Abstract
We compared the performance characteristics of a real-time PCR method, the LightCycler Strep-A assay (Roche Applied Science, Indianapolis, Ind.), to those of a rapid antigen immunoassay, the Directigen 1-2-3 Group A Strep Test kit (BD Diagnostic Systems, Sparks, Md.), and a standard culture method for detection of group A streptococci (GAS) from 384 throat swabs. The LightCycler PCR produced more positive results (n = 58) than either culture (n = 55) or the Directigen immunoassay (n = 31). The results of the LightCycler PCR and the Directigen method were independently compared to the results of the accepted “gold standard,” bacterial culture. The sensitivities, specificities, and positive and negative predictive values for this comparison were as follows: for the Directigen method, 55, 99, 97, and 93%, respectively; for the LightCycler PCR, 93, 98, 88, and 99%, respectively. In no case was a throat swab positive by both the LightCycler PCR and the Directigen method but negative by culture. The medical histories of patients whose throat swabs were negative by culture but positive by either the LightCycler PCR (n = 7) or the Directigen method (n = 1) were reviewed. All of these patients had signs or symptoms compatible with GAS disease, and therefore, all of these discordant positive results (along with positive results by either the Directigen method or the LightCycler PCR that agreed with the culture results) were counted as true positives for statistical analysis. For this analysis, the LightCycler PCR detected more true-positive results than the culture method (58 versus 55 swabs); however, this difference was not statistically significant (P = 0.5465). In contrast, statistically significantly more true-positive results occurred by culture than by the Directigen method (55 versus 31 swabs; P < 0.0001) and by the LightCycler PCR than by the Directigen method (58 versus 31 swabs; P < 0.0001). The LightCycler PCR is a suitable stand-alone method for the detection of GAS from throat swabs. Additionally, this method requires less than half the personnel time and the procedure can be completed in considerably less time (∼1 h) than our standard approach (up to 2 days) for detection of GAS in throat swabs (i.e., testing by the Directigen method with negative results verified by culture).
Group A streptococci (GAS) are the most common cause of acute pharyngitis and account for 15 to 30% of cases of acute pharyngitis in children and 5 to 10% percent of cases in adults (4). Rapid direct detection of GAS is preferred by patients and physicians. If the diagnosis can be provided on the same day as an office visit, antibiotic therapy can be provided immediately so that symptoms can be relieved sooner, sequelae, including rheumatic fever, can be prevented, and absenteeism from work, day care, or school can be decreased.
Over the past decade, streptococcal antigen immunoassays, also referred to as “rapid antigen assays,” have been available for point-of-care testing for GAS in physicians' offices. Although these tests provide a rapid (same-day) test result, they are expensive and sometimes challenging to perform. Frequently, these assays require multiple manual steps and interpretation, the latter of which requires visual inspection and is sometimes difficult. Furthermore, these assays lack sensitivity and, in some instances, specificity compared to the “gold standard” test for detection of GAS, bacterial culture (14, 22). Several U.S. national advisory committees recommend that, because rapid immunoassays lack sensitivity, all specimens with negative results by these tests be cultured (1, 5, 8). A major drawback of culture is that it requires a minimum of 16 to 18 h for detection of a positive result.
Recently, rapid-cycle real-time PCR methods which are highly sensitive and specific for the detection and quantification of infectious disease agents have become available (7). These assays are easy to perform and can be completed in as little as 30 to 60 min. We prospectively evaluated the efficacy of one rapid-cycle real-time PCR method, the LightCycler Strep-A assay (Roche Applied Science, Indianapolis, Ind.), for the detection GAS from throat swabs. We compared the accuracy, time for completion, and labor requirements of this rapid PCR method to those of both a rapid antigen immunoassay, the Directigen 1-2-3 Group A Strep Test kit (BD Diagnostic Systems, Sparks, Md.), and a standard culture method.
MATERIALS AND METHODS
Study design.
The study was approved by the Institutional Review Board of the Mayo Foundation. Consecutive throat swab specimens for culture were collected from 8 August 2001 through 25 September 2001 from 423 adult and pediatric patients attending a large ambulatory-care clinic. Thirty-nine of 423 (9.2%) patients were excluded from the evaluation because these patients or their guardians declined to provide permission to use their specimens and medical histories for evaluation (Minnesota Statute 144.335). A total of 384 throat swab cultures for 382 patients were available for analysis.
Collection of specimens.
All specimens were collected by physicians, nurse practitioners, or registered nurses by using a double-swab collection and transport system (CultureSwab; Becton Dickinson Microbiology Systems, Cockeysville, Md.). One swab was used for the Directigen rapid antigen immunoassay and culture and the other was used for the LightCycler PCR assay.
Culture.
The first swab was inoculated onto Strep Selective Agar (SSA; BBL Microbiology Systems Division, Becton Dickinson, Baltimore, Md.) plate. SSA plates were incubated at 35°C in ambient air for 24 to 48 h. Beta-hemolytic colonies were confirmed to be GAS by a direct fluorescent-antibody technique (Difco Laboratories, Detroit, Mich.) The quantities of GAS colonies were scored as follows: 1+, <10 colonies in the first quadrant streak area; 2+, ≥10 colonies in the first quadrant streak area and <5 colonies in the second streak area; 3+, >10 colonies in the first quadrant streak area, ≥5 colonies in the second quadrant streak area, and less than 5 colonies in the third quadrant streak area; and 4+, >10 colonies in the first quadrant streak area, >5 colonies in the second quadrant streak area, and ≥5 colonies in the third quadrant streak area.
Rapid antigen immunoassay.
After inoculation of the SSA plate, the same swab was used for evaluation with the Directigen 1-2-3 Group A Strep Test kit. The procedure was performed according to the instructions provided by the manufacturer.
LightCycler PCR assay.
The LightCycler technology combines the features of rapid PCR and real-time detection of an amplification product (7). Rapid PCR is possible by virtue of rapid thermocycling, which is a feature of the LightCycler instrument. Detection of the amplification product occurs after each PCR cycle, hence the designation “real-time PCR.” Real-time detection is performed by using dual fluorescent resonance energy transfer (FRET) hybridization probes that anneal in a head-to-tail configuration to single-stranded target DNA. During FRET detection of the product there is an energy transfer between the donor fluorophore, fluorescein, on the 3′ end of the first hybridization probe and the acceptor fluorophore, LightCycler Red640 (LC-Red640), on the 5′ end of the second hybridization probe. This energy transfer occurs when the probes hybridize to their adjacent positions on the target DNA molecule and an external light source excites the donor fluorescein that in turn transfers its energy to the LC-Red640 molecule. This energy transfer causes the LC-Red640 molecule to emit a signal that can be detected by the LightCycler instrument.
For each specimen the exposed shaft of the CultureSwab was secured by using a gloved hand and Bio-Screen biohazard wipe (Fisher Scientific, Pittsburgh, Pa.), and the swab portion was inserted into the inner tube of the Swab Extraction Tube System (S.E.T.S.; part no. 3 315 568; Roche Applied Science). The shaft was then broken above the swab portion contained in the inner tube. The S.E.T.S. with the swab was capped and centrifuged at 20,800 × g for 3 min in a microcentrifuge with an aerosol-tight fixed-angle rotor (Eppendorf 5417C; Brinkman Instruments, Westburg, N.Y.) to extract the bacteria from the swab and pellet them into the bottom of the outer tube of the S.E.T.S. The S.E.T.S. was removed from the centrifuge without tilting it, and the inner tube containing the swab was removed and discarded. The supernatant was aspirated from the outer tube of the S.E.T.S. with a fine-tip plastic transfer pipette. The pellet was resuspended in 100 μl of sterile irrigation water, and the screw cap was placed tightly on this tube, which was heated in a 100°C heating block for 10 min and then centrifuged at 20,800 × g for 3 min. Five microliters of the supernatant was combined with 15 μl of the PCR reagent mixture. The PCR reagent mixture was produced by using the following amounts and types of reagents: 2 μl of LightCycler Strep-A Primer/Hybridization Probes (catalog no. 3 272 940; Roche Applied Science), 2 μl of LightCycler FastStart Master Hybridization Probes mix with enzyme, 9 μl of H2O supplied with the mixture (catalog no. 3-003-248; Roche Applied Science), and 2 μl of LightCycler Strep-A Recovery Template (internal control; catalog no. 3 272 958; Roche Applied Science). The PCR mixture was pipetted into a LightCycler cuvette, and 5 μl of specimen was added. The cuvettes were sealed by capping, centrifuged at 1,000 × g for 30 s in a LightCycler centrifuge, and placed in a LightCycler instrument. The LightCycler instrument in which color compensation was installed was programmed as detailed in Table 1. Version 3.5 of the LightCycler software was used for most experiments. When older software was used, the gains were set at 1, 5, and 15 for channels F1, F2, and F3, respectively. Sterile water was used as a negative control. A positive control, LightCycler Strep-A Template DNA (catalog no. 3 272 966; Roche Applied Science), and one or two negative controls were included in each run. A 198-bp fragment of the ptsI (phosphotransferase) gene (GenBank accession no. AE004092) of GAS is amplified and detected with the aforementioned primers and probes.
TABLE 1.
Programming of the LightCycler instrument
| Program name | Analysis mode | No. of cycles | Temp (°C) | Time (s) | Temperature transition rate (°C/s) | Signal acquisition |
|---|---|---|---|---|---|---|
| Denature | None | 1 | 95 | 600 | 20 | None |
| PCR | Quantification | 45 | 95 | 10 | 20 | None |
| 55 | 10 | 20 | Single | |||
| 72 | 8 | 20 | None | |||
| Melt analysis | Melt | 1 | 95 | 0 | 20 | None |
| 45 | 10 | 20 | None | |||
| 80 | 0 | 0.2 | Continuous | |||
| Cool | None | 1 | 35 | 0 | 20 | None |
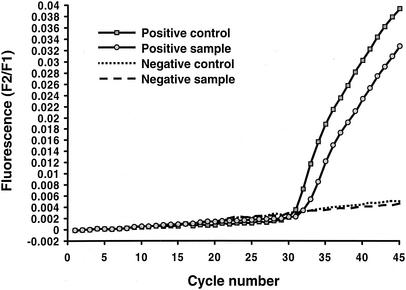
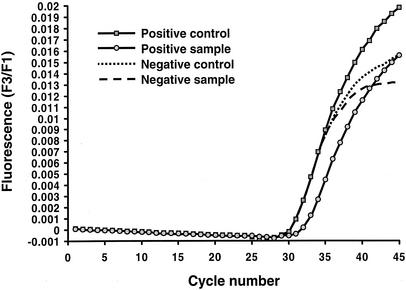
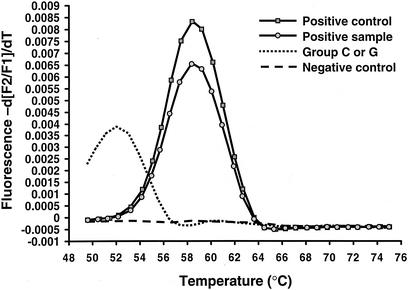
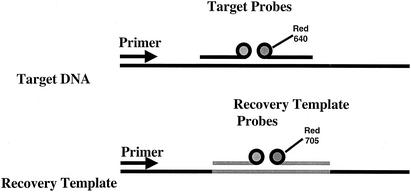
Representative results for analysis of the real-time quantification data are shown in Fig. 1 and 2. A representative melting curve analysis which confirms the identification of the amplicon as belonging to GAS by comparison of the melting temperatures (Tms) of the controls to those of the samples is shown in Fig. 3. For this analysis, at the conclusion of thermocycling the temperature of the reaction vessel is slowly increased, with the result being a loss of the FRET signal due to separation of the FRET probes from the target DNA. The negative derivative of fluorescence over time is compared with the temperature. This quality control step confirms that the target DNA amplified is the DNA of GAS. For this assay the Tm for GAS by melting curve analysis is 57.3 ± 2.5°C. Large-colony-forming group C and group G streptococci can cause human pharyngitis and have recently been classified in the same species (Streptococcus dysgalactiae subsp. equisimilis) (21). These streptococci have a single-base difference in the segment of DNA which is amplified by the LightCycler PCR assay. The cycling curves produced for GAS (Fig. 1) are not produced for group C or group G streptococci, as the annealing temperature used in that analysis (∼55°C) is too high to permit attachment of one of the FRET probes to the target DNA for group C or group G streptococci. However, for the melting curve analysis a lower initial (annealing) temperature (∼45°C) is used so that partial hybridization of the FRET probe with target DNA for group C or group G streptococci occurs. Therefore, a melting curve is generated for group C or group G streptococci with a Tm of 51.5 ± 2.5°C. A recovery template (internal control) is amplified in the same reaction (Fig. 4). The recovery template amplification product is detected in channel F3 and should be present for samples in which there are no GAS or group C or group G streptococci. The absence of the F3 signal from the replication template indicates PCR inhibition of the sample. Strongly positive samples sometimes compete with the amplification of the recovery template. As the recovery template indicates the efficiency (inhibition) of the PCR, amplification of the recovery template coincident with a positive result for the sample is unnecessary for interpretation of the results of the test.
FIG. 1.
Detection of GAS DNA with the LightCycler instrument and LightCycler Strep-A Primer/Hybridization Probes. In this quantitative analysis, positive results are indicated by an upwardly deflecting curve (cycle number, ∼31) for both the positive control and a positive sample. F2 refers to the fluorescence emission for the LC-Red640 fluorophore, and F1 is fluorescence emission for the fluorescein fluorophore.
FIG. 2.
Quantitative representation or cycling curve analysis of recovery template (internal control) for the LightCycler PCR assay. The recovery template FRET probe has a different reporter dye (LC-Red705) than the FRET probe used to detect target DNA in the sample (LC-Red640 dye) and is detected in channel F3 of the LightCycler instrument (see Fig. 4). F3 is the fluorescence emission for the LC-Red705 fluorophore, and F1 is the fluorescence emission for the fluorescein fluorophore. This quality control step indicates whether inhibition of the PCR occurred in any of the patient samples or the positive or negative controls. Amplification of the recovery template should occur with each of these analyses except when the amount of target DNA in the patient's sample significantly exceeds that of the recovery template DNA. In this case, the target DNA competes with the recovery template DNA, but because the sample is positive, assessment of PCR inhibition by means of recovery template DNA amplification is unnecessary.
FIG. 3.
Melting curve analysis for LightCycler PCR assays. The designation −d[F2/F1]/dT on the y axis refers to the negative derivative of fluorescence. F2 is the fluorescence emission for the LC-Red640 fluorophore, and F1 is the fluorescence emission for the fluorescein fluorophore. The Tm of GAS by melting curve analysis is 57.3 ± 2.5°C. Large-colony-forming group C and group G streptococci of human origin, which have recently been classified as the same species (S. dysgalactiae subsp. equisimilis), produce identical melting curves, with a Tm of 51.5 ± 2.5°C, but are not detected during the real-time quantification portion of the PCR.
FIG. 4.
Recovery template. The recovery template (internal control) has the same sequence as the PCR product, except that the probe region has been replaced with a sequence complementary to recovery template probes. FRET detection of the target DNA is with a probe labeled with the LC-Red640 dye in channel 2 of the LightCycler instrument, while the recovery template is detected with a probe labeled with the LC-Red705 dye in channel 3. A small amount of the recovery template is added to the PCR mixture and is amplified along with the target DNA by the same primers. Thus, the two reactions compete for the primers. Normally, the recovery template is amplified from all samples, including the negative control. If neither the recovery template nor the target DNA is amplified, it is assumed that inhibition of the PCR has occurred and the test for that sample is not valid. However, if the target DNA is amplified but the recovery DNA template is not, it is assumed that the target DNA is present in a proportionally greater amount. In this situation, partial inhibition of the PCR may be present but the target DNA is successfully amplified or the recovery template may not be able to compete for primers and the recovery template signal may be weak or not present. When this occurs, the positive result is valid because the recovery template amplification is unnecessary.
Specificity panel evaluation of LightCycler PCR assay.
The preclinical specificity of the LightCycler PCR assay was determined by evaluation of DNA extracted from pure cultures of a variety of gram-positive and gram-negative bacteria. These bacteria included a wide range of gram-positive streptococci as well as other gram-positive and gram-negative bacteria which can colonize or cause infection in the respiratory tract.
Clinical evaluation.
Each of the results for the Directigen rapid antigen immunoassay and LightCycler PCR were compared to the results for culture to determine sensitivities, specificities, and positive and negative predictive values. Therefore, culture was considered the gold standard for this comparison. Discordant positive results for either the Directigen immunoassay or the LightCycler PCR versus the results of culture were further evaluated by a review of the patient's medical history (see below).
Medical history reviews.
One of the coauthors of this article (S.C.A.) reviewed the medical histories of all patients for whom a positive result occurred by only one of three methods, the Directigen immunoassay, culture, or the LightCycler PCR assay. That reviewer is a physician who works in the outpatient clinic from which the throat swab specimens for the study were obtained and was not involved in the laboratory testing of specimens. Patient histories were reviewed for evidence of signs and symptoms compatible with GAS infection in an effort to distinguish GAS carriers from patients with GAS disease. For the purposes of the present study, we assumed that a true-positive result should correlate with GAS disease and not asymptomatic carriage of GAS. These signs and symptoms were summarized by Dajani and colleagues (8) and include the following: the characteristic symptoms sudden onset of sore throat, pain on swallowing, fever, headache, abdominal pain, or nausea and vomiting; the uncharacteristic symptoms coryza, hoarseness, cough, or diarrhea; the characteristic signs tonsillopharyngeal erythema, tonsillopharyngeal exudate, soft-palate petechiae, beefy red and swollen uvula, anterior cervical lymphadenitis, or scarlatiniform rash; and the uncharacteristic signs conjunctivitis, anterior stomatitis, or discrete ulcerative lesions. At the time that the study was performed, the results of standard testing (Directigen immunoassay and culture) but not the results of the LightCycler PCR were available to clinicians.
Statistical analysis.
Confidence intervals for sensitivity, specificity, and positive and negative predictive values were based on exact binomial probabilities. The differences between sensitivities were evaluated by McNemar's test (19).
Assessment of assay time and labor requirements.
The approximate time required to complete each test, including specimen preanalytical processing and assay time, was recorded for each assay. Additionally, the amount of personnel time required for each test method was assessed by using approved guidelines for workload recording published by the National Committee for Clinical Laboratory Standards (16).
RESULTS
Specificity panel evaluation for LightCycler PCR assay.
Archived clinical strains of the following nonstreptococcal bacteria, some of which can colonize and/or produce disease in the respiratory tract of humans, were tested and were negative by the LightCycler PCR assay: Acinetobacter baumannii, Acinetobacter lwoffii, Aeromonas hydrophila, Arcanobacterium haemolyticum, Bordetella bronchiseptica, Bordetella holmesii, Bordetella parapertussis, Bordetella pertussis, Burkholderia cepacia, Campylobacter jejuni, Chlamydia pneumoniae, Citrobacter freundii, Corynebacterium diphtheriae, Corynebacterium pseudodiphtheriticum, Escherichia coli, Haemophilus influenzae, Klebsiella oxytoca, Klebsiella pneumoniae, Legionella jordanis, Legionella micdadei, Legionella pneumophila, Listeria monocytogenes, Moraxella catarrhalis, Morganella morganii, Mycoplasma pneumoniae, Neisseria gonorrhoeae, Neisseria meningitidis, Proteus mirabilis, Proteus vulgaris, Pseudomonas aeruginosa, Pseudomonas fluorescens, Staphylococcus aureus, Staphylococcus epidermidis, and Stenotrophomonas maltophilia. A sample of human DNA (MRC-5 cells) was also tested and was found to be negative.
The following gram-positive streptococci were evaluated and were negative by the LightCycler PCR assay: Streptococcus suis, Streptococcus viridans, Streptococcus anginosus, Streptococcus equi, Streptococcus uberis, Streptococcus intermedius, Streptococcus mutans, Streptococcus bovis, Streptococcus mitis, Streptococcus canis, Streptococcus salivarius, Streptococcus equinus, Streptococcus pneumoniae, group F streptococci, group B streptococci, Enterococcus faecalis, Enterococcus faecium, and Lactococcus lactis. As expected, strains of group C and group G streptococci did not produce signals during the quantitative portion of the PCR but produced signals by melting curve analysis. The melting curves for these strains had clearly discernible Tm ranges below that of the melting curve peak for GAS.
Clinical evaluation.
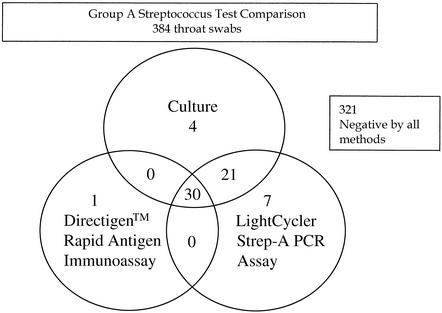
Among the 384 throat swab samples, 55 (14.3%) were identified as positive by conventional bacterial culture, 31 (8.1%) were identified as positive by the Directigen antigen immunoassay, and 58 (15.1%) were identified as positive by the LightCycler PCR (Fig. 5). PCR inhibition was detected by means of a lack of recovery template (internal control) amplification for 4 of 384 (1%) specimens evaluated by the LightCycler PCR method. Table 2 compares the results for each of the two rapid methods, the Directigen rapid antigen immunoassay and the LightCycler PCR, to those of the conventional gold standard method, bacterial culture. The sensitivities, specificities, and positive and negative predictive values were as follows: for the Directigen antigen immunoassay, 55, 99, 97, and 93%, respectively; for the LightCycler PCR assay, 93, 98, 88, and 99%, respectively. In no case was a throat swab positive by both the LightCycler PCR and the Directigen method but negative by culture. However, positive results occurred by either the LightCycler PCR or the Directigen method for some samples that were negative by culture. The results for swabs with discordant positive results by the LightCycler PCR versus the results of culture (n = 7) and by the Directigen method versus the results of culture (n = 1) were reconciled by a review of the medical history. All of the patients with discordant positive results by either of these methods had signs or symptoms compatible with GAS disease; therefore, these discordant positive results (along with positive results by either the Directigen method or the LightCycler PCR that agreed with the culture results) were considered true positives for statistical analysis (Table 3). For this analysis, the LightCycler PCR detected more true-positive results than the culture method (58 versus 55 swabs); however, this difference was not statistically significant (P = 0.5465). In contrast, statistically significantly more true-positive results occurred by culture than by the Directigen method (55 versus 31 swabs; P < 0.0001) and by the LightCycler PCR than by the Directigen method (58 versus 31 swabs; P < 0.0001).
FIG. 5.
Distribution of GAS-positive results for each of the assays tested. The positivity rate (any assay positive) was 63 of 384 samples (16.4%). For four samples (1.0%) inhibition of amplification of the recovery template (internal control) was detected by the LightCycler PCR assay. Of the 321 negative samples, 10 (3.1%) were found to contain DNA from group G or group C streptococci by the Light- Cycler PCR assay.
TABLE 2.
Sensitivities, specificities, and predictive values for Directigen rapid antigen immunoassay and LightCycler PCR assay compared to the results of culture for detection of GAS from throat swabsa
| Type of assay | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| Directgen antigen immunoassay | 55 (41, 68)b | 99 (98, 100) | 97 (83, 100) | 93 (90, 95) |
| LightCycler PCR assay | 93 (82, 98)b | 98 (96, 99) | 88 (77, 95) | 99 (97, 100) |
The prevalence rate of positive cultures was 14.3%. Values in parentheses are 95% confidence intervals.
Signficantly different by McNemar's test (P < 0.001).
TABLE 3.
Symptoms and signs for patients with discordant positive resultsa
| Symptom or sign | Directigen assay, patient 1 | LightCycler PCR
|
Culture
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 1b | Patient 2 | Patient 3 | Patient 4 | ||
| Symptoms | ||||||||||||
| Characteristic | ||||||||||||
| Sudden onset of sore throat | √ | √ | √ | √ | √ | √ | √ | √ | ||||
| Pain on swallowing | ||||||||||||
| Fever | √ | √ | √ | √ | √ | √ | ||||||
| Headache | √ | √ | ||||||||||
| Abdominal pain | ||||||||||||
| Nausea and vomiting | ||||||||||||
| Uncharacteristic | ||||||||||||
| Coryza | √ | √ | √ | |||||||||
| Hoarseness | √ | √ | ||||||||||
| Cough | √ | √ | √ | √ | ||||||||
| Diarrhea | ||||||||||||
| Signs | ||||||||||||
| Characteristic | ||||||||||||
| Tonsillopharyngeal erythema | √ | √ | √ | √ | √ | √ | √ | √ | √ | |||
| Tonsillopharyngeal exudate | √ | |||||||||||
| Soft-palate petechiae (“doughnut” lesions) | ||||||||||||
| Beefy red, swollen uvula | ||||||||||||
| Anterior cervical lymphadenitis | √ | √ | ||||||||||
| Scarlatiniform rash | ||||||||||||
| Uncharacteristic | ||||||||||||
| Conjunctivitis | ||||||||||||
| Anterior stomatitis | ||||||||||||
| Discrete ulcerative lesions | ||||||||||||
Symptoms and signs adapted from Dajani et al. (8).
This patient had a history of recurrent sore throat but was asymptomatic and had no clinical findings at the time the throat swab for culture was obtained.
From the review of the medical history, sore throat and/or tonsillopharyngeal erythema were recorded as a symptom and/or sign by the health care provider in the medical history for all seven patients positive only by the LightCycler PCR assay, for the single patient positive only by the Directigen antigen immunoassay, and for three of the four patients positive only by culture. Four of the seven patients positive only by the LightCycler PCR were treated with antibiotics effective against GAS, even though the health care providers responsible for these patients were unaware of the positive LightCycler PCR result. For the remaining patient positive only by culture, no signs or symptoms were present at the time that the specimen for culture was obtained. This patient had a history of recurrent sore throat. Although this result would be considered false positive, it was not discounted as such because for the purposes of this study culture was considered the gold standard.
A review of melting curve analyses for all LightCycler PCR assays negative by quantitative analysis for GAS demonstrated that 10 of 322 (3.1%) of the samples contained DNA compatible with either group C or group G streptococci.
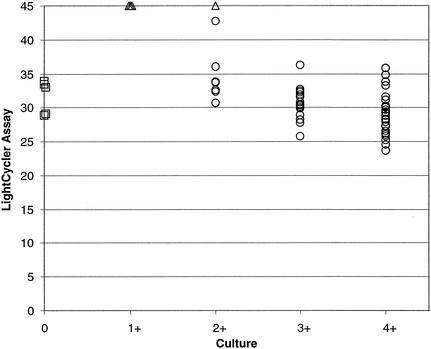
Figure 6 compares the LightCycler PCR crossing points (the cycle number at which the LightCycler PCR result is positive) with quantification (1+ to 4+) of colonies from the culture plate. As can be seen, as the quantity of colonies on the culture plate increases from 1+ to 4+, the cycle number (crossing point) decreases. This indicates that fewer thermocycles are required to amplify DNA from samples with higher initial quantities of target DNA, which correlates with higher colony counts on culture plates. As expected, lower colony counts were associated with negative LightCycler PCR results. Three of the four specimens positive only by culture had 1+ growth, and the one specimen positive only by culture had 2+ growth.
FIG. 6.
Comparison of LightCycler PCR crossing points with culture plate colony scores. ○, culture and LightCycler PCR positive; □, LightCycler PCR positive only; ▵, culture positive only. Refer to Materials and Methods for a description of culture quantitation.
Assessment of assay time and labor requirements.
The approximate times required to complete each test including preanalytical processing and analysis were as follows: Directigen rapid antigen immunoassay, 10 min; culture, 24 to 48 h; LightCycler PCR, 1.5 h. The personnel times (workloads) calculated for the Directigen rapid antigen immunoassay and culture, if culture was subsequently required, were combined as these methods reflect our present clinical practice. That is, the Directigen antigen immunoassay is performed, and if the result is negative, a culture is performed. The personnel time calculated for the LightCycler PCR was considered separately, as the results of the present study suggest that the LightCycler PCR can replace the sequential Directigen method-culture evaluation of a specimen. Unlike the Directigen method, which is performed on demand in the outpatient setting in our institution, the LightCycler PCR assays are performed in batches in the clinical microbiology laboratory, with up to 32 samples evaluated (specimens and controls) per run. For these processes the personnel time required for the combination of the Directigen antigen immunoassay and culture (7 min) was calculated to be more than twice that required for the LightCycler PCR method (3 min).
DISCUSSION
The gold standard test method for identification of GAS from throat swabs has been culture. Over the past two decades, we and others have compared a variety of rapid antigen immunoassays to culture (2, 12, 13, 18, 22). The sensitivities of these rapid assays have never equaled that of culture and are frequently considerably less than that of culture. For example, Wegner and colleagues (22) reported sensitivities ranging from 31 to 50% for five rapid antigen assays for GAS when they were compared with a two-plate culture method. This two-plate culture method (a 5% sheep blood Trypticase soy agar plate incubated aerobically and a 5% trimethoprim-sulfamethoxazole agar plate incubated anaerobically) was a rigorous gold standard. However, many subsequent studies that have used less rigorous culture-based gold standards (one agar plate with or without broth) have shown similar results (4).
Among antigen detection methods, an optical immunoassay (BioStar Strep A OIA; Thermo Biostar Inc., Boulder, Colo.) has had the best performance. In our hands the sensitivities of that method and our standard culture method compared to the results of a combined gold standard were 81.0 and 92%, respectively (9). The combined gold standard was defined as a positive culture result with 5% sheep blood agar or Todd-Hewitt broth or, if both of these culture methods were negative, a concomitant positive result by the BioStar optical immunoassay and another rapid antigen immunoassay, Pacific Biotech CARDS O.S. (Pacific Biotech, Inc., San Diego, Calif.).
A rapid molecular method that has gained popularity for detection of GAS is the Group A Streptococcus Direct Test (Gen-Probe, Inc., San Diego, Calif.). This method identifies specific rRNA sequences unique to GAS by using single-stranded DNA. Labeled DNA-RNA duplexes are formed. We noted a sensitivity of 89% for this method compared with the results of culture (5% sheep blood agar) (18).
Because studies have consistently demonstrated that rapid GAS detection methods, including those covered in the preceding discussion, are less sensitive than culture, several national advisory groups have recommended that for children and adolescents negative results of rapid tests be confirmed by culture (1, 5, 8). In contrast, the results of our study suggest that the LightCycler PCR assay is more sensitive than our standard culture method for detecting GAS in the throat swab samples of our patients. After discordant positive results for the LightCycler PCR and Directigen methods versus the results of culture were reconciled by a review of the patient's medical history, the LightCycler PCR detected more true-positive samples (n = 58) than either the Directigen method (n = 31) or culture (n = 55). This increase in sensitivity was statistically significant (P < 0.0001) for the LightCycler PCR versus the Directigen method but was not statistically significant for the LightCycler PCR versus culture (P = 0.5465). The enhanced detection of GAS by the LightCycler PCR may be related to both the inherent qualities of the real-time PCR assay and the DNA extraction technique used. In prior prospective clinical studies, we have consistently noted increases in sensitivities for assays that use the LightCycler PCR technology compared to the sensitivities of standard culture methods. These include assays for B. pertussis (219% increase) (20), herpes simplex virus (23% increase) (10), varicella-zoster virus (91% increase) (11), and cytomegalovirus (88% increase) (M. J. Espy and T. F. Smith, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. C-62, 2000). As a result of these clinical studies we have replaced standard culture-based methods with LightCycler PCR assays for direct detection of these organisms from clinical samples. We assume that the enhanced sensitivity of the LightCycler PCR assay is related in part to the S.E.T.S., which is used to prepare the specimen. This centrifugation appliance effectively concentrates pharyngeal secretions from throat swabs. Although we did not study this, higher rates of detection of GAS may occur if the concentrate from a tube in the S.E.T.S. is cultured or tested by the Directigen rapid antigen immunoassay.
The value of a real-time PCR assay for detection of group B streptococci directly from vaginal and/or anal swab specimens from women has recently been demonstrated by Bergeron and colleagues (3). To our knowledge, the present study is the first to demonstrate the utility of a real-time PCR method for detection of GAS directly from throat swab specimens. In both studies, the results were available on the same day that the test was ordered, allowing the initiation of appropriate antimicrobial therapy sooner and lessening the chance for untoward sequelae.
Using workload recording, an established National Committee for Clinical Laboratory Standards method for determination of the personnel time required to perform laboratory tests (16), we determined that the personnel time required for detection of GAS from throat swabs for the LightCycler PCR method (3 min) was less than half the time required to perform the combined rapid antigen detection and culture methods (7 min). In our practice, in accordance with practice guidelines provided by the Infectious Diseases Society of America (5), culture is performed for all specimens for which the rapid immunoassay is negative. Therefore, we counted the time required to perform the Directigen antigen immunoassay and culture if culture was subsequently required. This significant savings in personnel time is critically important in view of the diminishing pool of qualified laboratory technologists and the budgetary constraints that many laboratories are experiencing in the United States.
With the rapidity with which the LightCycler PCR assay can be performed, results can be relayed to health care providers and patients on the same day that the test is ordered. At the Mayo Clinic, we have found that it is convenient to perform three batches of tests per day, which permits a turnaround time for results of ≤8 h. This turnaround time competes favorably with that for culture, which requires up to 48 h before a final report can be issued. Also, throat swab specimens can be obtained from our ambulatory patients with pharyngitis by a nurse practitioner without the need for a physician visit. The results from analyses are entered electronically into a computerized telephone callback system. To access their results, patients call a toll-free telephone number and listen to a recorded message. The test results are provided and the patient is notified that a prescription can be picked up at a pharmacy preselected by the patient. Such rapid turnaround times and convenient reporting of results are welcomed by our patients and should have a significant effect on the appropriate and expedient delivery of antimicrobics for streptococcal pharyngitis.
Although the time required to complete the Directigen method is less than the time required to complete the LightCycler PCR (10 min versus 1.5 h), the lack of sensitivity of the Directigen method precludes a “rapid” result for the vast majority of our patients. Because cultures are performed for throat swabs with negative results by the Directigen method, at our institution the proportions of throat swabs that must be cultured and that therefore require up to 2 days for final results are approximately 85% in the winter months and 95% in the summer months. This is because in winter months the frequency of positive samples is nearly 30% and in summer months the frequency is 10%. Since the Directigen rapid antigen immunoassay has a sensitivity of approximately 50%, only half of the 30 of 100 and 10 of 100 true-positive samples will be detected during these periods, respectively. Besides accuracy, the LightCycler PCR assay (including the S.E.T.S. extraction step) has the advantage of a very rapid analytical time (∼1 h) compared to that of culture (1 to 2 days). Although the Directigen immunoassay that we tested can be performed within a few minutes, it was considerably less sensitive than either the LightCycler PCR method or culture. A potential drawback of enhanced sensitivity, as may occur with LightCycler PCR technology, is that low colony counts for GAS associated with carriage and not disease may be detected. However, in the present study all discordant positive results for the LightCycler PCR method versus the results of culture were believed to be associated with disease.
A recent publication suggests that because sequelae like rheumatic fever are less of a concern in adult patients, rapid antigen testing alone is adequate for diagnosis (6). We question the validity of this recommendation for several reasons. Because many rapid antigen tests are observed to have such low sensitivities, many adult cases of active disease and colonization may be underdiagnosed. These infected adults may serve as a reservoir for the spread of GAS to children. Although rarely a complication, invasive streptococcal disease can result in significant morbidity and mortality in adults. Antecedent GAS pharyngitis or GAS colonization likely precedes GAS invasive disease. Finally, the authors of this paper suggest that the high specificities but lower sensitivities of rapid antigen tests compared with the results of culture should minimize overprescription of antibiotics in adults. A counterargument might be that because rapid antigen tests miss so many GAS pharyngitis cases, physicians may discount the results and may more frequently prescribe antibiotics on an empirical basis.
The LightCycler PCR assay is relatively easy to perform and should be adaptable to smaller laboratories. The footprint of the instrument and the associated computer is relatively small compared to those of other laboratory instruments. As the present study suggests that the sensitivity of the LightCycler PCR assay for the detection of GAS in throat swabs exceeded that of culture, we believe that it is unnecessary to perform cultures when the results of the LightCycler PCR assay are negative. Furthermore, as susceptibility testing is not recommended for GAS (all strains remain susceptible to beta-lactam antimicrobics, although some strains are resistant to erythromycin [15]), culture, which is necessary for susceptibility testing, is not required. If culture is required for antibiotic resistance surveillance or for DNA genomic fingerprint analysis, as may be necessary during outbreaks of invasive streptococcal disease, specimens that are positive by the LightCycler PCR could be cultured.
An additional potential advantage of the LightCycler PCR assay is that it can distinguish among the three beta-hemolytic streptococci that have been associated with pharyngitis: GAS and group C and group G streptococci (17). This is possible by virtue of melting curve analysis with FRET probes. In the target nucleic acid used for the assay, group C and group G streptococci share the same sequence, which is 1 bp different from the sequence of GAS. Therefore, when the temperature is slowly increased, the separation (melting) of FRET hybridization probes from the target DNA occurs over a temperature range for GAS different from that for group C and group G streptococci (Fig. 3). This finding is expected, as present data suggest that large-colony-forming group C and group G beta-hemolytic streptococci which are associated with human disease are related at the species level and are classified as S. dysgalactiae subsp. equisimilis (21). In the present study, 10 (3%) of 322 throat swab specimens that were negative for GAS had melting curves compatible with group C or group G streptococci. In the present study, in accordance with our present practice, beta-hemolytic streptococci isolated in culture were screened only for the GAS antigen. In a prior study in which GAS and group C and group G streptococci were identified from throat swabs, we noted that 2.8% of all swabs negative for GAS had either group C or group G streptococci (15), which is similar to the percentage noted in the present study. Nevertheless, further clinical studies are required to correlate the melting curves for group C and group G streptococci produced by the LightCycler PCR assay for GAS with confirmation of the presence of group C and group G streptococci by culture identification methods.
In conclusion, the Roche LightCycler PCR assay is a reliable method for detection of GAS from throat swabs. The high sensitivity and specificity of the method make it a suitable stand-alone test. The advantages of the LightCycler PCR assay over the conventional diagnostic approach for detection of GAS from throat swabs (i.e., rapid antigen immunoassay and culture) include ease of use, less personnel time, and a considerably shorter analytical time (∼1 h versus up to 2 days), allowing the more rapid reporting of results to clinicians.
Acknowledgments
JoAnn Brunette is thanked for her efforts in preparing the manuscript.
REFERENCES
- 1.American Academy of Pediatrics. 2000. Group A streptococcal infections, p. 526-536. In L. K. Pickering (ed.), 2000 Redbook; report of the Committee on Infectious Diseases, 25th ed. American Academy of Pediatrics, Elk Grove Village, Ill.
- 2.Anhalt, J. P., B. J. Heiter, D. W. Naumovitz, and R. D. Bourbeau. 1992. Comparison of three methods for detection of group A streptococci in throat swabs. J. Clin. Microbiol. 30:2135-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron, M. G., D. Ke, C. Menard, F. J. Picard, M. Gagnon, M. Bernier, M. Ouellette, R. H. Roy, S. Marcoux, and W. D. Fraser. 2000. Rapid detection of group B streptococci in pregnant women after delivery. N. Engl. J. Med. 343:175-179. [DOI] [PubMed] [Google Scholar]
- 4.Bisno, A. L. 2001. Acute pharyngitis. N. Engl. J. Med. 344:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Bisno, A. L., M. A. Gerber, J. M. Gwaltney, Jr., E. L. Kaplan, and R. H. Schwartz. 1997. Diagnosis and management of group A streptococcal pharyngitis: a practice guideline. Clin. Infect. Dis. 25:574-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisno, A. L., M. A. Gerber, J. M. Gwaltney, Jr., E. L. Kaplan, and R. H. Schwartz. 2002. Practice guidelines for the diagnosis and management of group a streptococcal pharyngitis. Clin. Infect. Dis. 35:113-125. [DOI] [PubMed] [Google Scholar]
- 7.Cockerill, F. R., III, and J. R. Uhl. 2002. Applications and challenges of real-time PCR for the clinical microbiology laboratory, p. 3-27. In U. Reischl, C. Wittwer, and F. Cockerill (ed.), Rapid cycle real-time PCR—methods and applications: microbiology and food analysis. Springer-Verlag, Berlin, Germany.
- 8.Dajani, A., K. Taubert, P. Ferrieri, G. Peter, and S. Shulman. 1995. Treatment of acute streptococcal pharyngitis and prevention of rheumatic fever: a statement for health professionals. Pediatrics 96:758-764. [PubMed] [Google Scholar]
- 9.Dale, J. C., E. A. Vetter, J. M. Contezac, P. C. Iverson, P. C. Wollan, and F. R. Cockerill III. 1994. Evaluation of two rapid antigen assays, BioStar Strep A OIA and Pacific Biotech CARDS O.S., and culture for detection of group A streptococci in throat swabs. J. Clin. Microbiol. 32:2698-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espy, M. J., R. Teo, K. Ross, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espy, M. J., R. Teo, K. Ross, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Diagnosis of varicella-zoster virus infection in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:3187-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber, M. A. 1986. Diagnosis of group A beta-hemolytic streptococcal pharyngitis. Use of antigen tests. Diagn. Microbiol. Infect. Dis. 4:5S-15S. [DOI] [PubMed]
- 13.Hoffman, S. 1990. Detection of group A streptococcal antigen from throat swabs with five diagnostic kits in general practice. Diagn. Microbiol. Infect. Dis. 13:209-215. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. R., and E. L. Kaplan. 2001. False-positive rapid antigen detection test results: reduced specificity in the absence of group A streptococci in the upper respiratory tract. J. Infect. Dis. 193:1135-1137. [DOI] [PubMed] [Google Scholar]
- 15.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1998. Basic cost accounting for clinical services; approved guideline GP11-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Oster, H. R., and A. L. Bisno. 2000. Group A and group G streptococcal infections: epidemiology and clinical aspects, p. 190-194. In V. A. Fischetti, R. P. Novick, J. J. Ferreti, D. A. Portnog, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 18.Pakorski, S. J., E. A. Vetter, P. C. Wollan, and F. R. Cockerill III. 1994. Comparison of Gen-Probe group A streptococcus direct test with culture for diagnosing streptococcal pharyngitis. J. Clin. Microbiol. 32:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel, S. 1956. Nonparametric statistics for the behavioral sciences, p. 68-75. McGraw-Hill, New York, N.Y.
- 20.Sloan, L. M., M. K. Hopkins, P. S. Mitchell, E. A. Vetter, J. E. Rosenblatt, W. S. Harmsen, F. R. Cockerill, and R. Patel. 2002. Multiplex LightCycler PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis in nasopharyngeal specimens. J. Clin. Microbiol. 40:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandamme, P., B. Pot, E. Falsen, K. Kersters, and L. A. DeVries. 1996. Taxonomic study of Lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis. Int. J. Syst. Bacteriol. 46:774-781. [DOI] [PubMed] [Google Scholar]
- 22.Wegner, D. L., D. L. Witte, and R. D. Schrantz. 1992. Insensitivity of rapid antigen detection methods and single blood agar plate culture for diagnosing streptococcal pharyngitis. JAMA 267:695-697. [PubMed] [Google Scholar]