myc detected by immunoblotting. To provide standards for the mobility of the reduced and oxidized forms of Ero1p, cells expressing wild-type Ero1p-myc were treated with 5 mM DTT, or not treated. Lanes are labeled by the position of the alanine substitution in Ero1p-myc.
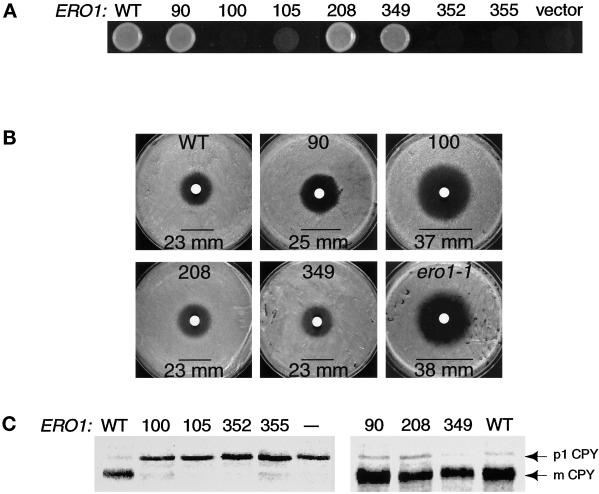
Each alanine substitution mutant of ERO1-myc was tested for complementation of the temperature-sensitive ero1-1 mutant (Frand and Kaiser, 1998). One complication in assessing the functionality of ERO1 mutants is that when protein oxidation in the ER is compromised, ERO1 expression is highly induced by the unfolded protein response (UPR), a pathway dependent upon the ER transmembrane kinase Ire1p (Cox et al., 1993). To control for the possible compensatory induction of ERO1 mutants by the UPR, episomal alleles of ERO1 that exhibited complementation also were tested in an ire1-Δ ero1-1 double mutant (CKY605). The alleles ero1-A100-myc, ero1-A105-myc, ero1-A352-myc, and ero1-A355-myc failed to restore growth to ero1-1 cells at restrictive temperature (38°C), whereas ero1-A90-myc, ero1-A208-myc, ero1-A349-myc, and wild-type ERO1-myc rescued the conditional growth defect of ero1-1 cells (Figure 2A). Inactivation of the UPR was needed to reveal the full phenotype associated with ero1-A100-myc because this allele partially rescued the temperature-sensitive growth defect of ero1-1 cells in an IRE1+ strain (CKY559), but rescue was not observed in the ire1Δ genetic background. The ero1-A105-myc allele also displayed partial rescuing activity when overexpressed from a high-copy plasmid (pAF126) in ero1-1 IRE1+ cells (data not shown).
Figure 2.
Cys100, Cys105, Cys352, and Cys355 of Ero1p are important for cell viability and for oxidative protein folding in the ER. (A) Complementation of the temperature-sensitive ero1-1 mutant by alanine substitution mutants of ERO1-myc. Cells were plated on rich medium and incubated overnight at restrictive temperature (38°C). Alleles of ERO1-myc with detectable rescuing activity were expressed in an ire1Δ genetic background to prevent induction of ERO1 by the UPR. Transformants of CKY605 (ero1-1 ire1Δ::URA3) carried the following plasmids: wild type (WT), pAF85 [CEN ERO1-myc]; 90, pAF153 [CEN ero1-A90-myc]; 100, pAF154 [CEN ero1-A100-myc]; 208, pAF155 [CEN ero1-A208-myc]; or 349, pAF156 [CEN ero1-A349-myc]. Transformants of CKY559 (ero1-1) hosted the following: 105, pAF122 [CEN ero1-A105-myc]; 352, pAF96 [CEN ero1-A352-myc]; 355, pAF95 [CEN ero1-A355-myc]; or vector, pRS316 [CEN URA3]. (B) Assays for the DTT sensitivity of cells with a chromosomal deletion of ERO1 rescued by an alanine substitution mutant of ERO1-myc. Three lawns of each strain were plated on rich medium and 10 μl of 3 M DTT applied to each lawn in a filter disk. The average diameter of the zone of growth inhibition (mm) was measured after incubation at 30°C for 1.5 days. The strains shown are as follows: WT, CKY600 (ero1-Δ1-500::LEU2 p[ERO1-myc]); 90, CKY601 (ero1-Δ1-500::LEU2 p[ero1-A90-myc]); 100, CKY602 (ero1-Δ1-500::LEU2 p[ero1-A100-myc]); 208, CKY603 (ero1-Δ1-500::LEU2 p[ero1-A208-myc]); 349, CKY604 (ero1-Δ1-500::LEU2 p[ero1-A349-myc]); and ero1-1, CKY559. (C) Processing of newly synthesized CPY in ero1-1 cells expressing each alanine substitution mutant of ERO1. Cells were pulse-labeled with [35S]methionine and cysteine at 38°C for 20 min. CPY was immunoprecipitated and the samples resolved by SDS-PAGE. The ER (p1) and vacuolar (mature) forms of CPY are indicated. Samples were prepared from the same strains as in A, except that “— ” refers to untransformed CKY559.
The alanine substitution mutants of ERO1-myc were further tested for their ability to rescue the inviability associated with a chromosomal deletion of ERO1. A diploid heterozygous for a disruption allele of ERO1 marked by LEU2 (ero1-Δ1-500::LEU2) was constructed lacking other functional alleles of LEU2 or URA3. This diploid (CKY599) was independently transformed with each allele of ERO1 on a plasmid marked by URA3. The transformants were sporulated, and the ability of a given episomal allele of ERO1 to confer viability to ero1-Δ1-500::LEU2 spores was scored by the recovery of Leu+ Ura+ spore clones. The alleles ero1-A105-myc, ero1-A352-myc, and ero1-A355-myc could not rescue the inviability of spores with a chromosomal deletion of ERO1, whereas ero1-A90-myc, ero1-A208-myc, ero1-A349-myc, and ERO1-myc could complement (our unpublished observations). The ero1-A100-myc allele failed to restore viability to ero1-Δ1-500::LEU2 spores in an ire1Δ background, but could partially restore growth to such cells in an IRE1+ strain. These results show that Cys352 and Cys355 of Ero1p are essential for yeast viability. Cys100 and Cys105 of Ero1p are also important for cell viability, but overproduction of Ero1p can circumvent the requirement for these residues (see DISCUSSION).
To assess the oxidizing capacity of the CA90, CA100, CA208, and CA349 mutants of Ero1p-myc, we examined the sensitivity to exogenous reductant of yeast strains that expressed ero1-A90-myc, ero1-A100-myc, ero1-A208-myc, or ero1-A349-myc covering a chromosomal deletion of ERO1. Lawns of each strain were plated on rich medium and 30 μmol of DTT was applied to each lawn on a sterile filter disk. Cells expressing ero1-A100-myc (CKY602) were unable to grow inside a zone 37 mm in diameter surrounding the DTT source (Figure 2B). A similar zone of growth inhibition was observed for ero1-1 cells grown at semipermissive temperature (Figure 2B; Frand and Kaiser, 1998). In contrast, cells expressing ero1-A90-myc, ero1-A208-myc, ero1-A349-myc, or wild-type ERO1-myc could grow to within 23–25 mm of the DTT source (Figure 2B). These results show that Cys100 is required for the oxidative activity of Ero1p, even though induction of the UPR permits CA100 Ero1p to support viability. In contrast, Cys90, Cys208, and Cys349 appear largely dispensable for the function of Ero1p in maintaining cellular oxidizing capacity.
If two cysteines in Ero1p normally reside in a disulfide-bonded pair, mutation of one of these cysteines could in theory free the other cysteine of the pair to form disulfide bonds with other proteins. This possibility raised the concern that substitution mutants of Ero1p associated with loss-of-function phenotypes might actually dominantly interfere with protein oxidation in the ER. To control for this possibility, we examined the DTT sensitivity of wild-type strains (CKY263) overexpressing ero1-A100-myc, ero1-A105-myc, ero1-A352-myc, or ero1-A355-myc from a high-copy plasmid under selective growth conditions. These strains displayed equivalent sensitivity to DTT as wild-type cells, showing that these ERO1 alleles are genetically recessive and therefore probably do not specify proteins that actively interfere with the ER-oxidizing machinery (our unpublished observations).
Cys100-Cys105 and Cys352-Cys355 of Ero1p Are Required for Efficient Oxidative Protein Folding in the ER
The vacuolar protease CPY undergoes oxidative protein folding in the ER to achieve a native structure with five intramolecular disulfide bonds (Endrizzi et al., 1994, Jämsäet al., 1994). In the ero1-1 conditional mutant, the formation of these disulfide bonds is disrupted, and reduced pro-CPY is consequently retained in the ER in the characteristic p1 form (Frand and Kaiser, 1998).
To determine the importance of each conserved cysteine residue in Ero1p for oxidative protein folding in the ER, the processing of newly synthesized CPY was monitored in ero1-1 cells expressing each alanine substitution mutant of ERO1. Episomal alleles of ERO1 with detectable rescuing activity were expressed in the ire1Δ genetic background to prevent induction of ERO1 with the UPR. Newly synthesized CPY was fully retained in the ER when ero1-1 cells expressing ero1-A100-myc, ero1-A105-myc, ero1-A352-myc, or ero1-A355-myc were pulse labeled for 30 min at restrictive temperature (Figure 2C). In contrast, CPY was processed to the mature, vacuolar form in ero1-1 cells expressing ero1-A90-myc, ero1-A208-myc, ero1-A349-myc, or wild-type ERO1-myc (Figure 2C). These results confirm that Cys100, Cys105, Cys352, and Cys355 of Ero1p are required for efficient disulfide bond formation in the ER.
Capture of Mixed-Disulfide Complexes between Alanine Substitution Mutants of Ero1p-myc and CGHS-CGHS Pdi1p
The capture of mixed disulfides between Ero1p-myc and Pdi1p has recently provided evidence that Ero1p engages directly in thiol–disulfide exchange with Pdi1p in vivo (Frand and Kaiser, 1999). These mixed-disulfide complexes can be detected after cells overproducing Ero1p-myc as well as Pdi1p are treated with TCA, a reagent that rapidly lowers intracellular pH, and thereby inhibits further thiol–disulfide exchange in vivo. Production of a CGHS–CGHS active-site mutant of Pdi1p enhances the detection of these complexes (Frand and Kaiser, 1999), presumably because this form of Pdi1p is defective in the resolution of mixed disulfides in vivo (Walker and Gilbert, 1997).
To evaluate the role of the conserved cysteines of Ero1p in thiol–disulfide exchange with Pdi1p, we assessed the efficiency of mixed-disulfide capture between each alanine substitution mutant of Ero1p-myc and CGHS–CGHS Pdi1p. Cells expressing these proteins were radiolabeled with [35S]methionine and cysteine, and cellular proteins were then precipitated with TCA. To isolate the mixed-disulfide complexes, free thiols were covalently modified with N-ethylmaleimide (NEM) prior to immunoprecipitation with anti-myc antibody under nonreducing but denaturing conditions. The anti-myc immunoprecipitates, which would contain any Ero1p-myc-Pdi1p mixed disulfides as well as free Ero1p-myc (Frand and Kaiser, 1999) were then reduced with DTT prior to reimmunoprecipitation with either anti-Pdi1p or anti-myc antibody. The efficiency of mixed-disulfide capture is expressed as the ratio of reimmunoprecipitated Pdi1p to Ero1p-myc, normalized to the value obtained for wild-type Ero1p-myc.
Mixed-disulfide formation was first examined upon expression of each alanine substitution allele of ERO1-myc in the conditional ero1-1 mutant (CKY598) at restrictive temperature (38°C). Substitution of Cys100, Cys105, Cys352, or Cys355 of Ero1p-myc with alanine decreased the efficiency of trapping mixed disulfides with CGHS–CGHS Pdi1p, respectively, to 15, 6, 8, and 4% that of wild-type Ero1p-myc (Figure 3A). Mutation of Cys208 decreased the efficiency of mixed-disulfide capture to 62% that of wild-type Ero1p-myc, whereas mutation of Cys90 or Cys349 actually increased the efficiency of mixed-disulfide capture. These results show that Cys100, Cys105, Cys352, and Cys355 are required for the efficient capture of Ero1p-myc-Pdi1p mixed-disulfide complexes when the corresponding mutants are expressed in ero1-1 cells. In this genetic background, an overall decrease in the redox potential of the ER lumen caused by the loss of ERO1 function could account for the diminished efficiency of trapping mixed disulfides with CA100, CA105, CA352, or CA355 Ero1p-myc. Alternatively, mutation of Cys100, Cys105, Cys352, or Cys355 could specifically impede the nucleophilic attack of Ero1p by the active-site cysteines of CGHS–CGHS Pdi1p.
To help distinguish between these possibilities, we next examined the efficiency of trapping mixed disulfides containing the Ero1p-myc mutants in wild-type cells. In this context, the oxidized redox state of the ER lumen should be sustained by the activity of endogenous Ero1p, so defects in mixed-disulfide formation associated with an overall decrease in the redox potential of the ER should not be observed. Mixed-disulfide complexes containing the CA352, CA355, or CA105 mutants of Ero1p-myc were now captured, respectively, 62, 76, or 54% as efficiently as those containing wild-type Ero1p-myc (Figure 3B), indicating that the CA352 and CA355 mutants of Ero1p-myc can readily form mixed disulfides with CGHS–CGHS Pdi1p when oxidizing equivalents are supplied to the ER lumen. In contrast, the defect in mixed-disulfide formation associated with mutation of Cys100 was not substantially improved by expression of the mutant protein in wild-type cells because mixed disulfides containing the CA100 mutant of Ero1p-myc were captured only 29% as efficiently as those containing wild-type Ero1p-myc (Figure 3 B). Together, these observations suggest that the active-site cysteines of Pdi1p preferentially attack Cys100 of Ero1p, and thereby implicate Cys100 and Cys105 in the formation of a redox-active disulfide bond. Cys352 and Cys355 play an auxiliary role in thiol–disulfide exchange with Pdi1p, consistent with the model that these residues form a second redox-active disulfide bond that participates in electron transfer by Ero1p. One explanation for the enhanced capture of mixed disulfides observed for the CA352 and CA355 mutants of Ero1p-myc in wild-type cells is that oxidation of Cys100 and Cys105 enables CGHS-CGHS Pdi1p to attack these mutants.
The analysis of mixed-disulfide formation was extended by examining the interaction of each mutant of Ero1p-myc with Mpd2p, a PDI homologue that is also a substrate of Ero1p (Tachikawa et al., 1997; Frand and Kaiser, 1999). Ero1p-Mpd2p mixed disulfides (125 kDa) are readily detected after the TCA-treatment of cells overproducing a CQHA active-site mutant of Mpd2p along with Ero1p-myc (Figure 3C; Frand and Kaiser, 1999). Substitution of Cys100 of Ero1p with alanine abolished the capture of mixed-disulfide complexes between CQHA Mpd2p and Ero1p-myc in wild-type cells. Substitution of Cys105, Cys352, or Cys355 decreased the efficiency of mixed-disulfide capture to 13, 16, or 35% that of wild-type Ero1p-myc, respectively, whereas substitution of Cys90 or Cys349 increased the efficiency of mixed-disulfide capture 170 or 180%, respectively (Figure 3C). These observations corroborate the results with Pdi1p. The higher sensitivity of trapping mixed disulfides with Mpd2p may be attributable to the specific biochemical properties of this enzyme.
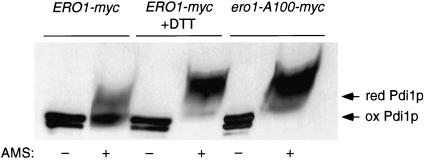
It has been proposed that mixed-disulfide complexes between Ero1p and Pdi1p correspond to physiological intermediates in the oxidation of Pdi1p by Ero1p (Frand and Kaiser, 1999). This hypothesis predicts that mutants of Ero1p specifically defective in mixed-disulfide formation with Pdi1p also should be defective in the oxidation of Pdi1p in vivo. The capacity of the ero1-A100-myc mutant to restore viability to cells with a chromosomal deletion of ERO1 allowed us to test this prediction by examining the oxidation state of Pdi1p in living cells. The oxidation state of Pdi1p was assessed through the modification of free thiols with the thiol-conjugating reagent AMS after the precipitation of cellular proteins with TCA. When Pdi1p was isolated from cells producing only the CA100 mutant of Ero1p-myc (CKY686), the molecular mass of Pdi1p increased by 8 kDa upon modification with AMS (Figure 4). Pdi1p that had been reduced in vivo by the treatment of cells with 10 mM DTT reacted with AMS to the same extent (Figure 4; Frand and Kaiser, 1999). In contrast, the majority of Pdi1p isolated from ero1-Δ1-500::LEU2 cells expressing wild-type Ero1p-myc could not be modified with AMS, indicating that the protein was largely oxidized in vivo (Figure 4). These results show that mutation of Cys100 of Ero1p disrupts oxidation of Pdi1p in vivo, consistent with the model that Ero1p-Pdi1p mixed disulfides serve as obligate intermediates in the efficient transfer of oxidizing equivalents from Ero1p to Pdi1p.
Figure 4.
Mutation of Cys100 of Ero1p blocks oxidation of Pdi1p in vivo. The in vivo oxidation state of Pdi1p was assessed by the modification of free thiols with AMS after the precipitation of cellular proteins with TCA. Samples were resolved by nonreducing SDS-PAGE and Pdi1p detected by immunoblotting. Samples were prepared from cells with a chromosomal deletion of ERO1 that expressed either ero1-A100-myc (CKY686) or ERO1-myc (CKY600). To provide a standard for the mobility of reduced Pdi1p, one sample of CKY600 was treated with 10 mM DTT. The AMS-modified reduced and oxidized forms of Pdi1p are indicated.
Cys352 and Cys355 Are Required for Complete Oxidation of Ero1p
The oxidation state of Ero1p in vivo can be assessed by the modification of free thiols with AMS after the precipitation of cellular proteins with TCA (Frand and Kaiser, 1999). In this assay, the apparent molecular mass of wild-type Ero1p-myc does not increase upon reaction with AMS, indicating that the protein is fully oxidized in vivo. In contrast, the apparent molecular mass of Ero1p-myc that has been reduced in vivo by the treatment of cells with 5 mM DTT increases by 16 kDa upon AMS modification (Figure 5; Frand and Kaiser, 1999). This increase in apparent molecular mass corresponds to alkylation of the redox-active cysteines in Ero1p, and also may reflect conformational changes in the reduced protein or the alkylation of additional cysteines. We have not observed forms of Ero1p-myc that display an intermediate shift in mobility after the AMS-modification of Ero1p-myc isolated from cells treated with 0.5–5 mM DTT, indicating that partially oxidized species of Ero1p-myc are not readily detected under steady-state conditions by this assay (Frand and Kaiser, 1999).
Because our results implicated Cys100, Cys105, Cys352, and Cys355 of Ero1p in the formation of redox-active disulfide bonds, we next examined the oxidation state of the CA100, CA105, CA352, and CA355 mutants of Ero1p-myc in wild-type cells. All of the CA355 mutant of Ero1p-myc and the majority of the CA352 mutant of Ero1p-myc were found in reduced form in vivo (Figure 5). Approximately one-half of the molecules of CA100 and CA105 Ero1p-myc also was found in reduced form in vivo, whereas the other half was captured in an oxidized state (Figure 5). The apparent molecular mass of the reduced form of each of these four mutants increased slightly less than that of reduced, wild-type Ero1p-myc after modification with AMS, as expected for mutants lacking one cysteine thiol normally reactive toward AMS. The CA208 mutant of Ero1p-myc appeared fully oxidized in vivo (data not shown). Interestingly, the molecular mass of both the CA90 and CA349 mutants of Ero1p-myc increased by ∼6 kDa after AMS-modification (Cuozzo, personal communication), suggesting that these proteins contained nonnative or uncommon intramolecular disulfide bonds. Together, these results show that Cys352 and Cys355 are required to maintain the oxidized redox state of Ero1p in vivo, and therefore suggest that reoxidation of Ero1p could proceed via oxidation of the Cys352-Cys355 cysteine pair. Cys100 and Cys105 facilitate maintenance of the fully oxidized redox state of Ero1p, consistent with a role for these residues in the cyclic reduction and reoxidation of Ero1p.
DISCUSSION
We have identified two pairs of conserved cysteines essential for the oxidative activity of Ero1p through a mutational analysis of the seven cysteine residues that are absolutely conserved in the eukaryotic sequence homologues of Ero1p. Cys100–Cys105 and Cys352–Cys355 of yeast Ero1p are critical for protein disulfide bond formation in the ER and for cell viability. Substitution of Cys100 of Ero1p with alanine impedes the capture of mixed-disulfide complexes with Pdi1p or Mpd2p in wild-type cells, and also blocks oxidation of Pdi1p in vivo. Substitution of Cys352 or Cys355 with alanine prevents reoxidation of Ero1p in vivo, and also decreases the efficiency of trapping mixed-disulfide complexes with Pdi1p in ero1-1 cells. The observation that those cysteine residues directly or indirectly participating in efficient mixed-disulfide formation with Pdi1p also are required for oxidative protein folding in the ER supports the model that thiol–disulfide exchange between Ero1p and Pdi1p drives the major pathway for protein oxidation in eukaryotic cells (Frand et al., 2000).
The observation that Cys100 is required for efficient thiol–disulfide exchange with Pdi1p implicated Cys100 in the formation of a redox-active disulfide bond in Ero1p. The CA100 and CA105 mutants of Ero1p-myc display similar properties, suggesting that Cys100 and Cys105 form this bond together. A precedent for a similar configuration of a redox-active disulfide bond is provided by glutathione disulfide reductase, an enzyme catalyzing the NADPH-dependent reduction of oxidized glutathione through an FAD cofactor and an enzymic disulfide found in the motif GTCVNVGCVP (Kuriyan et al., 1991). However, the formal possibility that Cys100 forms a disulfide bond with a cysteine residue other than Cys105 cannot be excluded at this time.
The residues corresponding to Cys352 through Cys355 of yeast Ero1p appear in the consensus sequence C(D/E)(K/R)C (Figure 1B), a site resembling the Cys-Xa-Xb-Cys motif that is a hallmark of the redox-active disulfide bond in thiol–disulfide oxidoreductases of the thioredoxin superfamily (Martin, 1995; Chivers et al., 1997). Although Ero1p does not display obvious sequence homology to thioredoxin, the protein could nevertheless contain a thioredoxin-like fold because primary sequence data alone may be insufficient to identify this domain (Ellis et al., 1992). If the conserved C(D/E)(K/R)C sequence of Ero1p does indeed define a redox-active Cys-Xa-Xb-Cys motif, then the internal (D/E)(K/R) residues would be expected to influence the redox potential of this disulfide bond (Grauschopf et al., 1995; Chivers et al., 1997).
Because the motif CXXCXXC can specify part of an iron–sulfur cluster in iron-binding proteins (Beinert, 1990), it has been suggested that the conjugation of iron through Cys349, Cys352, and Cys355 would be important for Ero1p to function as a redox sensor (Pollard et al., 1998). The observation that Cys349 is dispensable for the oxidative function of Ero1p disfavors, but does not exclude, this possibility. The residue corresponding to Cys349 of yeast Ero1p also is not required for the activity of human ERO1 in complementing the conditional ero1-1 mutant of yeast, whereas the residues corresponding to Cys352 and Cys355 are essential for the function of human Ero1 (Cabibbo et al., 2000).
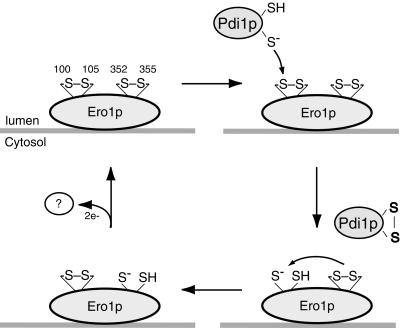
The results presented herein suggest the following model for electron transfer by Ero1p (Figure 6). Cys100 and Cys105 form a redox-active disulfide bond that preferentially engages in thiol–disulfide exchange with Pdi1p and Mpd2p. Cys352 and Cys355 form a second redox-active disulfide bond that serves to reoxidize the Cys100–Cys105 cysteine pair, possibly through an intramolecular thiol–disulfide exchange reaction. Reoxidation of Ero1p could proceed via the transfer of electrons from the Cys352–Cys355 cysteine pair to an as yet unidentified electron acceptor. In theory, a redox cofactor associated with Ero1p could serve as the immediate electron acceptor or donor for Cys352–Cys355.
Figure 6.
Model for the catalytic mechanism of Ero1p. Two redox-active disulfide bonds are present in Ero1p (upper left). The Cys100-Cys105 disulfide engages in thiol–disulfide exchange with Pdi1p, whereas the Cys352–Cys355 disulfide serves to reoxidize the Cys100–Cys105 cysteine pair, possibly through an intramolecular thiol–disulfide exchange reaction. Reoxidation of Ero1p could proceed via oxidation of the Cys352–Cys355 cysteine pair by an as yet unidentified electron acceptor. Only one thioredoxin-like domain of Pdi1p is shown.
Although these data suggest that oxidizing equivalents flow most efficiently from Ero1p to ER oxidoreductases via the Cys100–Cys105 disulfide bond, alternative, relatively inefficient pathways may conduct the flow of oxidizing equivalents away from Ero1p in the absence of Cys100 or Cys105. In theory, oxidizing equivalents introduced at the Cys352–Cys355 site of the CA100 or the CA105 mutant of Ero1p-myc could ultimately be transferred to secretory proteins through a series of thiol–disulfide exchange reactions involving the ER oxidoreductases or glutathione. These alternative pathways could account for the ability of an active unfolded protein response to bypass the requirement for Cys100 for cell viability because induction of Ero1p and the ER oxidoreductases by the UPR could increase the efficiency of protein oxidation through these alternative pathways. Consistent with this idea, small quantities of mixed disulfides have been detected between the CA100 mutant of Ero1p-myc and CGHS–CGHS Pdi1p in both wild-type and ero1-1 cells. One possible explanation for the formation of these complexes is that CGHS–CGHS Pdi1p can, albeit with relatively low efficiency, attack mixed-disulfide bonds formed between Cys105 and glutathione when Cys100 is not available. Similar complexes have been isolated between an active-site mutant of thioredoxin and an active-site mutant of thioredoxin reductase (Wang et al., 1996). CGHS–CGHS Pdi1p also could attack other intramolecular disulfide bonds in the CA100 mutant of Ero1p, such as Cys352–Cys355. The extent if any to which such bypass reactions occur under normal conditions remains to be determined.
Interestingly, mutation of either Cys90 or Cys349 of Ero1p increases the efficiency of capturing mixed-disulfide complexes with CGHS–CGHS Pdi1p or CQHA Mpd2p, indicating that Cys90 and Cys349 may normally expedite the dissolution of mixed-disulfide complexes or slow their formation. In theory, Cys90 and Cys349 could perform these functions independently, by influencing thiol–disulfide exchange reactions at the Cys100–Cys105 and Cys352–Cys355 sites, respectively. Alternatively, Cys90 and Cys349 could perform a common function because they form a disulfide bond with one another.
The pathway for protein disulfide-bond formation in the Escherichia coli periplasm provides a useful analogy for the pathway for protein oxidation in the eukaryotic ER. In E. coli, the thioredoxin-like oxidoreductase DsbA transfers disulfide bonds directly to secretory proteins (Bardwell et al., 1991). DsbA is then reoxidized via thiol–disulfide exchange with the cytoplasmic membrane protein DsbB (Bardwell et al., 1993; Dailey and Berg, 1993; Missiakas et al., 1993). The properties of yeast ERO1 are strikingly similar to those of E. coli dsbB (Frand et al., 2000). Both the ero1-1 mutant of yeast and the dsbB mutant of E. coli display a defect in protein disulfide-bond formation that can be suppressed by the addition of a thiol oxidant to the growth medium. Moreover, the overexpression of ERO1 in yeast or of dsbB in E. coli confers resistance to otherwise toxic levels of the reductant DTT. Most importantly, both Ero1p and DsbB directly oxidize soluble, thioredoxin-like enzymes (Bardwell et al., 1993; Bader et al., 1998; Frand and Kaiser, 1999).
The oxidative activity of DsbB also depends on two redox-active disulfide bonds (Jander et al., 1994; Bader et al., 1999a). Although DsbB is not likely to contain a thioredoxin-like fold, the residues Cys41 and Cys44 nevertheless define a Cys-Xa-Xb-Cys motif in DsbB, and the residues Cys104 and Cys130 comprise a second redox-active disulfide bond in the enzyme (Jander et al., 1994). The Cys104–Cys130 disulfide bond engages directly in thiol–disulfide exchange with DsbA because Cys104 of DsbB is specifically required for the capture of mixed-disulfide complexes with DsbA in bacterial cells (Guilhot et al., 1995; Kishigami et al., 1995; Kishigami and Ito, 1996). The Cys41–Cys44 disulfide bond serves to reoxidize the Cys104–Cys130 cysteine pair, and this reaction is thought to proceed via an intramolecular thiol–disulfide exchange reaction (Guilhot et al., 1995; Kishigami and Ito, 1996). Consistent with this model, mutational inactivation of the Cys41–Cys44 disulfide leads to reduction of the Cys104–Cys130 disulfide bond in vivo (Kobayashi and Ito, 1999). Moreover, substitution mutants of DsbB lacking Cys41 or Cys44 display defects in the formation of mixed disulfides with DsbA33S when bacterial cells are grown in the absence of small-molecule oxidants such as oxidized glutathione (Guilhot et al., 1995; Kishigami and Ito, 1996). The catalytic mechanism of DsbB thus provides a precedent for the use of an internal thiol–disulfide exchange reaction as an intermediate step in the oxidation of a thioredoxin-like oxidoreductase.
The results presented herein show that the properties of the CA100 and CA105 mutants of Ero1p-myc resemble those of DsbB mutants lacking Cys104 or Cys130, whereas the properties of the CA352 and CA355 mutants of Ero1p-myc resemble those of DsbB mutants lacking Cys41 or Cys44. These similarities suggest that key steps in the mechanism of electron transfer by Ero1p correlate to steps in the catalytic mechanism of DsbB. However, further investigation is likely to reveal unique aspects of Ero1p function because Ero1p is a novel protein and is expected to possess a substantially different domain structure from that of DsbB. Moreover, the substrates of Ero1p and DsbB possess different redox potentials (Lundstrom and Holmgren, 1993; Grauschopf et al., 1995), and the immediate electron acceptor for Ero1p has not been identified.
Recently, pathways for oxidation of E. coli DsbB have been elucidated, and this work may expedite the search for electron acceptors for protein–disulfide bond formation in eukaryotic cells (Debarbieux and Beckwith, 1999). In bacteria, DsbB is reoxidized predominantly through the transfer of electrons to molecular oxygen via later stages of the respiratory electron transport chain (Kobayashi et al., 1997; Bader et al., 1998, 1999a; Kobayashi and Ito, 1999). Electrons flow directly from DsbB to ubiquinone associated with cytochrome bd or cytochrome bo oxidase, two heme-containing terminal oxidases that shuttle electrons directly to molecular oxygen (Bader et al., 1999). Under anaerobic growth conditions, electrons may flow from DsbB to alternative acceptors via menaquinone (Bader et al., 1999).
In yeast, at least two cytochrome-based systems reside in the ER membrane that are responsible for sterol and unsaturated fatty acid biosynthesis (Daum et al., 1998). Both systems transfer electrons directly to molecular oxygen and are thought to be oriented to the cytosolic face of the ER membrane (Daum et al., 1998). These electron transport chains might serve as electron acceptors for Ero1p. If so, a lipid-soluble, small-molecule electron acceptor may be needed to shuttle electrons between the lumenal and cytosolic leaflets of the ER membrane. Alternatively, because Ero1p is an integral membrane protein (Frand and Kaiser, 1998, Cabbibo et al., 2000), a portion of Ero1p could directly contact the prosthetic groups of the proteins comprising these electron transport chains. The identification of the redox-active sites of Ero1p enables further study of the structure and function of Ero1p in vitro and in vivo.
ACKNOWLEDGMENTS
We thank Tom Stevens, Hiroyuki Tachikawa, and Hidde Ploegh for providing antibodies, and Jakob Winther for supplying alleles of PDI1. We also thank Peter Chivers, John Cuozzo, and Carolyn Sevier for technical assistance and for critical reading of this manuscript. This work was supported by grants from the National Institute of General Medical Sciences (to C.A.K.), and by a National Institutes of Health predoctoral traineeship (to A.F.).
REFERENCES
- Bader M, Muse W, Ballou DP, Gassner C, Bardwell JCA. Oxidative protein folding is driven by the electron transport system. Cell. 1999a;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- Bader M, Muse W, Zander T, Bardwell J. Reconstitution of a protein disulfide catalytic system. J Biol Chem. 1998;273:10302–10307. doi: 10.1074/jbc.273.17.10302. [DOI] [PubMed] [Google Scholar]
- Bader M, Winther JR, Bardwell JCA. Protein oxidation: prime suspect found “not guilty.”. Nat Cell Biol. 1999b;1:E57–E58. doi: 10.1038/11025. [DOI] [PubMed] [Google Scholar]
- Bardwell JCA, Lee J-O, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JCA, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Beinert H. Recent developments in the field of iron-sulfur proteins. FASEB J. 1990;4:2483–2491. doi: 10.1096/fasebj.4.8.2185975. [DOI] [PubMed] [Google Scholar]
- Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cuozzo JW, Kaiser CA. Competition between glutathione and protein thiols for disulphide-bond formation. Nat Cell Biol. 1999;1:130–135. doi: 10.1038/11047. [DOI] [PubMed] [Google Scholar]
- Dailey FE, Berg HC. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Debarbieux L, Beckwith J. Electron avenue: pathways of disulfide bond formation and isomerization. Cell. 1999;99:117–119. doi: 10.1016/s0092-8674(00)81642-6. [DOI] [PubMed] [Google Scholar]
- Ellis LB, Saurugger P, Woodward C. Identification of the three-dimensional thioredoxin motif: related structure in the ORF3 protein of the Staphylococcus aureus mer operon. Biochemistry. 1992;31:4882–4891. doi: 10.1021/bi00135a020. [DOI] [PubMed] [Google Scholar]
- Endrizzi JA, Breddam K, Remington SJ. 2.8-Å structure of yeast serine carboxypeptidase. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- Frand AR, Cuozzo J, Kaiser CA. Pathways for protein disulfide bond formation in the endoplasmic reticulum. Trends Cell Biol. 2000;5:203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. Ero1p oxidizes PDI in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA. such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- Guilhot C, Jander G, Martin NL, Beckwith J. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci USA. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst B, Tachibana C, Winther JR. Active site mutations in yeast protein disulfide isomerase cause dithiothreitol sensitivity and a reduced rate of protein folding in the endoplasmic reticulum. J Cell Biol. 1997;138:1229–1238. doi: 10.1083/jcb.138.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jämsä E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Jander G, Martin NL, Beckwith J. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 1994;13:5121–5127. doi: 10.1002/j.1460-2075.1994.tb06841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1994. [Google Scholar]
- Kishigami S, Ito K. Roles of cysteine residues of DsbB in its activity to reoxidize DsbA, the protein disulfide bond catalyst of Escherichia coli. Genes Cells. 1996;1:201–208. doi: 10.1046/j.1365-2443.1996.d01-233.x. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Kanaya E, Kikuchi M, Ito K. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J. 1999;18:1192–1198. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Krishna TSR, Wong L, Guenther B, Williams CH, Jr, Model P. Convergent evolution of similar function in two structurally divergent enzymes. Nature. 1991;352:172–174. doi: 10.1038/352172a0. [DOI] [PubMed] [Google Scholar]
- Laboissière MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J Biol Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- LaMantia M, Lennarz WJ. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell. 1993;74:899–908. doi: 10.1016/0092-8674(93)90469-7. [DOI] [PubMed] [Google Scholar]
- Lundstrom J, Holmgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- Martin JL. Thioredoxin. A fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Scherens B, Dubois E, Messenguy F. Determination of the sequence of the yeast YCL313 gene localized on chromosome III. Homology with the protein disulfide isomerase (PDI gene product) of other organisms. Yeast. 1991;7:185–193. doi: 10.1002/yea.320070212. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H, Funahashi W, Takeuchi Y, Nakanishi H, Nishihara R, Katoh S, Gao XD, Mizunaga T, Fujimoto D. Overproduction of Mpd2p suppresses the lethality of protein disulfide isomerase depletion in a CXXC sequence dependent manner. Biochem Biophys Res Commun. 1997;239:710–714. doi: 10.1006/bbrc.1997.7426. [DOI] [PubMed] [Google Scholar]
- Walker KW, Gilbert HF. Scanning and escape during protein-disulfide isomerase-assisted protein folding. J Biol Chem. 1997;272:8845–8848. doi: 10.1074/jbc.272.14.8845. [DOI] [PubMed] [Google Scholar]
- Wang PF, Veine DM, Ahn SH, Williams CH., Jr A stable mixed disulfide between thioredoxin reductase and its substrate, thioredoxin: preparation and characterization. Biochemistry. 1996;35:4812–4819. doi: 10.1021/bi9526793. [DOI] [PubMed] [Google Scholar]