Abstract
A fluorogenic-probe hydrolysis (TaqMan)-reverse transcriptase PCR assay for classical swine fever virus (CSFV) was developed and evaluated in experimentally infected swine. The assay detected CSFV, representing different phylogenetic groupings, but did not amplify viral RNA from related pestiviruses. The assay met or exceeded the sensitivity (1 to 100 50% tissue culture infective doses per ml) of viral cultures of samples from experimentally infected animals. Viral RNA was detected in nasal and tonsil scraping samples 2 to 4 days prior to the onset of clinical disease. The assay can be performed in 2 h or less, thus providing a rapid method for the diagnosis of classical swine fever.
Classical swine fever (CSF) is a highly contagious and often fatal disease of swine, affecting domestic and wild pig populations. Classical swine fever virus (CSFV), the causative agent of CSF, is a member of the genus Pestivirus, which belongs to the Flaviviridae family (37). Other important animal pathogens within the genus Pestivirus are bovine viral diarrhea virus (BVDV) of cattle and border disease virus (BDV) of sheep (26), both of which can naturally infect pigs. Antibodies generated during these infections cross-react with CSFV in serologic assays, making a CSF diagnosis problematic (5, 22, 23, 34).
CSFV is an enveloped virus with a 12.5-kb single-stranded RNA genome of positive polarity (12, 38). The genome comprises a single open reading frame that encodes a 4,000-amino-acid polyprotein that is co- and posttranslationally processed by viral and cellular proteases into 12 polypeptides (19). Both ends of the genome contain untranslated regions (UTR), which are highly conserved among all virus isolates (3, 16, 18, 31, 39).
Clinical signs of CSF can remain undetected, particularly during infections with CSFV strains of low virulence (33). Moreover, gross lesions observed at necropsy are diverse and often not pathognomonic (4, 7, 8, 14, 20, 21, 29, 33, 35, 36). Rapid and precise detection of CSFV is critical for disease containment. Current diagnostic methods, including detection of viral antigens in tonsils by using fluorescent antibodies (28) or antigen capture enzyme-linked immunosorbent assay (6, 32) and detection of genomic RNA by reverse transcription-PCR (3, 11, 13, 15, 39), are relatively rapid diagnostic tests; however, these techniques require centralized laboratory facilities and clinical specimen submissions that might delay disease diagnosis, thus affecting the efficiency of emergency disease management measures.
A rapid, presumptive diagnosis at the site of a suspected disease outbreak would be extremely useful for controlling CSF. To address this need, a fluorogenic-probe hydrolysis (TaqMan)-reverse transcriptase (RT) PCR assay for CSFV was developed and evaluated in experimentally infected swine.
The assay was designed as a probe hydrolysis (TaqMan)-RT-PCR single-tube assay. Available CSFV nucleotide sequences were aligned by using BioEdit sequence alignment software. Specific oligonucleotide primers and the fluorogenic probe were designed to target a highly conserved region within the 5′ UTR of the CSFV genome (GenBank accession number NC_000294.1). The locations and sequences of the primers and probes were as follows: forward primer, starting at base position 180, 5′ CCCTGGGTGGTCTAAG; reverse primer, starting at base 247, 5′ CATGCCCTCGTCCAC; and probe, starting at base 139, 5′ CCTGAGTACAGGACAGTCGTCAGTAGTT. The Taqman probe was labeled with a 5′ reporter dye, 6-carboxyfluorescein, and a 3′ quencher, 6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA) (PE Biosystems, Foster City, Calif.).
Reagents from Perkin-Elmer (EZ-RT-PCR; PE Biosystems) were used to prepare master mixture recipes according to the guidelines of the manufacturer for individual component concentrations. The final RT-PCR mixture for a 25-μl-volume assay consisted of the 5× buffer solution supplied with the kit with the addition of 5 mM Mn2 acetate, a 0.3 M concentration of each primer, 0.1 mM dATP, dCTP, or dGTP, 0.2 mM dUTP, 0.1 U of recombinant Tth DNA polymerase per μl, 0.1 μg of bovine serum albumin per μl, and 0.5 M trehalose. The RT-PCR mixture was then dried within the reaction tubes (Fisher Scientific, Atlanta, Ga.). For sample testing, dried reagents were rehydrated with 22.5 μl of 1× buffer (see above) and 2.5 μl of RNA sample material. Cycling consisted of an RT step at 60°C for 10 min, followed by 45 amplification cycles (95°C for 2 s and 60°C for 30 s). The assay was optimized for use on a SmartCycler (Cepheid, Inc., Sunnyvale, Calif.), a 22-lb portable instrument that is operated by a laptop computer. Positive and negative controls consisting of viral RNA and a nontemplate reaction mixture were included with each RT-PCR run.
For all experiments, RNA was extracted from 150 μl of tissue culture supernatant, nasal swab samples, tonsil scrapings, or blood samples with an RNeasy kit or an RNA blood extraction kit (Qiagen, Stanford, Calif.) and resuspended in 40 μl of RNase-free water.
Viral RNAs extracted from CSFV-infected cell supernatants with approximate viral titers of 106 50% tissue culture infectious doses (TCID50) per ml were tested with the real-time RT-PCR assay. All CSFVs were detected with cycle threshold (Ct) values ranging from 25 to 30 (data not shown). Positive fluorogenic signals were observed when testing RNA obtained from tissue culture supernatant of SK6 cells infected with CSFV isolates Haiti-96, DR-1, Brescia, CS, and Shimen, CSF 901 (Alfort), CSF 906 UD (Bergen), CSF 216 V1081/94, CSF 0309 Kanagawa, CSF 0410, CSF 123, CSF 277, CSF 573, and CSF 634 (Table 1). Importantly, the assay detected geographically and temporally distinct viruses belonging to different phylogenetic groups (17) (i.e., Brescia, 1945, Italy; Haiti-96, 1996, Haiti; and recent isolates from Germany, from 1995, 1997, and 1999).
TABLE 1.
Pestiviruses used in testing specificity of CSF real-time RT-PCR
| Pestivirus isolate (original source, yr) | Sourcea |
|---|---|
| CSFV | |
| Haiti-96 (Haiti) | PIADC |
| DR-1 (Dominican Republic) | PIADC |
| Bavaro (Dominican Republic) | PIADC |
| CS vaccine strain (Russia) | Ivanovsky Institute |
| Shimen (China) | Ivanovsky Institute |
| Brescia (Italy) | Ivanovsky Institute |
| CSF 0901 (Alfort) | EU Reference Lab |
| CSF 0906 UD (Bergen) | EU Reference Lab |
| CSF 0216 V1081/94 | EU Reference Lab for CSFV |
| CSF 0410 (congenital tremor) | EU Reference Lab for CSFV |
| CSF 0309 (Kanagawa) | EU Reference Lab for CSFV |
| CSF 123 (Germany, 1995) | EU Reference Lab for CSFV |
| CSF 277 (Germany, 1997) | EU Reference Lab for CSFV |
| CSF 573 (Italy, 1998) | EU Reference Lab for CSFV |
| CSF 634 (Germany, 1999) | EU Reference Lab for CSFV |
| BVDV | |
| NADL | NADC |
| Singer | NADC |
| NY-1 | NADC |
| 890 | NADC |
| SD-1 | NADC |
| 1373 | NADC |
| BDV | |
| Idaho 207 | NADC |
| CB02 | NADC |
| CB05 | NADC |
PIADC, J. Lubroth, Plum Island Animal Disease Center, Animal Plant Health Inspection Service, U.S. Department of Agriculture, Greenport, N.Y.; Ivanovsky Institute, Alexei Zaberezhny, Ivanovsky Institute, Moscow, Russia; EU Reference Lab for CSFV, G. Floegel-Niesmann, Institute of Virology, Hannover Veterinary School, Hannover, Germany; NADC, J. Ridpath, National Animal Disease Center, Agriculture Research Service, U.S. Department of Agriculture, Ames, Iowa.
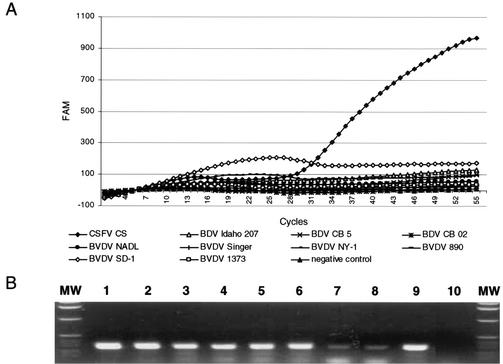
No fluorogenic signal was observed in the CSFV assay with samples containing viral RNA, detected by the classic RT-PCR that is described elsewhere (30), of BVDV strains NADL, Singer, NY-1, 890, SD-1, and 1373 and BDV strains Idaho 207, CB02, and CB05 (Fig. 1), which indicated 100% specificity for the selected panel. Additionally, no false-positive results were observed for nasal swab samples taken from 100 clinically healthy pigs from a farm in the United States (data not shown).
FIG. 1.
Specificity of CSFV RT-PCR assay. (A) Genomic RNAs obtained from CSFV strain CS, BDV strains Idaho 207, CB02, and CB05, and BVDV strains NADL, Singer, NY-1, 890, SD-1, and 1373 were assayed by CSFV-specific real-time RT-PCR. (B) RT-PCR amplification of genomic RNA obtained from the pestiviruses listed above by a general pestivirus RT-PCR assay. Amplicons of the 5′ UTR of viral genomic templates were resolved in a 1% agarose gel stained with ethidium bromide. Shown are results for BVDV strains NADL, Singer, NY-1, 890, SD-1, and 1373 (lanes 1 to 6); BDV strains CB02 and Idaho 207 (lanes 7 and 8); CSFV strain CS (lane 9); and the nontemplate control (lane 10). Lanes MW contain molecular weight standards. FAM, 6-carboxyfluorescein.
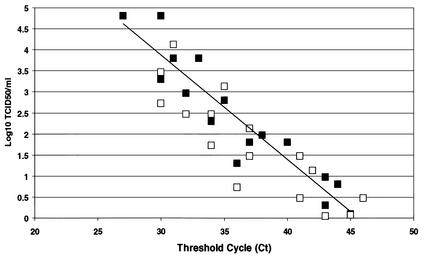
To assess assay sensitivity, viral RNA was extracted from tissue culture supernatants of CSFV-infected SK6 cells and from clinical samples (nasal swabs and tonsil scrapings) obtained from experimentally infected pigs. The samples were diluted in log-10 steps in Dulbecco's minimal essential medium, and RNA was extracted and tested to determine the end-point dilution at which a positive amplification signal could be obtained. The results demonstrated that, regardless of sample origin, the sensitivity of the assay ranged between 1 and 100 TCID50 per ml of the sample. In eight independent determinations, the assay range of linearity was at least 5 log dilutions of extracted CSFV RNA (Fig. 2). After the sample volume used was adjusted for the RNA extraction (140 μl), the yield of purified RNA (40 μl), and the volume tested (2.5 μl), assay sensitivity could be estimated to be between 0.01 and 1 TCID50. It must be emphasized that this is an estimate based on RNA extraction, reverse transcription, and PCR efficiencies that are less than 100%.
FIG. 2.
Sensitivity of CSFV real-time RT-PCR assay was calculated by using CSFV samples obtained from tissue culture (filled squares) or clinical samples (open squares). An inverse linear relationship (r = 0.89) exists between cycle threshold and the logarithm of TCID50 per milliliter originally present in the sample.
The ability of the assay to detect CSFV during acute infection was assessed in experimentally infected pigs. Two 40- to 50-lb pigs were inoculated intramuscularly in the neck with 106 TCID50 of the moderately virulent CSFV isolate Haiti-96 and placed in contact with four naïve animals 24 h postinfection. Nasal swabs, tonsil scrapings, and blood samples were obtained daily from all animals after contact and examined by RT-PCR and virus isolation. CSFV titers in clinical samples were determined on 96-well plates (Costar, Cambridge, Mass.) with SK6 cells. After 4 days of culture, virus infection was determined by an immunoperoxidase assay (1) using CSFV monoclonal antibody 303 (10) and a Vectastain ABC kit (Vector Laboratories, Burlingame, Calif.). Titers were calculated by the method of Reed and Muench and expressed as TCID50 per milliliter (27).
The contact pigs developed fever (body temperature above 104°F) and anorexia 11 to 13 days postexposure. Nasal swabs and tonsil scraping samples were positive by RT-PCR for all contact-exposed pigs at 7 to 8 days postcontact, and the samples were positive for virus isolation, beginning at 11 to 13 days postcontact (Fig. 3).
FIG. 3.
Preclinical detection of CSFV in tonsil scraping (A) and nasal swab (B) samples. Four pigs (animals 10167, 10168, 10169, and 10170) were contact-exposed to experimentally infected pigs. The bars represent body temperature after contact exposure. Empty bars represent samples negative by both real-time RT-PCR and virus isolation, hatched bars represent samples positive by PCR and negative by virus isolation, and filled bars represent nasal swabs samples positive by PCR and virus isolation. Asterisks indicate the time of appearance of CSF clinical symptoms.
Thus, the CSF RT-PCR assay outperformed virus isolation, which is considered the diagnostic standard for CSF (8, 24, 28). CSFV Haiti-96 was detected in the nasal swabs and tonsil scrapings of contact-infected pigs (n = 4) 3 to 5 days before detection by virus isolation (Fig. 3). This enhanced sensitivity suggests the suitability of the assay for early detection of CSF. During contact infection, Haiti-96, a moderately virulent CSFV strain, apparently replicated at very low levels in the tonsils and nasal cavities of contact-infected pigs (Fig. 3) at early stages of infection and was undetectable by virus isolation with SK6 cells for 3 to 5 days. The failure to isolate the virus early in infection could be due to poor replication of moderately virulent CSFV strains in SK6 cells. Given the normal growth kinetics of Haiti-96 in SK6 cells, this does not appear to be the case (G. Risatti, unpublished data). Alternatively, it may be the case that little to no infectious virus is present during the earliest stages of oronasal infection (2). Haiti-96 virus titers observed in oronasal samples tend to be low, indicating that early replication events might be impaired, resulting in less infectious virus. Thus, the amount of infectious virus present in the oronasal cavity early after contact infection is not enough to be detected by virus isolation; however, viral RNA, which is likely to be present in much greater quantities, is detectable by the RT-PCR assay. Although virus isolation is the “gold standard” for CSFV detection, its use as a screening test, with complete blood samples, in clinically healthy pig herds showed little predictive value, with 4.5% of infected herds detected during the 1997 to 1998 CSF epizootics in The Netherlands (9, 25). In the experiment described here, CSFV Haiti-96 was isolated from blood samples at the same time (9 to 10 days postcontact) that it was detected by PCR (Fig. 4). These data clearly indicate that nasal swabs and tonsil scrapings are the samples of choice during the early stages of infection, with blood becoming the sample of choice at later times. Nasal and tonsil samples may have a higher predictive value of detecting CSF infection during disease surveillance screening.
FIG. 4.
Detection of CSFV in blood samples from contact-exposed pigs (animals 10167, 10168, 10169, and 10170) by virus isolation and real time-RT-PCR. The bars represent viremia titers expressed as TCID50 per milliliter. RT-PCR results are expressed as positive (+) or negative (−).
In summary, the assay described here, a fluorogenic-probe hydrolysis (TaqMan)-RT-PCR single-tube assay for rapid detection of CSFV, is performed in a single tube that contains all RT-PCR reagents dried, and test results are obtained by using a portable detection instrument in real time, thus simplifying all pre- and post-PCR operating procedures. The simplicity and portability of the assay allow for its use as a pen-side assay for rapid on-site detection of CSFV.
Importantly, the assay detected the presence of the virus before the appearance of the disease. Virus was detected in the oronasal area 3 to 5 days before the appearance of clinical symptoms and in blood samples 0 to 3 days before the appearance of clinical disease (Fig. 3 and 4). The early detection of CSFV suggests two potential uses in disease control for the assay: as a surveillance tool in areas free of the disease and as a screening assay for monitoring a disease outbreak in real time.
Acknowledgments
We thank Julia Ridpath (National Animal Disease Center, Agriculture Research Service, U.S. Department of Agriculture, Ames, Iowa), Gundula Floegel-Niessel (Institute of Virology, Hannover Veterinary School, Hannover, Germany), and Juan Lubroth (Plum Island Animal Disease Center, Animal Plant Health Inspection Service, U.S. Department of Agriculture, Greenport, N.Y.) for providing virus strains used in this work.
REFERENCES
- 1.Afshar, A., G. C. Dulac, and A. Bouffard. 1989. Application of peroxidase labeled antibody assays for detection of porcine IgG antibodies to hog cholera and bovine viral diarrhea viruses. J. Virol. Methods 23:253-254. [DOI] [PubMed] [Google Scholar]
- 2.Biront, P., J. Leumen, and J. Vandeputte. 1987. Inhibition of virus replication in the tonsils of pigs previously vaccinated with a Chinese strain vaccine and challenged oronasally with a virulent strain of classical swine fever virus. Vet. Microbiol. 14:105-113. [DOI] [PubMed] [Google Scholar]
- 3.Boye, M., S. Kamstrup, and K. Dalsgaard. 1991. Specific sequence amplification of bovine virus diarrhea virus (BVDV) and hog cholera virus and sequencing of BVDV nucleic acid. Vet. Microbiol. 29:1-13. [DOI] [PubMed] [Google Scholar]
- 4.Carbrey, E. A., W. C. Stewart, and S. H. Young. 1966. The changing picture of hog cholera: case studies. J. Am. Vet. Med. Assoc. 149:1724. [Google Scholar]
- 5.Carbrey, E. A., W. C. Stewart, J. I. Kresse, and M. L. Snyder. 1976. Natural infection of pigs with bovine viral diarrhea virus and its differential diagnosis from hog cholera. J. Am. Vet. Med. Assoc. 169:1217-1219. [PubMed] [Google Scholar]
- 6.Clavijo, A., E. M. Zhou, S. Vydelingum, and R. Heckert. 1998. Development and evaluation of a novel antigen capture assay for the detection of classical swine fever virus antigens. Vet. Microbiol. 60:155-168. [DOI] [PubMed] [Google Scholar]
- 7.Dahle, J., and B. A. Liess. 1992. A review on classical swine fever infections in pigs: epizootiology, clinical disease, and pathology. Comp. Immunol. Microbiol. Infect. Dis. 15:203-211. [DOI] [PubMed] [Google Scholar]
- 8.De Smit, A. J., P. L. Eble, E. P. de Kluijver, M. Bloemraad, and A. Bouma. 1999. Laboratory decision-making during the classical swine fever epidemic of 1997-1998 in The Netherlands. Vet. Prev. Med. 42:185-199. [DOI] [PubMed] [Google Scholar]
- 9.De Smit, A. J. 2000. Classical swine fever. Efficacy of marker vaccines and laboratory diagnosis. Ph.D. thesis. University of Utrecht, Utrecht, The Netherlands.
- 10.Edwards, S., V. Moenning, and G. Wensvoort. 1991. The development of an international reference panel of monoclonal antibodies for the differentiation of hog cholera virus from other pestiviruses. Vet. Microbiol. 29:101-108. [DOI] [PubMed] [Google Scholar]
- 11.Harding, M., C. Lutze-Wallace, I. Prud'Homme, X. Zhong, and J. Rola. 1994. Reverse transcriptase-PCR assay for detection of hog cholera virus. J. Clin. Microbiol. 32:2600-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horzinek, M. C. 1991. Pestivirus-taxonomic perspectives. Arch. Virol. Suppl. 3:1-5. [PubMed] [Google Scholar]
- 13.Katz, J. B., J. F. Ridpath, and S. R. Bolin. 1993. Presumptive diagnostic differentiation of hog cholera virus from bovine viral diarrhea and border disease viruses by using cDNA nested-amplification approach. J. Clin. Microbiol. 31:565-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, T. C., Y. Shimizu, T. Kumagai, and J. Sasahara. 1969. Pathogenesis of hog cholera infection in experimentally inoculated swine. Natl. Inst. Anim. Health Q. Tokyo 9:20-27. [PubMed] [Google Scholar]
- 15.Liu, S. T., S. N. Li, D. C. Wang, S. F. Chang, S. C. Chiang, W. C. Ho, Y. S. Chang, and S. S. Lai. 1991. Rapid detection of hog cholera virus in tissues by the polymerase chain reaction. J. Virol. Methods 35:227-236. [DOI] [PubMed] [Google Scholar]
- 16.Lowings, P., D. J. Paton, J. J. Sands, and G. Ibata. 1996. Detection of classical swine fever in blood: a comparison between virus isolation, antigen ELISA, and RT-PCR, p. 130-134. In S. Edwards et al. (ed.), Proceedings of the Third ESVV Symposium on Pestivirus Infections. Institute for Animal Science and Health (ID-DLO), Lelystad, The Netherlands.
- 17.Lowings, P., G. Ibata, J. Needham, and D. Paton. 1996. Classical swine fever virus diversity and evolution. J. Gen. Virol. 77:1311-1321. [DOI] [PubMed] [Google Scholar]
- 18.McGoldrick, A., J. P. Lowings, G. Ibata, J. J. Sands, S. Belak, and D. J. Paton. 1998. A novel approach to the detection of classical swine fever virus by RT-PCR with a fluorogenic probe (TaqMan®). J. Virol. Methods 72:125-135. [DOI] [PubMed] [Google Scholar]
- 19.Meyer, G., H. J. Thiel, and T. Rumenapf. 1996. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J. Virol. 70:1588-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moennig, V. 1992. The hog cholera virus. Comp. Immunol. Microbiol. Infect. Dis. 15:189-201. [DOI] [PubMed] [Google Scholar]
- 21.Moennig, V., and P. G. Plagemann. 1992. The pestiviruses. Adv. Virus Res. 41:53-98. [DOI] [PubMed] [Google Scholar]
- 22.Paton, D. J., V. Simpson, and S. H. Done. 1992. Infection of pig and cattle with bovine viral diarrhea virus on a farm in England. Vet. Res. 131:185-188. [DOI] [PubMed] [Google Scholar]
- 23.Paton, D. J., and S. H. Done. 1994. Congenital infection of pig with ruminant-type pestiviruses. J. Comp. Pathol. 11:151-163. [DOI] [PubMed] [Google Scholar]
- 24.Pearson, J. E. 1992. Hog cholera diagnostic techniques. Comp. Immunol. Microbiol. Infect. Dis. 15:231-239. [DOI] [PubMed] [Google Scholar]
- 25.Pluimers, F. H., P. W. de Leeuw, J. A. Smak, A. R. W. Elbers, and J. A. Stegeman. 1999. Classical swine fever in The Netherlands 1997-1998: a description of organization and measures to eradicate the disease. Prev. Vet. Med. 42:139-155. [DOI] [PubMed] [Google Scholar]
- 26.Pringle, C. R. 1999. The universal system of virus taxonomy, updated to include the new proposal ratified by the International Committee on Taxonomy of Virus during 1998. Arch. Virol. 144:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:709-716. [Google Scholar]
- 28.Ressang, A. A., and G. F. de Boer. 1968. A comparison between the cell culture, frozen tissue section, impression and mucosal smear techniques for fluorescent antibody in the diagnosis of hog cholera. Neth. J. Vet. Sci. 1:72. [Google Scholar]
- 29.Ressang, A. A. 1973. Studies of the pathogenesis of hog cholera. I. Demonstration of hog cholera virus subsequent to oral exposure. Zentbl. Vetmed. Reihe B 20:256-271. [PubMed] [Google Scholar]
- 30.Ridpath, J. F., and S. R. Bolin. 1998. Differentiation of types 1a, 1b, and 2 bovine viral diarrhea virus (BVDV) by PCR. Mol. Cell. Probes 12:237-243. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder, B. A., and T. C. Balassu Chan. 1990. Specific sequence amplification of bovine viral diarrhea virus nucleic acid. Arch. Virol. 111:239-246. [DOI] [PubMed] [Google Scholar]
- 32.Shannon, A. D., C. Morrissy, S. G. Mackintosh, and H. A. Westbury. 1993. Detection of hog cholera virus antigens in experimentally infected pigs using an antigen-capture ELISA. Vet. Microbiol. 34:233-248. [DOI] [PubMed] [Google Scholar]
- 33.Terpstra, C. 1991. Hog cholera: an update of present knowledge. Br. Vet. J. 147:397-406. [DOI] [PubMed] [Google Scholar]
- 34.Terpstra, C., and G. A. Wensvoort. 1997. A congenital persistent infection of bovine virus diarrhea virus in pigs: clinical virological and immunological observations. Vet. Q. 19:97-101. [DOI] [PubMed] [Google Scholar]
- 35.Van Oirschot, J. T., and C. Terptra. 1977. A congenital persistent swine fever infection. I. Clinical and virological observations. Vet. Microbiol. 2:121-132. [Google Scholar]
- 36.Van Oirschot, J. T. 1999. Hog cholera, p. 159-172. In S. D'Allaire. (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 37.Wengler, G., D. W. Bradley, M. S. Colett, F. X. Heinz, R. W. Schlesinger, and J. H. Strauss. 1995. Flaviviridae. Arch. Virol. Suppl. 10:415-427.
- 38.Wensvoort, G., C. Terpstra, E. P. Kluijver, C. de Kragten, and J. C. Warmaar. 1989. Antigenic differences of pestivirus strains with monoclonal antibodies against hog cholera virus. Vet. Microbiol. 21:9-20. [DOI] [PubMed] [Google Scholar]
- 39.Wirtz, B., J.-D. Tratchin, H. K. Muller, and D. B. Mitchell. 1993. Detection of hog cholera virus and differentiation from other pestiviruses by polymerase chain reaction. J. Clin. Microbiol. 31:1148-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]