Abstract
In order to develop a species-specific PCR for the detection of Mycoplasma genitalium, the sequence of 1,490 bases of the 16S rRNA gene was determined for M. genitalium G37 (type strain) and four Danish isolates of M. genitalium. The sequences of the four Danish strains, mutually different with respect to their MgPa gene, were 100% homologous, although they carried a single common base substitution compared to the type strain. Among members of the Mycoplasma pneumoniae phylogenetic cluster, M. genitalium showed the most-prominent homology to the 16S rRNA sequence of M. pneumoniae (98% homology). From regions showing the least homology to the M. pneumoniae 16S rRNA gene sequence, primers were chosen to amplify DNA from M. genitalium only. Two sets of primers were selected for their ability to detect <10 to 50 M. genitalium genome copies without cross-reactions with M. pneumoniae. The performance of these primers was compared to the performance of two pairs of primers amplifying parts of the MgPa adhesin gene; 1,030 randomly selected specimens submitted for Chlamydia trachomatis culture were screened with one of the 16S rRNA gene primer sets. A total of 41 specimens were found to be positive for this gene; 40 of these could be confirmed by one of the MgPa primer sets, whereas the other MgPa primer set detected only 21 positive specimens out of 40. These results indicate that estimates of the prevalence of M. genitalium in various populations using MgPa PCR primers could be incorrectly low if the PCR primers are located in variable regions of the MgPa gene.
Two Mycoplasma genitalium strains were originally isolated in 1980 from the urogenital tracts of 2 of 13 men with nongonococcal urethritis (NGU) (27). Despite repeated attempts with conventional culture techniques, no other urogenital isolates have been reported (21, 24). Using a cell-culture-based method, however, we succeeded in isolating four new strains from the urethras of male patients with NGU who were PCR positive for M. genitalium (11). M. genitalium and Mycoplasma pneumoniae share several structural properties, such as the flask shape and the terminal tip-like structure, and a significant antigenic relationship between the two mycoplasma species has hampered diagnostic serology (15; K. Lind, Letter, Lancet ii:1158-1159, 1982).
Because traditional diagnostic procedures for M. genitalium such as culture and serology have failed, other methods have been investigated. The development of a DNA probe provided some evidence for the presence of M. genitalium in the male urogenital tract (8), but data indicating that M. genitalium is a potential cause of NGU have only recently been demonstrated by the use of PCR (2, 9, 10, 13, 26).
M. genitalium strains recently isolated from Danish patients (11) show a significant degree of sequence diversity of the main adhesin (MgPa) gene. This variability has not yet been sufficiently characterized to determine the presence of conserved regions in the gene so that PCR primers covering all M. genitalium strains can be designed. Consequently, we decided to develop a species-specific PCR based on amplification of rRNA gene sequences.
(Part of this study was presented at the 10th International Congress of the International Organization for Mycoplasmology, Bordeaux, France, July 1994. The abstract has been previously published [Int. Org. Mycoplasmol. Lett. 3:332-333, 1994].)
MATERIALS AND METHODS
Organisms and growth conditions.
M. genitalium G-37T and four new Danish M. genitalium strains designated M 2298, M 2300, M 2321, and M 2341 were grown in modified Friis' FF medium (11) containing horse serum. The following species (strains) were grown in modified Hayflick's medium (15) and harvested by centrifugation in the late log phase: M. pneumoniae (FHT, Mac, M129-B8, M129-B170, and two clinical isolates), Mycoplasma hominis (PG21T, H34, H27, and three clinical isolates), Mycoplasma salivarium (PG20T), Mycoplasma buccale (CH 20247T), Mycoplasma orale (Patt and one clinical isolate), Mycoplasma fermentans (GT and S38), Mycoplasma faucium (DC 333T), Mycoplasma primatum (Navel), Mycoplasma pirum (Zeus), Mycoplasma lipophilum (Maby BT), Mycoplasma hyorhinis (GDL), Mycoplasma arginini (G230T), Mycoplasma gallisepticum (15302), Acholeplasma laidlawii (AT), and Ureaplasma urealyticum (serotypes I [F. Black 7] and VIII [F. Black 960T]). All were grown in U10C medium (20).
DNA extraction.
DNA from M. genitalium G-37T and from the M. pneumoniae Mac strain was extracted with chloroform as previously described (12). DNA was quantified spectrophotometrically and by visual comparison after gel electrophoresis and ethidium bromide staining. DNA from the other mycoplasma species was released by resuspending the pellet from 2 ml of broth culture obtained after centrifugation at 30,000 × g for 15 min at 4°C in 100 μl of lysis buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA, 0.5% Tween 20, and 0.5% Nonidet P-40) containing proteinase K (200 μg/ml). The samples were incubated at 55°C for 30 min, the proteinase was inactivated at 94°C for 15 min, and the tubes were briefly centrifuged to collect condensation droplets. Clinical specimens were treated similarly, except that only 200 μl of the specimen was used; 10 μl was used for PCR.
Sequence determination of the 16S rRNA gene.
Before we began development of the present assays, the 16S rRNA gene sequence was not available in public databases. Furthermore, four Danish M. genitalium strains were found to have hypervariable parts in their MgPa gene (18). Consequently, it was decided to determine the 16S rRNA gene sequence of the type strain G37 of M. genitalium as well as that of the four Danish strains in order to document the conserved nature of this gene. The 16S rRNA gene was amplified by PCR with primers binding to sequences conserved in most eubacteria. The PCR products were cloned in the pBluescript II SK vector (Stratagene, La Jolla, Calif.), and the sequencing reactions were performed using the Applied Biosystems cycle sequencing kit with dye-dideoxy terminators. Sequences were read by an Applied Biosystems model 373A automatic sequencer. Both strands were sequenced from at least two clones from each strain.
Sequence analysis.
Sequence data were analyzed with the Genetics Computer Group (Madison, Wis.) program package. The program Pileup was used for multiple sequence alignments.
Design of primers.
Primers were selected from regions of the 16S rRNA gene showing the least sequence homology with the corresponding M. pneumoniae sequence. Since the two sequences were very similar, a maximum of three mismatches could be incorporated in each primer. Care was taken to avoid primers having a 3′-terminal T (14). The primers were examined for melting temperature (Tm), secondary structure, and tendency to primer-dimer formation using the Oligo primer analysis software (version 4.0; MedProbe, Olso, Norway). Three forward and 10 reverse primers with predicted Tm between 58.3 and 60.2°C (Table 1) were selected, allowing for 17 possible combinations. Eleven combinations were examined for their limit of detection (LOD) with purified M. genitalium DNA and for their specificity with M. pneumoniae DNA, corresponding to 3,500 copies and a pool of urogenital specimens PCR negative for M. genitalium.
TABLE 1.
Primers for PCR deduced from M. genitalium 16S rRNA gene sequence used in initial evaluation
| Primer | Sequence |
|---|---|
| MG16-45F | 5′ TAC ATG CAA GTC GAT CGG AAG TAG C |
| MG16-181R | 5′ ACC CTT GCA GGT CCT TTC AAC TTT A |
| MG16-252R | 5′ CGT CAT TGC CTT GGT AGG CCA |
| MG16-411R | 5′ TTC TTC CCA AAT AAA AGA ACT TTA CAA TCA A |
| MG16-447R | 5′ AAA CTC CAG CCA TTG CCT GCT AG |
| MG16-455R | 5′ GGT ACA GTC AAA CTC CAG CCA TTG |
| MG16-439F | 5′ GAA TGA CTC TAG CAG GCA ATG GCT G |
| MG16-636R | 5′ TCC AAA ACT CCC TAC CAC ACT CTA GAC TG |
| MG16-1117R | 5′ TCC TCC AAT TTA CAT TAG CAG TCT CGT TAA |
| MG16-1204F | 5′ CAA TGG CCA ATA CAA ACA GTA GCC AA |
| MG16-1301R | 5′ CTG ATT CGC GAT TAC TAG TGA TTC CAG |
| MG16-1428R | 5′ ACC GGT GCT ATC CTT GAC ATG CA |
| MG16-1421R | 5′ GTG CTA TCC TTG ACA TGC ACT TCC AA |
Primer combinations MG16S-45F-MG16S-447R and MG16S-1204F-MG16S-1301R (underlined primers) were M. genitalium specific and used in the clinical evaluation.
PCR assay.
The primer combinations were screened under the following conditions. A final reaction volume of 100 μl containing 1× PCR buffer (Super Taq; HT Biotechnology, Cambridge, United Kingdom) (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 0.01% [wt/vol] gelatin, 0.1% Triton X-100) with 1.5 mM MgCl2; a 0.4 μM concentration of each primer; a 125 μM concentration (each) of dATP, dGTP, and dCTP; and 250 μM dUTP. After preheating to 80°C (hot start) (3) 2 U of Taq DNA polymerase (Amplitaq; Perkin-Elmer, Allerød, Denmark) diluted in 10 μl of 1× reaction buffer (withheld from the master mix) was added to initiate the reaction. Forty cycles were performed in an OmniGene Thermal Cycler (Hybaid Ltd., Teddington, United Kingdom), each consisting of a 94°C 30-s denaturation, a 55°C 30-s annealing, and a 72°C 1-min extension. Amplicons were visualized after electrophoresis on 2% agarose gels, which were stained with ethidium bromide and examined by UV transillumination.
The optimized assay using primers MG16-45F and MG16-447R was performed as described above except that the MgCl2 concentration was 2.0 mM and that 40 two-step cycles were performed, each consisting of 94°C for 30 s and 60° for 60 s. The optimized MG16-1204F-MG16-1301R assay used 2.5 mM MgCl2, and 50 cycles of 94°C for 30 s and 58°C for 60 s were performed. The MgPa-476-MgPa-903 assay previously described (13) was slightly modified, as 1 U of Super Taq polymerase (HT Technology) was used and 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s were performed. The MgPa-1-MgPa-3 assay (12) was also slightly modified by running 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s with the reaction components described above except that the dUTP was replaced with dTTP at 125 μM and the MgCl2 concentration was 3.5 mM.
Construction of IPC for inhibition.
In order to detect the presence of Taq DNA polymerase inhibitors or suboptimal reaction conditions, an internal process control (IPC) was constructed. Primers amplifying parts of the phage lambda genome were selected based on the following criteria: (i) amplicon size at least 100 bp longer than the specific M. genitalium rRNA gene amplicon in order to allow preferential amplification of the rRNA gene amplicon, (ii) lack of secondary structure and primer-primer interaction as determined by the Gene Runner program (Hastings Software, Inc., Hastings, N.Y.), and (iii) a high Tm in order to render the IPC less efficient to amplify than the rRNA gene amplicon.
The primers included the sequence of each of the rRNA gene primers added to the 5′ end of the corresponding lambda primer (5′TACATGCAAGTCGATCGGAAGTAGCCTGACGGTTTCTAAC and 5′AAACTCCAGCCATTGCCTGCTAGGACATACGGAAATAG; sequence in boldface type corresponds to the phage lambda sequence at positions 13663 to 13677 and positions 14266 to 14280, respectively). PCR products thus containing the binding sites of the rRNA gene primers were obtained by amplification of 1 ng of purified lambda DNA with an annealing temperature of 40°C. After gel purification of the amplicon, a 10-fold titration was performed, and the dilution of the IPC producing no increase in the detection limit of purified M. genitalium DNA was used in the assay.
MTP-based hybridization assay.
Amplicons produced with primers MG16S-45F-MG16S-447R were labeled with digoxigenin (DIG) during PCR by addition of DIG-11-dUTP to the master mix. The reaction components and cycling conditions were as described above, except that the reaction mixture contained a 62.5 μM concentration (each) of dATP, dGTP, and dCTP; 125 μM dUTP; 0.4 μM DIG-11-dUTP; and 10 μl of the appropriate dilution of IPC. After PCR, 5 μl of the reaction products was added to wells of a heat-stable Microtiter plate (MTP) each containing 100 μl of hybridization solution (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 5 mM EDTA, and 20 pM biotinylated probe MG16S-240 Bio [5′ Bio-C-Bio-C-Bio-TTG GTA GGG TAA TGG CC]). The MTP was sealed with tape and placed in a 96-well combi thermal reactor TR2 (Hybaid) and subjected to a 2-min 95°C denaturation and 10 min 55°C hybridization. MaxiSorp MTPs (Nunc, Roskilde Denmark) were coated overnight at 4°C with streptavidin (5 μg/ml; Sigma, St. Louis, Mo.) in carbonate buffer, pH 9.6. The wells were blocked for 15 min with 1% blocking reagent (Boehringer Mannheim) in phosphate-buffered saline, pH 7.4, with 0.05% Tween 20 (PBST) prepared as a 1:10 dilution of 10% blocking reagent dissolved in maleic acid buffer, as described by the manufacturer. Hybrids were collected for 30 min at 37°C. After capture, three washes with PBST were performed, and the bound hybrids were detected by incubation at 37°C for 30 min with peroxidase-conjugated sheep anti-DIG immunoglobulin G Fab fragments (0.6 U/ml; Boehringer Mannheim) diluted in PBST with 1% blocking reagent. After three additional washes with PBST the hybrids were visualized with 1,2-phenylenediamine-hydrochloride (KemEnTec, Copenhagen, Denmark) in citrate buffer pH 5.0. The reactions were stopped by addition of H2SO4, and the A490 was read in an ELISA reader (Molecular Devices, Menlo Park, Calif.). M. pneumoniae 16S rRNA gene amplicon produced by low-stringency PCR was included as specificity control.
Precautions to avoid PCR product carryover.
Strict physical separation between PCR setup and analysis laboratories was maintained as previously described (13). Sterile filter tips (ART; SDS, Falkenberg, Sweden) were used in all manipulations with the samples. All surfaces in the PCR setup laboratory were regularly wiped with a 4% Diversol solution containing hypochlorite (19) and exposed to UV light between sessions with the purpose of destroying contaminating DNA.
Positive controls had low copy numbers, containing 10 and 100 genome copies of M. genitalium, respectively. At least two negative controls were included in each run. All M. genitalium 16S rRNA gene PCRs were performed with dUTP instead of dTTP, allowing for enzymatic prevention of PCR product carryover with uracil-N-glycosylase (16). Uracil-N-glycosylase was not used, however, since no carryover was observed.
Clinical specimens.
A total of 1,030 samples from 730 patients were randomly chosen from specimens submitted for culture of Chlamydia trachomatis. The only available information about the patients was age, sex, and sampling site. A total of 885 urogenital specimens from 595 women were submitted (302 urethral, 581 cervical, and 2 vaginal specimens). The median age of women who had urogenital specimens submitted was 28 years (range, 2 to 58 years). The remaining 13 women had 16 specimens submitted (8 conjunctival, 2 respiratory, and 6 unknown). For the 120 male patients, 129 specimens were examined: 73 urethral swabs, 23 semen specimens, 18 conjunctival specimens, 8 respiratory specimens, and 7 other specimens. The median age of the 73 patients who had urethral specimens submitted was 27 years (range, 18 to 55 years). For two patients (two specimens) the sex and age were not known. Specimens were collected and transported in 2SP medium (12) and stored at −80°C until tested in the PCR.
RESULTS
Sequencing.
A total of 1,490 bp of the 16S rRNA gene was sequenced from M. genitalium G37 and each of the four Danish M. genitalium strains M2288, M2300, M2321, and M2341. The sequence of the M. genitalium G37 type strain has been deposited in GenBank under the accession number X77334. Compared to the G37 type strain, one nucleotide was different in Danish M. genitalium strains (a T instead of a C in position 1430). A significant homology of 98% with the M. pneumoniae 16S rRNA gene sequence was found; only 28 nucleotides differed in the M. genitalium G37 sequence, and only 29 nucleotides differed in the Danish strains.
Design of primers.
Even if relatively relaxed criteria regarding general rules for design of primers were followed, only 13 different primers could be selected. These primers allowed for 17 possible combinations, but since some of the combinations would yield amplicons longer than 600 bp, only 11 combinations were actually tested. Two primer combinations were selected for further studies since they produced M. genitalium specific amplicons with a high efficiency even under the nonoptimized screening conditions. The MG16-45F-MG16-447R primers were located in the V1 and V3 hypervariable regions, respectively, and produced the expected 427-bp amplicon, whereas the MG16-1204F-MG16-1301R primers were in the V8 region and outside defined variable regions, respectively. The 124-bp amplicon produced by the 1204-1301 primers was more difficult to detect by visual inspection of ethidium bromide-stained agarose gels; hence, the MG16-45F-MG16-447R primers were selected for the primary assay.
LOD of assays.
Both primer sets had a LOD corresponding to <5 fg, equivalent to approximately six genome copies, when visual inspection of ethidium bromide-stained agarose gels was used for detection. The MTP-based hybridization assay did not improve the LOD, when purified DNA was tested; however, very faint bands in clinical specimens could be clearly positive in the hybridization assay. M. genitalium 16S rRNA gene PCR products visible by gel electrophoresis showed optical density (OD) readings of >0.5, whereas a large amount of M. pneumoniae 16S rRNA gene amplicon produced by low-stringency PCR showed OD readings of <0.05 (Fig. 1). The LOD was not increased when the M. genitalium DNA was added to clinical specimens PCR negative for M. genitalium (Fig. 2).
FIG. 1.
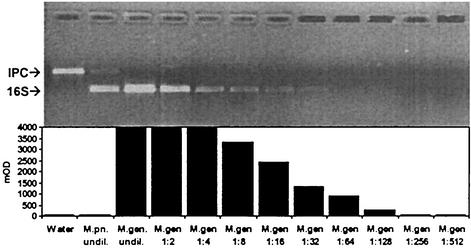
LOD of MTP-based hybridization assay performed at 50°C with negative controls and various dilutions of M. genitalium amplicon. The biotinylated probe Mg16S-240 Bio was used at 10 pM. The negative control contained the IPC amplicon. Amplicons from M. pneumoniae were produced by low-stringency annealing of the primers.
FIG. 2.
Ethidium bromide-stained agarose gel showing the LOD of the M. genitalium 16S rRNA gene PCR and the effect of addition of the IPC and human DNA. Lane 1, 100-bp marker (Pharmacia); lane 2, ∼500 genome copies of M. genitalium DNA; lane 3, ∼50 genome copies of M. genitalium DNA; lane 4, ∼5 genome copies of M. genitalium DNA; lane 5, negative control; lanes 6 to 9, same as lanes 2 to 5 but with addition of IPC; lanes 10 to 13, same as lanes 2 to 5 but with addition of IPC and 10 μl of clinical urogenital specimen.
Specificity.
The hot-start procedure was found to be crucial for the specificity of amplification; >103 copies of M. pneumoniae DNA produced faint bands with the MG16-45F-MG16-447R primer set without hot start, whereas 105 copies of M. pneumoniae DNA gave negative results when the hot-start procedure was used. The effect of hot start could also be shown in a recent modification of the protocol, where the Amplitaq was substituted with 2 U of Platinum Taq (Invitrogen, Tåstrup, Denmark). Furthermore, the specificity of the assay was ensured by the hybridization step, since even large amounts of M. pneumoniae 16S amplicon produced by decreasing the annealing temperature to 40°C did not hybridize with the internal probe. None of the other mollicutes tested gave rise to unspecific amplification.
The MG16-1204F-MG16-1301R primer set cross-reacted with DNA from M. gallisepticum even when the hot-start procedure was applied. This cross-reaction was not considered to be important since this species has never been isolated from humans. Due to the short sequence between the primers with only a single base substitution between the M. genitalium, the M. pneumoniae, and the M. gallisepticum sequences, respectively, construction of an M. genitalium-specific probe was not attempted. No amplification was observed when a pool of M. genitalium-negative urogenital specimens used in the initial screening was tested. However, when the specificity evaluation was extended with a larger number of clinical specimens, amplicons with the same length as the positive control appeared in some of the urogenital specimens showing negative reactions both with the MG16-45F-MG16-447R and the MgPa-1-MgPa-3 primer sets. Consequently, the MG16-1204F-MG16-1301R primer set was not used for further clinical studies.
IPC for inhibition.
The addition of approximately 10- to 50-fold the detection limit of the IPC produced the expected 666-bp fragment without increasing the LOD as shown in Fig. 2. As expected, preferential amplification of the shorter 16S rRNA gene amplicon occurred, resulting in the absence of the IPC from specimens with high amounts of M. genitalium DNA. It has been found that approximately 5% of clinical specimens contain inhibitors. No correlation as to the type of specimen has been shown, and in most specimens, the IPC can be amplified after a 1:2 or a 1:10 dilution of the specimen.
MTP-based hybridization assay.
The optimal concentration of the biotinylated probe was found to be crucial for the performance of the MTP-based hybridization assay, since the OD value dropped significantly after two- to fourfold dilution from the optimal concentration. The optimal concentration of the probe had to be determined for each new batch of probe, since differences were observed. The optimal hybridization temperature appeared to be of less importance since the OD of the M. pneumoniae amplicon control remained <0.05 even at 40°C.
Clinical specimens. (i) Urogenital specimens from women.
Among the specimens from 595 women for whom urogenital specimens were submitted for C. trachomatis culture, M. genitalium was found in 27 (4.5%). For comparison, C. trachomatis was detected by PCR in 34 (5.6%). The median age of those positive for M. genitalium was 25 years (range, 17 to 56 years) but not statistically different from the age of the women tested (median age, 28 years). Patients with a positive C. trachomatis PCR were significantly younger (median age, 22 years; range, 17 to 37 years) than all women tested. No woman was positive at the same time for M. genitalium and C. trachomatis. Nineteen women PCR positive for M. genitalium had both a urethral and a cervical specimen submitted for C. trachomatis culture. Nine (47%) were PCR positive for M. genitalium in both specimens, whereas seven (41%) were positive in the cervical specimen only and three (16%) were positive in the urethral specimen only. Thus, the sensitivity for urethral specimens was 63% (12 of 19), and that for cervical specimens was 84% (16 of 19).
(ii) Urogenital specimens from men.
Among the specimens from 73 men for whom urethral specimens were submitted for C. trachomatis culture, 7 (9.6%) were positive by PCR for M. genitalium. Ten (13.7%) were positive for C. trachomatis by PCR, but no specimen was positive for both organisms. None of the 23 semen specimens were PCR positive for M. genitalium.
(iii) Other clinical specimens.
M. genitalium was not detected in any of the 49 extragenital specimens.
Comparison between the M. genitalium 16S rRNA gene PCR and MgPa PCR.
The 41 specimens positive in the M. genitalium 16S rRNA gene PCR were subjected to PCR with the MgPa-1-MgPa-3 and the MgPa-476-MgPa-903 primer sets. In the MgPa-1-MgPa-3 assay, 40 of the positive specimens could be confirmed, whereas only 21 out of 40 available 16S rRNA gene PCR-positive specimens were also positive in the MgPa-476-MgPa-903 PCR assay. The urethral and the cervical swabs of eight women were both positive with the M. genitalium 16S rRNA gene PCR. When the urethral and cervical swabs of these women were examined with the MgPa-476-MgPa-903 primer set, the results for two women were concordant positive, the results for one woman were concordant negative, and the results for five women were discrepant.
DISCUSSION
In recent years, DNA amplification techniques with PCR for M. genitalium have been introduced. By using these methods several studies have demonstrated an association between M. genitalium and NGU in males (2, 9, 10, 13, 25, 26; T. Deguchi, H. Komeda, M. Yasuda, K. Tada, H. Iwata, M. Asano, T. Ezaki, and Y. Kawada, Letter, Int. J. STD AIDS 6:144-145, 1995). Also, M. genitalium has been proposed to be involved in cervicitis (28) and endometritis in females (4). At present, only a few studies have compared the efficiency of different PCR assays on clinical specimens (5, 13). Deguchi et al. (5) found results obtained with the MgPa-1-MgPa-3 primer set to be identical to those found with the primers described by Palmer et al. (17). This comparison was very important, since the majority of clinical studies have used one of these two assays. However, only 18 positive specimens were compared. We have previously found that the part of the MgPa gene amplified by the MgPa-1-MgPa-3 primer set contained mutations resulting in different restriction fragment length polymorphisms (12). Later, we observed that the amount of amplicons produced with the MgPa-1-MgPa-3 primer set compared with that produced with the MgPa-476-MgPa-903 primer set from patients enrolled in a clinical study of NGU was extremely variable. Furthermore, the sequence amplified with the MgPa-476-MgPa-903 primers revealed remarkable variation when subjected to restriction enzyme analysis (13). Despite this variation, a complete agreement between the two assays was found.
These observations led us to speculate whether the MgPa gene was an optimal target for a diagnostic PCR assay, and therefore, the 16S rRNA gene was selected as the target for development of a new assay. At that time, the sequence of the M. genitalium G37 16S rRNA gene was not available, and, furthermore, it was felt to be important to assure that the newly isolated M. genitalium strains had the same sequence as the type strain of M. genitalium. Consequently, the 16S rRNA gene sequences of five different strains were obtained. Only one nucleotide was found to differ between the M. genitalium G37 and the four Danish M. genitalium strains. The M. genitalium G37 16S rRNA gene sequence was later shown to be identical to that available from the genome sequence (6). Whether the single-nucleotide difference found among the Danish strains reflects geographical differences remains to be determined.
The two 16S rRNA gene primer combinations selected for further evaluation had an equal LOD when evaluated on dilutions of purified M. genitalium DNA, both alone and in simulated positive specimens. Using the hot-start protocol both primer sets were specific, when tested with M. pneumoniae DNA. A range of other mollicutes was tested to assure the specificity. Only DNA from M. gallisepticum cross-reacted in the MG16-1204F-MG16-1301R assay, even when the hot-start procedure was applied, but this was not regarded as a significant problem, since this species is not expected to be found in humans. Initially, a pool of M. genitalium MgPa PCR negative specimens were used for negative controls and for preparation of simulated positive specimens. No positive reactions were detected in the pooled specimens with the 16S PCRs. However, when individual clinical specimens were tested in the MG16-1204F-MG16-1301R assay sporadic positive reactions, which could not be confirmed with other primer sets, were observed. Therefore, these primers could not be recommended for use without an internal probe or confirmation with another PCR assay.
The MTP-based hybridization assay improved the specificity against M. pneumoniae since amplicons produced by low-stringency annealing were consistently negative even though only two nucleotides differed between the M. genitalium and the M. pneumoniae sequences. The PCR assay was adjusted to detect very few genome copies during the amplification. This allowed the rapid detection of the specific product and the internal processing control by agarose gel electrophoresis. The MTP hybridization assay did not add additional sensitivity, and in a routine screening setting, only those specimens positive by gel electrophoresis would need hybridization in order to keep the specificity high. Many authors argue that gel electrophoresis is unsuitable for large-scale amplicon detection. In our hands, however, gel electrophoresis was found to be very time- and cost-efficient, since the result of 35 clinical specimens can be recorded within 30 min after completion of the PCR. On the other hand, the objective reading of a spectrophotometer and the possibility of automation are important advantages of the MTP-based hybridization assay.
The MG16-45F-MG16-447R primer set was not evaluated on many nonurogenital specimens during this study. M. pneumoniae has been reported in a few publications to be present in the urogenital tract (7, 23), and M. genitalium has been detected in respiratory tract specimens (1). Therefore, the mere detection of an amplicon on agarose gel electrophoresis should not be taken as evidence of an M. genitalium infection without a hybridization step or some kind of confirmatory PCR assay. For a confirmatory assay, the results of the present study clearly indicate that the MgPa-1-MgPa-3 primers are the best alternative primers of those tested. The single specimen positive only by the MG16-45F-MG16-447R primer set, could not be confirmed by any of the other primer sets, and therefore, it seems likely that it was a false positive in that assay.
The discrepancy between the low clinical sensitivity of the MgPa-476-MgPa-903 primer set found in the present study and the good agreement between the two MgPa-based assays in a previous study could be explained by the slightly changed reaction conditions used in this study. We increased the annealing temperature to 55 from 50°C, and the number of cycles was decreased from 50 to 40. These changes were justified by experiments using dilution series of M. genitalium G37 DNA, where it was found that the SuperTaq enzyme produced a significantly higher yield of amplicon than the AmpliTaq used in the original assay. These observations emphasize the importance of careful evaluations of PCR assays, whenever reaction parameters are changed. Due to lack of sample material, attempts to run the discrepant samples with the original protocol could not be undertaken. The fact that five of eight paired urethral and cervical specimens positive in the M. genitalium 16S rRNA gene PCR showed discrepant results in the MgPa-476-MgPa-903 assay indicates that the decreased clinical sensitivity could be due to a strain-dependent relative lack of sensitivity caused by sequence variation.
One 16S rRNA gene PCR assay has been published previously (22), but its application on clinical specimens has not been reported. The LOD was stated to be 1,000 M. genitalium cells, which is insufficient for clinical diagnostic work. Recently, a TaqMan real-time PCR assay based on amplification of the M. genitalium 16S rRNA gene was published (29). The LOD was reported to be 10 copies/reaction, which is marginally higher than that of the present assay. However, since external reference standards are not available, a head-to-head comparison between assays would be needed to determine the clinical sensitivity.
M. genitalium was found only in the urogenital specimens examined both from men and women. The rate of positive findings was significantly higher in men than in women; however, this could easily be explained by the different healthcare-seeking behavior of men and women and by the different criteria for taking a specimen for C. trachomatis testing. Symptomatic men are more likely to be examined since screening among asymptomatic men is less widely used than screening among asymptomatic women. A corresponding higher proportion of C. trachomatis positive men further substantiate this explanation.
In both men and women, the rate of detection of M. genitalium was only slightly lower than that of C. trachomatis—4.5% compared to 5.6% in women and 9.6% compared to 13.7% in men—and the two organisms were not detected in the same patients, indicating that they may act as separate causes of urogenital tract illness. This is in agreement with most clinical studies published (2, 9, 10, 13, 25, 26; Deguchi et al., letter).
In conclusion, the PCR assay presented here could be a valuable supplement to the MgPa-based assays most widely used (12, 17), which appear to have a comparable performance. On the other hand, the results also demonstrate that other MgPa gene-based assays should be carefully evaluated before they can be used with confidence.
REFERENCES
- 1.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björnelius, E., P. Lidbrink, and J. S. Jensen. 2000. Mycoplasma genitalium in non-gonococcal urethritis—a study in Swedish male STD patients. Int. J. STD AIDS 11:292-296. [DOI] [PubMed] [Google Scholar]
- 3.Chou, Q., M. Russell, D. E. Birch, J. Raymond, and W. Bloch. 1992. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 20:1717-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, C. R., L. E. Manhart, E. A. Bukusi, S. Astete, R. C. Brunham, K. K. Holmes, S. K. Sinei, J. J. Bwayo, and P. A. Totten. 2002. Association between Mycoplasma genitalium and acute endometritis. Lancet 359:765-766. [DOI] [PubMed] [Google Scholar]
- 5.Deguchi, T., C. B. Gilroy, and D. Taylor Robinson. 1995. Comparison of two PCR-based assays for detecting Mycoplasma genitalium in clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 14:629-631. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 7.Goulet, M., R. Dular, J. G. Tully, G. Billowes, and S. Kasatiya. 1995. Isolation of Mycoplasma pneumoniae from the human urogenital tract. J. Clin. Microbiol. 33:2823-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooton, T. M., M. C. Roberts, P. L. Roberts, K. K. Holmes, W. E. Stamm, and G. E. Kenny. 1988. Prevalence of Mycoplasma genitalium determined by DNA probe in men with urethritis. Lancet i:266-268. [DOI] [PubMed]
- 9.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582-585. [DOI] [PubMed] [Google Scholar]
- 10.Janier, M., F. Lassau, I. Casin, P. Grillot, C. Scieux, A. Zavaro, C. Chastang, A. Bianchi, and P. Morel. 1995. Male urethritis with and without discharge: a clinical and microbiological study. Sex. Transm. Dis. 22:244-252. [DOI] [PubMed] [Google Scholar]
- 11.Jensen, J. S., H. T. Hansen, and K. Lind. 1996. Isolation of Mycoplasma genitalium strains from the male urethra. J. Clin. Microbiol. 34:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, J. S., S. A. Uldum, J. Søndergård-Andersen, J. Vuust, and K. Lind. 1991. Polymerase chain reaction for detection of Mycoplasma genitalium in clinical samples. J. Clin. Microbiol. 29:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, J. S., R. Ørsum, B. Dohn, S. Uldum, A. M. Worm, and K. Lind. 1993. Mycoplasma genitalium: a cause of male urethritis? Genitourin. Med. 69:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok, S., D. E. Kellogg, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind, K., B. Ø. Lindhardt, H. J. Schütten, J. Blom, and C. Christiansen. 1984. Serological cross-reactions between Mycoplasma genitalium and Mycoplasma pneumoniae. J. Clin. Microbiol. 20:1036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, H. M., C. B. Gilroy, P. M. Furr, and D. Taylor-Robinson. 1991. Development and evaluation of the polymerase chain reaction to detect Mycoplasma genitalium. FEMS Microbiol. Lett. 61:199-203. [DOI] [PubMed] [Google Scholar]
- 18.Peterson, S. N., C. C. Bailey, J. S. Jensen, M. B. Borre, E. S. King, K. F. Bott, and C. A. Hutchison. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. USA 92:11829-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince, A. M., and L. Andrus. 1992. PCR—how to kill unwanted DNA. BioTechniques 12:358.. [PubMed] [Google Scholar]
- 20.Razin, S., and J. G. Tully. 1983. Methods in mycoplasmology, vol. 1. Mycoplasma characterization. Academic Press, Inc., New York, N.Y.
- 21.Samra, Z., M. Borin, Y. Bukowsky, Y. Lipshitz, and D. Sompolinsky. 1988. Non-occurrence of Mycoplasma genitalium in clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 7:49-51. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki, Y., M. Shintani, T. Shimada, H. Watanabe, and T. Sasaki. 1992. Detection and Discrimination of Mycoplasma pneumoniae and Mycoplasma genitalium by the in vitro DNA amplification. Microbiol. Immunol. 36:21-27. [DOI] [PubMed] [Google Scholar]
- 23.Sharma, S., R. Brousseau, and S. Kasatiya. 1998. Detection and confirmation of Mycoplasma pneumoniae in urogenital specimens by PCR. J. Clin. Microbiol. 36:277-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor-Robinson, D., P. M. Furr, and N. F. Hanna. 1985. Microbiological and serological study of non-gonococcal urethritis with special reference to Mycoplasma genitalium. Genitourin. Med. 61:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor-Robinson, D., J. S. Jensen, G. Fehler, F. Radebe, and R. C. Ballard. 2002. Observations on the microbiology of urethritis in black South African men. Int. J. STD AIDS 13:323-325. [DOI] [PubMed] [Google Scholar]
- 26.Totten, P. A., M. A. Schwartz, K. E. Sjostrom, G. E. Kenny, H. H. Handsfield, J. B. Weiss, and W. L. Whittington. 2001. Association of Mycoplasma genitalium with nongonococcal urethritis in heterosexual men. J. Infect. Dis. 183:269-276. [DOI] [PubMed] [Google Scholar]
- 27.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet i:1288-1291. [DOI] [PubMed]
- 28.Uno, M., T. Deguchi, H. Komeda, M. Hayasaki, M. Iida, M. Nagatani, and Y. Kawada. 1997. Mycoplasma genitalium in the cervices of Japanese women. Sex. Transm. Dis. 24:284-286. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, T., T. Deguchi, M. Ito, S. Maeda, M. Tamaki, and H. Ishiko. 2002. Quantitative detection of Mycoplasma genitalium from first-pass urine of men with urethritis and asymptomatic men by real-time PCR. J. Clin. Microbiol. 40:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]