Abstract
Molecular typing of Neisseria gonorrhoeae strains is an important tool for epidemiological studies of gonococcal infection and transmission. The recently developed multilocus sequence typing (MLST) method is based on the genetic variation among housekeeping genes. As a preliminary investigation for the development of such a method, we characterized the genetic diversity at 18 gonococcal housekeeping gene loci. Approximately 17,500 nucleotides, spanning 18 loci, were sequenced from 24 isolates. Including strain FA 1090, which has been fully sequenced, and three unique glnA sequences from GenBank, the number of alleles identified for the 18 loci ranged from 2 to 18, with a mean of 8.3 alleles per locus. The majority of polymorphic sites were distributed randomly along the genes, consistent with evolution of DNA sequences by point mutation. In addition, several examples of clustered mutations and insertions or deletions were detected, which most likely occurred by recombinational events. While purifying selection is the dominant force driving the evolution of these housekeeping genes, positive selection also appeared to operate on the abcZ and gpdh loci. The 25 completely characterized strains each had a unique allelic profile with as few as three loci (pilA, abcZ, and pip or pgi2). Molecular typing based on the allelic profile of housekeeping genes resolved the isolates better than either porB nucleotide sequencing or typing of the opa gene. The allelic profiles for the pilA, abcZ, and serC loci of paired strains from 16 sexual contacts were identical. A potential MLST for N. gonorrhoeae, based on ∼500- to 600-bp gene fragments of seven housekeeping gene loci, would include the pilA, abcZ, serC, glnA, gdh, gnd, and pip loci.
Gonorrhea is a bacterial infection that is transmitted primarily by sexual contact (for review, see reference 10). Infection of the lower genital tract most commonly causes urethritis in men and cervicitis in women. Ascending infection in women leads to acute pelvic inflammatory disease, one of the most common causes of female infertility in the world. The causative agent of gonorrhea is Neisseria gonorrhoeae. At present, there is no effective vaccine against N. gonorrhoeae; therefore, control of gonococcal infections depends on surveillance of at risk populations, public health measures to limit the spread of infection, and early intervention to treat infected individuals. Knowledge of the gonococcal strains circulating in a community and of temporal changes in prevalent strains can identify patterns of transmission of gonorrhea and guide prevention and control efforts.
A number of typing methods have been developed for N. gonorrhoeae based on growth requirements for specific nutrients and cofactors (1), antibiotic sensitivity (3, 20), differences in electrophoretic mobility of bacterial enzymes (17), and serological reactivity against surface antigens (12). The introduction of PCR and high-throughput nucleotide sequencing has led to the development of several DNA-based typing methods. Two methods that are highly discriminating are the opa gene (OPA) typing method and sequencing of the porB gene (POR sequencing), which we as well as others have employed (11, 16, 18, 22). A disadvantage of the above two methods is that variation at these loci is evolving rapidly and for reasons that are not fully understood, although immune selection plays an important role. An alternative approach is to use variation that is accumulating slowly and that is likely to be selectively neutral. Although only limited variation may be observed at a single locus, high levels of discrimination can be achieved by analyzing many loci. Brian Spratt and colleagues recently developed a molecular typing method, multilocus sequence typing (MLST), which fulfills the above criteria (14). MLST is based on the principles of multilocus enzyme electrophoresis, but characterizes the alleles present at multiple housekeeping genes directly by nucleotide sequencing rather than indirectly by the electrophoretic mobility of their gene products. The advantages of MLST compared to typing methods that involve comparison of DNA fragments in gels are that the data are unambiguous, can be shared between laboratories, and lend themselves to analyses of the genetic relationships among isolates. MLST schemes have been developed for several bacterial species (4-7, 13, 15), demonstrating the broad applicability of the method, and MLST-generated data have been used to address aspects of the population genetics and evolutionary biology of bacterial species (2, 8, 9). As an investigation preliminary to the development of an MLST scheme for N. gonorrhoeae, we have characterized the genetic diversity at 18 gonococcal housekeeping gene loci.
MATERIALS AND METHODS
N. gonorrhoeae strains.
Thirty-two gonococcal isolates from 16 sexual contact pairs were obtained from clients of the Baltimore City Sexually Transmitted Diseases Clinic over a 2-year period from 1991 to 1992. Only one isolate of each pair was subjected to complete genetic analysis. Two additional isolates were obtained from individuals who reported sexual contact, but the isolates were shown to be genetically and serologically very distinct, raising concerns about laboratory misclassification or inaccurate reporting of sexual behavior (22). Six gonococcal isolates recovered between 1940 and 1941 from subjects living in the Washington, D.C., area, were obtained from the Bacterial Collection of the Walter Reed Army Institute. The isolates from Baltimore had previously been genotyped by POR sequencing and OPA typing, and those from the 1940s had been typed by POR sequencing (19, 22). The N. gonorrhoeae isolates from Baltimore were derived from single colonies picked from primary isolation plates, subcultured once or twice, and frozen. The strains had not been passaged prior to this study. No information was available on the passage history of the strains from Walter Reed Army Institute.
Preparation of chromosomal DNA.
For DNA analysis, strains were plated from frozen stocks onto chocolate II agar plates (Becton Dickinson Microbiology, Franklin Lakes, N.J.) and incubated overnight at 37°C in 5% CO2. Genomic DNA was prepared from N. gonorrhoeae cells scraped off an agar plate culture by using the Clontech Nucleopsin tissue kit (BD Biosciences, Clontech, Palo Alto, Calif.) according to the manufacturer's instructions.
Amplification and nucleotide sequence determination of housekeeping genes.
PCR products were amplified with oligonucleotide primer pairs designed from the genome sequence of N. gonorrhoeae strain FA1090 (www.genome.ou.edu/gono.html). The primers are shown in Table 1. Each 50-μl amplification reaction mixture contained ∼10 ng of gonococcal chromosomal DNA, 0.5 μM each PCR primer, 1× Expand PCR buffer (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), 0.2 mM deoxynucleotide triphospates, and 2 U of Expand High Fidelity (Boehringer Mannheim Biochemicals). The reaction conditions were 94°C for 1 min, primer annealing at 55°C for 30 s, and extension at 72°C for 30 s for 30 cycles and a final extension reaction for 10 min at 72°C. The amplification products were purified with the QIAaquick PCR purification kit (Qiagen, Valencia, Calif.), and their nucleotide sequences were determined on each DNA strand by using the amplification primers or sequencing primers (not shown) and half the amount of the manufacturer's recommended BigDye Ready Reaction mix (PE Biosystems, Foster City, Calif.). Unincorporated dye terminators were removed by the RapXtract II kit (Prolinx, Bothell, Wash.), and the reaction products were separated and detected with an ABI Prism 3700 automated DNA sequencer from PE Biosystems (Synthesis and Sequencing Facility, Department of Biological Chemistry, Johns Hopkins University School of Medicine). Trace data were edited, and nucleotide sequences were assembled with the SeqMan software program (DNASTAR, Inc., Madison, Wis.).
TABLE 1.
PCR primers and sizes of gene fragments for 18 N. gonorrhoeae housekeeping genes
| Locus | Primer sequence
|
Amplicon | Size (bp) of:
|
% of ORF sequenced | |
|---|---|---|---|---|---|
| Forward | Reverse | Sequenced gene fragment | |||
| abcZ | 5′-CATCATTATTGTTTCCGTCCTGCC | 5′-GATGATTTCTTCAGGTCGTTTGA | 2,053 | 884 | 46.33 |
| adk | 5′-CAAGCCGTGTAGAATCGTAAACCA | 5′-CTGCCCGTGGGTACGACCTTTTC | 738 | 610 | 94.57 |
| aroE | 5′-CGCCCCGCCTTTATGGAATATGA | 5′-AGCCATTTGACGATGCGGAACAT | 861 | 796 | 98.64 |
| fumC | 5′-GAGAAACGTATGAACACCCGCACC | 5′-CTTCGCATTGGGTCGGGTTGACTT | 999 | 856 | 61.76 |
| gdh | 5′-CTGCTTGCGCTGCGTTTTGCCAAT | 5′-TTGTGATTTCAGACGGCATATCCC | 1,029 | 861 | 100.00 |
| glnA | 5′-GTGTTCAGACGGCATTGCTACTGA | 5′-CCGCATATTTTGCCATTTCCCTTT | 1,491 | 1,356 | 95.76 |
| gnd | 5′-TTAAATCTATAACGGAGCTTCCTG | 5′-TCTATTTCCTGCAAAACAAATGCC | 1,533 | 1,446 | 100.00 |
| gpdh | 5′-CCAAACAGGAAGCCGCACAGAATA | 5′-CTGAAACTTGAAAACATCGGGTTT | 1,318 | 1,023 | 99.42 |
| gpdhC | 5′-CTTTATTTCACAACCGGGAGACAA | 5′-GCAATGTGTTCAGACGGCATTTAC | 1,078 | 992 | 99.00 |
| pdhC | 5′-GCAAACAGGCCGTCTGAAACATCA | 5′-GATGAACGAGAACTACACCCATC | 617 | 498 | 88.77 |
| pgi1 | 5′-GAAAACCTCAATCGGGAGCATACA | 5′-TCGATAAAAATGCCGTCTGAAAC | 1,728 | 954 | 58.03 |
| pgi2 | 5′-TACAGAAAGGCGGCGGTACTTTTA | 5′-GCCGTTTGTTAGACTACATTCTGC | 1,726 | 1,618 | 98.60 |
| pgm | 5′-TTTACAGAGGCCAGATTAAATGCG | 5′-GGCGAAGCCATTCAAGAACTTTTA | 1,017 | 906 | 65.65 |
| pilA | 5′-CGCTACAATACGCCCTATTTCAAG | 5′-TTTGCAGCACGCGCTTCACTTTTT | 1,045 | 932 | 73.79 |
| pip | 5′-AAAACAAAAGCAGCTTCCAAACA | 5′-GGATTGTCAGAAGCAATATGGGAG | 1,012 | 826 | 88.82 |
| ppk | 5′-AGCGTATGGGGGAAGTGCTTGTGC | 5′-CGTGGTCGATATGATACGCGAAGC | 1,029 | 906 | 44.09 |
| pyrD | 5′-TTCCAAATGACGGCTGATTG | 5′-CGTTTACGCCCAGTTGTTCA | 1,112 | 1,005 | 100.00 |
| serC | 5′-ACAATCTTACCACCTCCAAAACGC | 5′-AAGCCTGCTGCAAGCCTAAAATCG | 1,246 | 1,105 | 100.00 |
Sequence analysis.
For each of the housekeeping gene fragments, the sequences from all isolates were compared, and allele numbers were assigned to each unique sequence. No weighting was given to the degree of sequence divergence between alleles, since in the absence of knowledge of the proportion of allelic changes that are due to recombination rather than mutation, one cannot say that alleles that differ at many sites are any more distantly related than those that differ at a single site. The alleles present at the 18 loci define the allelic profile of a strain. The allelic profiles were entered into an Excel database. The data were loaded from Excel into the S.T.A.R.T. program (Keith Jolley, University of Oxford), which was used to produce a matrix of pairwise differences in the allelic profiles and to construct a dendrogram from the matrix by the unweighted pair group method with arithmetic means (UPGMA) method. The average frequencies of synonymous substitutions per potential synonymous site (ds) and nonsynonymous substitutions per potential nonsynonymous site (dn) were calculated by the method of Nei and Gojobori. The Sawyer's run test was performed with the S.T.A.R.T. program with 10,000 resamplings of the data.
Nucleotide sequence accession numbers.
Novel sequences determined in this study have been deposited in GenBank under accession no. AF520224 to AF520355.
RESULTS
Choice of loci.
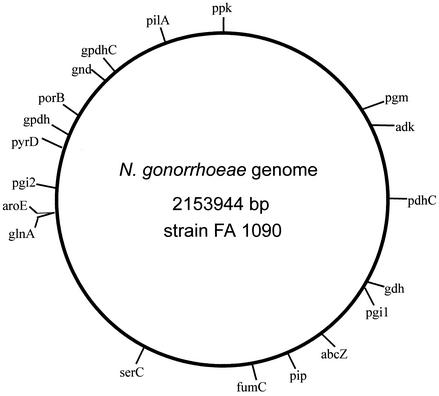
We identified the loci of 18 genes encoding enzymes responsible for intermediate metabolism by searching the N. gonorrhoeae (strain FA 1090) genome database with gene sequences from Neisseria meningitidis (21). Primers were designed to amplify and sequence segments of each gene ranging from ∼600 bp to ∼2 kb (Table 1). The resulting segments included more than 90% of the putative open reading frame (ORF) of 9 loci and from 44 to 89% of the ORF of the remaining loci. The locations of the 18 genes on a physical map of the N. gonorrhoeae genome (strain FA 1090) are shown in Fig. 1. For strain FA 1090, the median distance between loci was 99,194 bp (interquartile range, 27,207 to 147,739 bp). The most closely spaced genes, glnA and aroE, were separated by 310 bp.
FIG. 1.
Location of 18 housekeeping genes and porB on chromosomal map of N. gonorrhoeae strain FA 1090
Diversity of housekeeping genes.
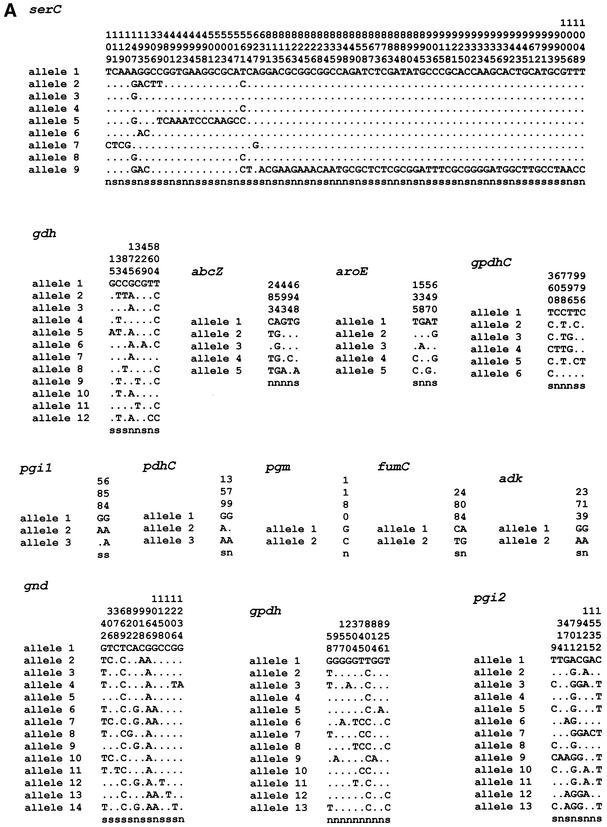
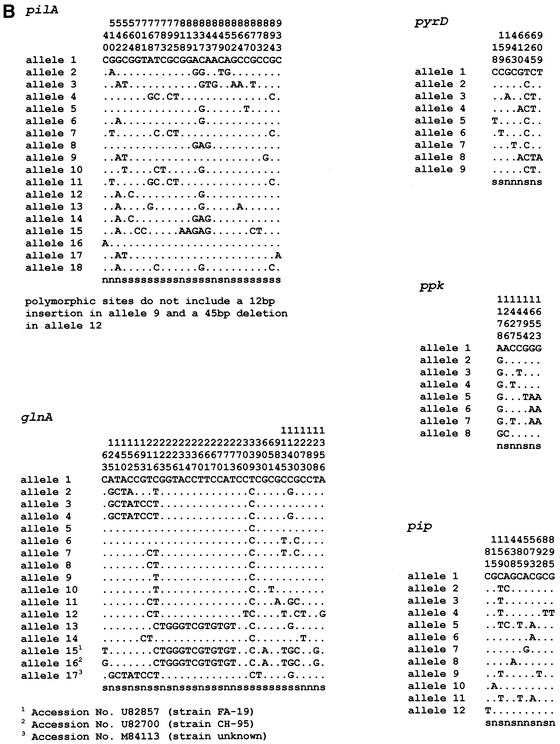
In all, ∼17,500 nucleotides spanning 18 loci were sequenced from 24 isolates. For each isolate, the sequences obtained at each of the 18 loci were compared with those of every other isolate, and sequences were assigned as distinct alleles if they differed at one or more nucleotide sites. A search of the GenBank database revealed three unique N. gonorrhoeae glnA sequences. No N. gonorrhoeae sequences were found in GenBank that matched the other 17 loci. The allele from strain FA 1090, whose genome has been completely sequenced, was arbitrarily assigned as allele 1. The number of alleles identified at the 18 loci ranged from 2 to 18 (Table 2). The mean number of alleles per locus was 8.3, which would potentially allow >3.7 × 1016 sequence types to be distinguished. The number of polymorphic (variable) sites at the 18 loci varied between 2 and 75. The positions of the polymorphic sites for all loci are shown in Fig. 2. Visual inspection of the sequences suggested that, with two exceptions, the distribution of polymorphic sites along each gene was random. An identical cluster of 13 polymorphic sites occurred in three alleles of glnA between positions 216 and 273. Three other alleles of glnA shared eight identical polymorphic sites over a region spanning positions 125 to 216. Two alleles of serC contained clusters of polymorphic sites; allele 5 had 12 polymorphic sites between positions 480 and 507, and allele 9 had 51 polymorphic sites distributed over the region from positions 639 to 1049. The visual impression was confirmed by the results of Sawyer's run test, which provided strong support for recombination (P < 0.0001 and P = 0.0003 for glnA and serC, respectively). Allele 1 of pilA contained a 12-bp in-frame insertion at position 250, and allele 12 had a 45-bp in-frame deletion from positions 286 to 330 (data not shown in Fig. 2). The average percent nucleotide difference between pairs of alleles was ≤2.0% at 17 of the 18 loci (Table 2). For serC, the interallelic differences ranged from 0.1 to 6.1%. The high value was due to the presence of a large number of unique polymorphic sites in allele 9. The mean percentage of nucleotide differences between alleles of N. gonorrhaoeae and the most closely related N. meningitidis allele at the 18 loci was 5.3%, with a range of 1.55% to 9.50% (Table 2). The dn/ds ratio was calculated as a measure of the degree of selection in the population of sequences (Table 2). As expected for evolutionarily conserved genes, the ratio was less than 1.0 for 14 housekeeping gene loci. For abcZ and gpdh, the dn/ds ratios were 1.3132 and 1.5239, respectively.
TABLE 2.
Nucleotide sequence variation in 18 N. gonorrhoeae housekeeping gene fragments
| Locus | Putative function | No. of alleles | No. of polymorphic sites | % Nucleotide difference
|
dn/ds ratio | |
|---|---|---|---|---|---|---|
| Between pairs of alleles (range) | vs N. meningitidis (mean) | |||||
| abcZ | ATP-binding protein | 5 | 5 | 0.1-0.5 | 6.24 | 1.3132 |
| adk | Adenylate kinase | 2 | 2 | 0.3 | 4.80 | 0.3035 |
| aroE | Shikimate dehydrogenase | 5 | 4 | 0.1-0.4 | 7.82 | 0.2168 |
| fumC | Fumarate hydratase class II | 2 | 2 | 0.2 | 5.90 | 0.3323 |
| gdh | Glucose-6-phosphate 1-dehydrogenase | 12 | 8 | 0.1-0.5 | 9.33 | 0.1857 |
| glnA | Glutamine synthetase | 14 | 35 | 0.1-2.0 | 4.22 | 0.1191 |
| gnd | 6-Phosphogluconate dehydrogenase | 14 | 13 | 0.1-0.4 | 1.55 | 0.1586 |
| gpdh | Glyceraldehyde 3-phosphate dehydrogenase | 13 | 10 | 0.1-0.6 | 5.98 | 1.5239 |
| gpdhC | Glyceraldehyde 3-phosphate dehydrogenase | 6 | 6 | 0.1-0.4 | 3.02 | 0.3849 |
| pdhC | Pyruvate dehydrogenase | 3 | 2 | 0.2-0.4 | 4.73 | 0.3274 |
| pgi1 | Glucose-6-phosphate isomerase | 3 | 2 | 0.1-0.2 | 2.57 | 0 |
| pgi2 | Glucose-6-phosphate isomerase | 13 | 8 | 0.1-0.4 | 4.75 | 0.1533 |
| pgm | Phosphoglucomutase | 2 | 1 | 0.1 | 6.15 | 0 |
| pilA | Regulatory protein PilA | 18 | 28 | 0.1-1.7 | 5.89 | 0.0722 |
| pip | Proline iminopeptidase | 12 | 11 | 0.1-0.6 | 3.08 | 0.3108 |
| ppk | Polyphosphate kinase | 8 | 7 | 0.1-0.4 | 3.31 | 0.4626 |
| pyrD | Dihydroorotate dehydrogenase | 9 | 8 | 0.1-0.4 | 9.50 | 0.5009 |
| serC | 3-Phosphoserine aminotransferase | 9 | 75 | 0.1-6.1 | 5.20 | 0.2323 |
FIG. 2.
The variable sites in each of the unique sequences (alleles) of 18 gonococcal housekeeping genes. The nucleotide present at each variable site among the 25 isolates is shown for strain FA 1090 (allele 1). For the other alleles, only those sites that differ are shown. Sites that are the same as those in allele 1 are shown by dots. Nucleotide sites that are the same in all alleles are not shown. The sites are numbered above in vertical format based on the nucleotide numbering of N. gonorrhoeae strain FA 1090. The polymorphisms that are synonymous (s) and nonsynonymous (n) are shown below.
Genetic relatedness of gonococcal isolates.
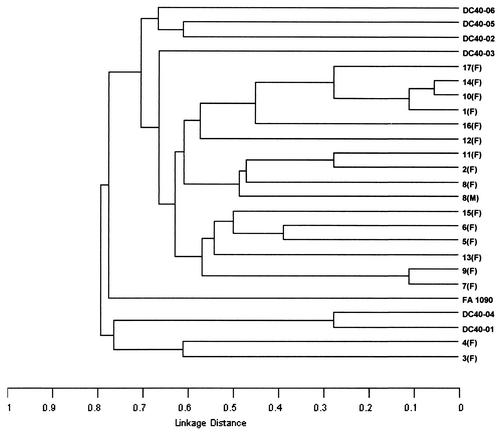
The allelic profiles obtained for 25 strains are shown in Table 3, and a dendrogram based on a matrix of pairwise differences between the allelic profiles is shown in Fig. 3. Each of the 25 strains had a unique allelic profile, and the profile from as few as three loci (pilA, abcZ, and pip or pgi2) was sufficient to distinguish all strains.
TABLE 3.
Allelic profile of 25 N. gonorrhoeae strains
| Isolate | Allele no.
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fumC | gdh | pdhC | pilA | pip | ppk | glnA | gpdh | adk | aroE | pgm | gpdhC | pyrD | abcZ | serC | pgi1 | pgi2 | gnd | |
| DC40-06 | 1 | 2 | 2 | 17 | 12 | 6 | 12 | 13 | 1 | 5 | 1 | 5 | 4 | 5 | 9 | 3 | 3 | 12 |
| DC40-05 | 1 | 2 | 3 | 16 | 11 | 6 | 11 | 12 | 1 | 1 | 1 | 4 | 4 | 3 | 7 | 2 | 4 | 11 |
| DC40-02 | 1 | 2 | 2 | 14 | 9 | 6 | 4 | 10 | 1 | 1 | 1 | 3 | 8 | 1 | 3 | 2 | 2 | 13 |
| DC40-03 | 1 | 10 | 2 | 15 | 2 | 6 | 13 | 11 | 1 | 1 | 1 | 6 | 9 | 4 | 7 | 3 | 12 | 10 |
| 17(F) | 1 | 12 | 2 | 3 | 8 | 8 | 14 | 4 | 1 | 1 | 1 | 2 | 3 | 2 | 4 | 3 | 2 | 14 |
| 14(F) | 1 | 12 | 2 | 3 | 2 | 8 | 14 | 4 | 1 | 1 | 1 | 6 | 3 | 3 | 7 | 3 | 13 | 14 |
| 10(F) | 1 | 12 | 2 | 3 | 2 | 8 | 14 | 4 | 1 | 1 | 1 | 6 | 3 | 1 | 7 | 3 | 13 | 14 |
| 1(F) | 1 | 12 | 1 | 3 | 2 | 8 | 14 | 4 | 1 | 1 | 1 | 6 | 3 | 2 | 7 | 3 | 13 | 14 |
| 16(F) | 1 | 2 | 1 | 12 | 8 | 6 | 3 | 4 | 1 | 1 | 1 | 6 | 3 | 3 | 4 | 3 | 9 | 14 |
| 12(F) | 1 | 4 | 1 | 10 | 3 | 4 | 7 | 7 | 1 | 1 | 1 | 6 | 7 | 3 | 4 | 3 | 7 | 7 |
| 11(F) | 1 | 2 | 2 | 9 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 6 | 2 | 2 | 6 | 3 | 6 | 6 |
| 2(F) | 1 | 2 | 2 | 5 | 1 | 2 | 2 | 1 | 1 | 3 | 1 | 6 | 2 | 1 | 6 | 3 | 2 | 2 |
| 8(F) | 1 | 3 | 2 | 7 | 1 | 2 | 4 | 2 | 1 | 2 | 1 | 6 | 2 | 3 | 8 | 3 | 10 | 6 |
| 8(M) | 1 | 2 | 1 | 8 | 3 | 2 | 6 | 4 | 1 | 1 | 1 | 6 | 2 | 2 | 8 | 3 | 3 | 6 |
| 15(F) | 1 | 8 | 2 | 4 | 7 | 5 | 8 | 8 | 1 | 1 | 1 | 6 | 1 | 3 | 2 | 3 | 3 | 8 |
| 6(F) | 1 | 4 | 2 | 18 | 5 | 2 | 8 | 2 | 1 | 1 | 1 | 6 | 4 | 2 | 3 | 3 | 5 | 5 |
| 5(F) | 1 | 6 | 2 | 18 | 6 | 2 | 8 | 6 | 1 | 1 | 1 | 6 | 1 | 3 | 3 | 3 | 2 | 4 |
| 13(F) | 2 | 7 | 2 | 11 | 7 | 8 | 8 | 1 | 1 | 1 | 1 | 6 | 6 | 3 | 5 | 3 | 8 | 3 |
| 9(F) | 2 | 5 | 1 | 2 | 4 | 2 | 8 | 3 | 1 | 1 | 1 | 6 | 5 | 3 | 6 | 3 | 3 | 7 |
| 7(F) | 2 | 5 | 1 | 2 | 4 | 2 | 8 | 3 | 1 | 1 | 1 | 6 | 5 | 2 | 6 | 3 | 3 | 3 |
| FA 1090 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| DC40-04 | 1 | 11 | 1 | 18 | 10 | 7 | 10 | 4 | 1 | 4 | 2 | 6 | 2 | 3 | 3 | 2 | 11 | 9 |
| DC40-01 | 1 | 9 | 1 | 13 | 1 | 7 | 9 | 9 | 1 | 4 | 2 | 6 | 2 | 3 | 3 | 2 | 11 | 9 |
| 4(F) | 1 | 3 | 2 | 4 | 1 | 2 | 5 | 1 | 2 | 1 | 2 | 3 | 1 | 3 | 8 | 3 | 4 | 3 |
| 3(F) | 1 | 3 | 2 | 6 | 6 | 3 | 3 | 5 | 1 | 2 | 2 | 2 | 2 | 3 | 1 | 3 | 4 | 2 |
FIG. 3.
Dendrogram of 25 N. gonorrhoeae strains. For each strain, the allele at 18 housekeeping gene loci defined an allelic profile. The dendrogram was constructed by the UPGMA method from a matrix of pairwise differences in the allelic profiles of the 25 strains.
Comparison of MLST, POR sequencing, and OPA typing.
The 18 strains obtained from clients of the Baltimore City Sexually Transmitted Diseases Clinic had previously been typed by POR sequencing and OPA typing, and the 6 strains from the 1940s had previously been typed by POR sequencing. Strains 7(F) and 9(F), which had identical OPA types, differed at two housekeeping loci (abcZ and gnd). Several pairs of strains had the same porB sequence, but differed at one or more housekeeping gene loci. Strains 10(F) and 14(F) differed at 1 of the 18 housekeeping gene loci (abcZ); strains DC40-01 and DC40-04 differed at 5 loci; and strains 8(F) and 15(F) differed at 11 loci. Seven of the 25 strains fell within the porB PIA homology group, and 18 strains fell within the PIB homology group. There was no clustering of strains according to porB homology group in the dendrogram based on the allelic profile of the 18 housekeeping genes (data not shown). For 16 study subjects, a gonococcal isolate was available from an epidemiologically linked sex partner. The allelic profiles derived from the pilA, abcZ, and serC loci of paired strains from sexual contacts were identical.
DISCUSSION
A bacterial typing system that is used for transmission studies has to be highly discriminating in order to distinguish epidemiologically unrelated infections. For MLST, which is based on genetic variation of housekeeping loci, multiple loci must be used because the variation at any single locus is low. The number and choice of loci have to be determined empirically. We identified in the genome sequence of N. gonorrhoeae strain FA 1090 homologues of 11 housekeeping genes described in the published literature on an MLST scheme for N. meningitidis. We selected an additional seven N. gonorrhoeae housekeeping gene loci that showed modest nucleotide sequence divergence compared to their homologues in N. meningitidis. The locations of the genes on the chromosomal map of N. gonorrhoeae strain FA 1090 showed that the loci were spread around the genome, with a median distance between loci of ∼100,000 bp. Ideally, the loci should be spaced far enough apart that pairs of alleles are unlikely to be inherited together by recombinational events, because this would diminish the independent discriminatory power of each locus. Approximately 300 bp separated aroE from glnA, and ∼3,000 bp separated gdh and pgi1. Thus, the use of both loci of these pairs would be inadvisable in designing an MLST for N. gonorrhoeae. One caveat to this recommendation is that, for other strains, the genomic locations of the loci may differ.
Although only 25 gonococcal strains were studied, and 18 were obtained from a temporally and geographically restricted population, eight or more alleles were identified at 10 of the 18 loci examined, and from two to six alleles were identified at the remaining 8 loci. It is difficult to compare the diversity of gonococcal housekeeping genes to that of other bacterial species, because the loci and the numbers and sources of the strains studied vary. The distribution of polymorphic sites along each gene fragment was generally random and characterized by single nucleotide replacements, consistent with evolution of DNA sequences by point mutation. However, clusters of polymorphic sites were observed in some alleles of glnA and serC. Clustered polymorphisms may be indicative of a recombinational event. Two alleles of pilA contained either an insertion or deletion. Insertions and deletions may arise through recombination or replication slippage. In general, pairwise differences in nucleotide substitutions between alleles of N. gonorrhoeae were lower (average, ∼2.0%) than those between gonococcal and meningococcal alleles (average, 5.3%). However, for allele 9 of serC, the average intraspecies nucleotide difference (6.1%) was greater than the average nucleotide difference between gonococcal and meningococcal alleles (5.2%). This may be an example of an interspecies recombinational event at the serC locus. Housekeeping genes are believed to be either selectively neutral or subject to purifying selection. Thus, the rate of synonymous substitutions (ds) should be equal to or slightly greater than the rate of nonsynonymous substitutions (dn), giving a dn/ds ratio of ≤1. For 14 of the 16 housekeeping gene loci, for which sufficient data were available to calculate dn and ds, the ratio was <1. However the ratios were ∼1.3 and ∼1.5 for abcZ and gpdh, respectively, suggesting that these genes may be subject to positive Darwinian selection. If ratios >1 are confirmed with a larger data set, possible explanations for positive selection acting on these loci, such as differences in pathogenic potential, transmissibility or tissue tropism, should be investigated.
How many and which loci should be used in an MLST scheme depend on the cost and level of discrimination desired. The MLST schemes for N. meningitidis, Streptococcus pneumoniae, Streptococcus pyogenes, Staphylococcus aureus, and Campylobacter jejuni are all based on seven loci (http://www.mlst.net/). Knowledge of the number and position of polymorphic sites at a locus is essential for selecting the most informative set of loci. Another consideration is the size of the gene fragment. Because of the technical limitations of PCR product sequencing and the need to sequence both strands for maximal accuracy, gene fragments should be no longer than ∼600 bp. The data from our study can be used to help design an MLST scheme for N. gonorrhoeae. The 25 strains we studied were completely resolved by the allelic profiles of three housekeeping genes: pilA, abcZ, and pip or pgi2. The pgi2 locus is not ideal for an MLST scheme, because the polymorphic sites were distributed over ∼1,500 bp. As additional strains of N. gonorrhoeae are typed, it is likely that the list of loci will need to expand in order to achieve a high level of discrimination among strains. Other potentially informative loci include glnA from positions 125 to 651, gdh from positions 15 to 560, gnd from positions 669 to 1234, and pip from positions 81 to 573.
Typing of bacterial isolates can be used for clinical purposes to establish strain-specific correlates of microbial pathogenesis, for epidemiological purposes to investigate the endemic and epidemic spread of organisms, and for genetic purposes to better understand bacterial evolution. The choice of typing method will depend on the question that needs to be addressed. For clinical studies, it may be sufficient to study a single candidate virulence gene. MLST is the preferred method for evolutionary studies, because the variation detected is amenable to quantitative analysis and is believed to be selectively neutral. For epidemiological studies, the sometimes-conflicting demands of discriminatory power and clonal stability must be balanced. Short-term epidemiological studies require methods with high discriminatory power in order to identify recent transmission events. For long-term epidemiological studies, a genetic marker must exhibit sufficient clonal stability that ancestral relationships among strains can be discerned. N. gonorrhoeae poses a particular challenge, because the extent of recombination is very high. MLST is based on slowly accumulating genetic variation and thus is ideal for addressing long-term epidemiological questions. Previous investigations using MLST have been concerned primarily with the global molecular epidemiology of various bacterial species (4, 6,7, 14). However, by combining data from several genetic loci into an allelic profile, MLST can achieve high levels of discrimination. We studied strains obtained from a single sexually transmitted diseases clinic in Baltimore over a 2-year period and found that MLST could resolve the isolates better than either POR sequencing or OPA typing. The discriminatory power of MLST did not preclude the ability to identify the same strain in individuals who were known sexual contacts. Although the data set was small, the results suggest that MLST for N. gonorrhoeae may be useful for both short-term and long-term epidemiological studies. In future studies, we plan to characterize strains isolated in different years and from different subpopulations in Baltimore in order to assess changes in allele frequencies with time and in relation to the distribution of strains within core and peripheral transmission groups. In addition, we will examine strains recovered in association with local genital mucosal infection or systemic disseminated disease.
Acknowledgments
This work was supported by NIH grant RO1-AI50217 to R.P.V.
REFERENCES
- 1.Catlin, B. W. 1973. Nutritional profiles of Neisseria gonorrhoeae, Neisseria meningitidis, and Neisseria lactamica in chemically defined media and the use of growth requirements for gonococcal typing. J. Infect. Dis. 128:178-194. [DOI] [PubMed] [Google Scholar]
- 2.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 3.Dillon, J. R., S. M. Bygdeman, and E. G. Sandstrom. 1987. Serological ecology of Neisseria gonorrhoeae (PPNG and non-PPNG) strains: Canadian perspective. Genitourin. Med. 63:160-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 10.Handsfield, H. H., and P. F. Sparling. 1995. Neisseria gonorrhoeae, p. 1909-1926. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas and Bennett's principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 11.Hobbs, M. M., T. M. Alcorn, R. H. Davis, W. Fischer, J. C. Thomas, I. Martin, C. Ison, P. F. Sparling, and M. S. Cohen. 1999. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179:371-381. [DOI] [PubMed] [Google Scholar]
- 12.Knapp, J. S., M. R. Tam, R. C. Nowinski, K. K. Holmes, and E. G. Sandstrom. 1984. Serological classification of Neisseria gonorrhoeae with use of monoclonal antibodies to gonococcal outer membrane protein I. J. Infect. Dis. 150:44-48. [DOI] [PubMed] [Google Scholar]
- 13.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nallapareddy, S. R., R.-W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Rourke, M., C. A. Ison, A. M. Renton, and B. G. Spratt. 1995. Opa-typing: a high-resolution tool for studying the epidemiology of gonorrhoea. Mol. Microbiol. 17:865-875. [DOI] [PubMed] [Google Scholar]
- 17.O'Rourke, M., and E. Stevens. 1993. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J. Gen. Microbiol. 139:2603-2611. [DOI] [PubMed] [Google Scholar]
- 18.Poh, C. L., G. K. Loh, and J. W. Tapsall. 1995. Resolution of clonal subgroups among Neisseria gonorrhoeae IB-2 and IB-6 serovars by pulsed-field gel electrophoresis. Genitourin. Med. 71:145-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posada, D., K. A. Crandall, M. Nguyen, J. C. Demma, and R. P. Viscidi. 2000. Population genetics of the porB gene of Neisseria gonorrhoeae: different dynamics in different homology groups. Mol. Biol. Evol. 17:423-436. [DOI] [PubMed] [Google Scholar]
- 20.Schwarcz, S. K., J. M. Zenilman, D. Schnell, J. S. Knapp, E. W. Hook III, S. Thompson, F. N. Judson, and K. K. Holmes. 1990. National surveillance of antimicrobial resistance in Neisseria gonorrhoeae. The Gonococcal Isolate Surveillance Project. JAMA 264:1413-1417. [PubMed] [Google Scholar]
- 21.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 22.Viscidi, R. P., J. C. Demma, J. Gu, and J. Zenilman. 2000. Comparison of sequencing of the por gene and typing of the opa gene for discrimination of Neisseria gonorrhoeae strains from sexual contacts. J. Clin. Microbiol. 38:4430-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]