Abstract
In the fission yeast Schizosaccharomyces pombe, the “start” of the cell cycle is controlled by the two functionally redundant transcriptional regulator complexes, Res1p-Cdc10p and Res2p-Cdc10p, that activate genes essential for the onset and progression of S phase. The activity of the Res2p-Cdc10p complex is regulated at least by the availability of the Rep2 trans-activator subunit in the mitotic cell cycle. We have recently isolated the pas1+ gene as a multicopy suppressor of the res1 null mutant. This gene encodes a novel cyclin that shares homology with the Pho85 kinase–associated cyclins of the budding yeast Saccharomyces cerevisiae. Genetic analysis reveals that Pas1 cyclin is unrelated to phosphate metabolism and stimulates the G1-S transition by specifically activating the Res2p-Cdc10p complex independently of Rep2p. Pas1 cyclin also controls mating pheromone signaling. Cells lacking pas1+ are highly sensitive to mating pheromone, responding with facilitated G1 arrest and premature commitment to conjugation. Pas1 cyclin associates in vivo with both Cdc2 and Pef1 kinases, the latter of which is a fission yeast counterpart of the budding yeast Pho85 kinase, but genetic analysis indicates that the Pef1p-associated Pas1p is responsible for the activation of Res2p-Cdc10p during the G1-S transition.
INTRODUCTION
In virtually all eukaryotes, the onset of proliferation and differentiation is controlled at the point called “start” in the prestart G1 phase of the cell cycle. At least two distinct control elements are known to be required for passing through start to initiate the cell cycle. One includes Cdks, which regulate the activity of proteins crucial for the onset and progression of S phase, and the other is transcriptional factors that activate a subset of genes essential for the onset and progression of S phase (for review, see Okayama et al., 1996).
In higher eukaryotes, several cyclins associated with distinct kinases, such as Cdk4/6-cyclin D and Cdk2-cyclin E, control the G1-S transition, whereas Cdc2-cyclin B is used exclusively to control the G2-M transition (for review, see Nigg, 1995). Similarly, in yeast, different cyclins regulate the G1-S and G2-M transitions, although unlike their mammalian counterparts, Cdc2 (Cdc28) kinase is their common association partner. In the budding yeast Saccharomyces cerevisiae, one of the three G1 cyclins, Cln1p, Cln2p, or Cln3p, associated with Cdc28 kinase is essential for the cell cycle start. The critical target for these Cdc28p-Cln complexes is Sic1p, a Cdk inhibitor, because deletion of the SIC1 gene rescued the inviability of the cln1 cln2 cln3 triple mutant (Schneider et al., 1996; Tyers, 1996). Phosphorylation of Sic1p by Cdc28p-Cln complexes directs degradation, resulting in liberation from inhibition and consequently making available B-type cyclin (Clb)-bound Cdc28 kinase for the activation of the origins of replication (Feldman et al., 1997; Skowyra et al., 1997; Verma et al., 1997). In addition, Pho85p-Pcl1/2p complexes, which constitute another Cdk-G1 cyclin set, seemed to play some role in promoting start (Espinoza et al., 1994; Measday et al., 1994). Although inessential for cell viability in the wild-type background, they are required for G1 progression in the Δcln1 Δcln2 background. One possible target for their cell cycle start function is Sic1p (Nishizawa et al., 1998). Meanwhile, Pho85p associated with Pho80p, a member of the Pcl cyclin family, controls phosphate metabolism (Kaffman et al., 1994).
In the fission yeast Schizosaccharomyces pombe, Cig2 (also called Cyc17) B-type cyclin associated with Cdc2 kinase promotes the G1-S transition, although this role is shared by the Cig1 and Cdc13 mitotic B-type cyclins (Fisher and Nurse, 1996; Martin-Castellanos et al., 1996; Mondesert et al., 1996). A Cln-related cyclin, Puc1p, was isolated by phenotypic complementation of a cln3 mutant of S. cerevisiae and shown to associate with Cdc2 kinase (Forsburg and Nurse, 1991, 1994). This cyclin seems to regulate G1-phase progression in response to cell size (Martin-Castellanos et al., 2000).
In the budding yeast, the transcriptional factor complexes essential for the cell cycle start are two members of the Swi/Cdc10 family that are functionally distinct and are called Swi4p-Swi6p and Mbp1p-Swi6p (for review, see Koch and Nasmyth, 1994; Breeden, 1996). Swi4p-Swi6p activates the cis element called the Swi4/6-dependent cell cycle box (SCB) that is present in the promoters of the HO endonuclease and G1 cyclin genes (CLN1, CLN2, PCL1, and PCL2), whereas Mbp1p-Swi6p activates the cis element called the MluI cell cycle box (MCB) contained in the promoter of a subset of genes required for the onset and progression of S phase, such as CLB5 and CLB6. Activation of these transcriptional factor complexes at the G1-S boundary requires Cdc28 kinase associated with one of the three Cln cyclins (Cross and Tinkelenberg, 1991; Dirick and Nasmyth, 1991). In vitro band-shift and in vivo footprinting experiments have indicated that the Cln cyclin–associated Cdc28 kinase regulates the ability of Swi4p-Swi6p to bind SCB or the ability of previously bound Swi4p-Swi6p to activate transcription in a positive feedback manner (Taba et al., 1991; Koch et al., 1996). In fact, any one of the Cln cyclins is capable of activating late G1-specific transcription when ectopically expressed. However, recent studies have suggested that Cln3p is specialized to activate Swi4p-Swi6p and Mbp1p-Swi6p in the in vivo situation (Tyers et al., 1993; Dirick et al., 1995; Stuart and Wittenberg, 1995; Levine et al., 1996).
Fission yeast possesses similar yet slightly different transcriptional activator complexes essential for the cell cycle start. No system corresponding to Swi4p-Swi6p has been found in this organism. Instead, it has the two functionally redundant MCB-activating systems, Res1p-Cdc10p and Res2p-Cdc10p (Res1p and Res2p are also called Sct1p and Pct1p, respectively) (Lowndes et al., 1992; Tanaka et al., 1992; Caliguiri and Beach, 1993; Miyamoto et al., 1994; Zhu et al., 1994), the former of which functions predominantly for the start of the mitotic cycle and the latter of which functions predominantly for the start of meiosis (for review, see Woollard and Nurse, 1995; Okayama et al., 1996). Nonetheless, the latter set also plays an important role in the mitotic cycle. In the mitotic cycle, Res2p-Cdc10p forms a tertiary complex with Rep2p, a coactivator subunit, and drives the onset of S phase, whereas in meiosis, it seems to form a complex with Rep1p to drive the onset of premeiotic S phase (Sugiyama et al., 1994; Nakashima et al., 1995). Both rep2+ and rep1+ are under stringent transcriptional control by nutrient and/or pheromone availability. In the absence of the coactivator subunits, Res2p-Cdc10p binds and sequesters MCB, thereby acting as a strong inhibitor of the cell cycle start in response to external conditions. The target genes for these Res-Cdc10p-Rep complexes include cdc18+, cdc22+, and cdt1+, which are essential for S-phase onset (Lowndes et al., 1992; Kelly et al., 1993; Hofmann and Beach, 1994). The expression of those genes is periodic during the cell cycle, peaking at G1-S and reaching its nadir at G2. Res subunits are critically required for the periodic transcription (Baum et al., 1997). No coactivator subunits for Res1p-Cdc10p have been found.
The possible regulation of the Res-Cdc10p-Rep complexes by Cdks has been highly controversial. The MCB-binding activity exerted by Res-Cdc10p-Rep, which was detected by in vitro band-shift assays, similarly oscillated in a cell cycle–dependent manner with its reactivation at G1-S, depending on a G1 form of Cdc2 kinase (Reymond et al., 1993). Moreover, there was a report that the phosphorylation of Cdc10p by Cdc2 kinase was essential for the formation of the Res1p-Cdc10p complex (Connolly et al., 1997). However, the MCB-binding activity correlated with the inactive state of Res-Cdc10p-Rep–dependent transcription (Baum et al., 1997). In addition, Res-Cdc10p-Rep–dependent genes began to be expressed during mitosis, when the G1 form of Cdc2 kinase was absent (Baum et al., 1998). Furthermore, the Res-Cdc10p-Rep–dependent transcription could be activated without Cdc2 kinase activity (Baum et al., 1997).
In a search for new factors controlling the cell cycle start, we isolated a new cyclin named Pas1, which is structurally similar to the Pcl family members that associate with Pho85 kinase and regulate phosphate metabolism, glycogen biosynthesis, actin regulation, and cell cycle progression in S. cerevisiae (Moffat et al., 2000). Despite such structural homology, Pas1 cyclin resembles Cln3/1/2p of budding yeast in function and promotes the cell cycle start by specifically activating the Res2p-Cdc10p complex. Pas1 cyclin associates in vivo with both Cdc2p and a newly identified fission yeast counterpart of the budding yeast Pho85p, and genetic analysis indicates that Pas1p associated with the fission yeast Pho85p (Pef1p) is responsible for activating Res2p-Cdc10p. In this report, we present genetic and functional data demonstrating the properties and biological role of Pas1 cyclin.
MATERIALS AND METHODS
Fission Yeast Strains, Media, and Genetic Methods
The strains of S. pombe used for this study are listed in Table 1. Multiple gene deletions were obtained by crossing strains with the use of tetrad analysis and confirmed by PCR analysis and/or by checking genetic markers. Strains were cultured in the complete medium YE or in the minimal medium MM (also called EMM2 or PM) (Alfa et al., 1993). When necessary, carbon (glucose, referred to as G), nitrogen (ammonium chloride, referred to as N), or phosphate (disodium hydrogen phosphate, referred to as P) concentrations were reduced in the MM. Transformations were performed according to the lithium acetate procedure as described previously (Okazaki et al., 1990). Transformed cells were spread on two minimal medium agar (MMA) plates (Gutz et al., 1974) and incubated at the permissive and the nonpermissive temperatures. The suppression efficiencies were calculated by dividing the number of colonies formed at the nonpermissive temperature by the number of colonies formed at the permissive temperature. Flow cytometry was performed as described previously (Tanaka et al., 1992). Cell numbers were determined with a particle counter (Z1, Beckman Coulter, Fullerton, CA). Northern blot analysis was performed as described (Nakashima et al., 1995). Other general genetic manipulations for S. pombe have been described (Moreno et al., 1991; Alfa et al., 1993)
Table 1.
Strain list
| Strain | Genotype |
|---|---|
| L972 | h− prototroph |
| L968 | h90 prototroph |
| EV3A | h−leu1-32 |
| K156-D1 | h−ura4-D18 res1∷ura4+leu1-32 |
| K165-A12 | h−ura4-D18 res2∷ura4+ |
| M222 | h−ura4-D18 res2∷ura4+leu1-32 |
| K166-A5 | h−ura4-D18 rep2∷ura4+ |
| K166-A1 | h90 ura4-D18 rep2∷ura4+ |
| N3-141S | h−ura4-D18 rep2∷ura4+leu1-32 |
| K193-A1 | h−ura4-D18 pas1∷ura4+ |
| K207-A1 | h90 ura4-D18 pas1∷ura4+ |
| K182-A7 | h− ura4-D18 pas1∷ura4+leu1-32 |
| K182-A7-P1 | h−ura4-D18 pas1∷ura4+ ≪ pas1+-kanMX6 leu1-32 |
| K205-A1 | h−ura4-D18 sxa2∷ura4+ |
| 14-1 | h90 ura4-D18 nrd1∷ura4+ |
| NP2-461 | h−ura4-D18 res2∷ura4+rep2∷ura4+leu1-32 |
| NPP-12D | h−ura4-D18 rep1∷ura4+rep2∷ura4+leu1-32 |
| K189-3A | h+s ura4-D18 res2∷ura4+pas1∷ura4+ |
| K189-23C | h−ura4-D18 res2∷ura4+pas1∷ura4+leu1-32 |
| K190-22B | h+s ura4-D18 rep2∷ura4+pas1∷ura4+ |
| K190-5D | h−ura4-D18 rep2∷ura4+pas1∷ura4+leu1-32 |
| K539-1A | h+s ura4-D18 res2∷ura4+rep2∷ura4+pas1∷ura4+leu1-32 |
| K209-22A | h−ura4-D18 pas1∷ura4+sxa2∷ura4+ |
| K228-29A | h90 ura4-D18 nrd1∷ura4+pas1∷ura4+ |
| K224-4D | h−ura4-D18 cig1∷ura4+cig2∷ura4+puc1∷ura4+ |
| K224-51C | h−ura4-D18 cig1∷ura4+cig2∷ura4+pas1∷ura4+ |
| K224-59C | h−ura4-D18 cig1∷ura4+puc1∷ura4+pas1∷ura4+ |
| K224-35D | h+s ura4-D18 cig2∷ura4+puc1∷ura4+pas1∷ura4+ |
| K224-42D | h−ura4-D18 cig1∷ura4+cig2∷ura4+puc1∷ura4+pas1∷ura4+ |
| K226-42A | h−ura4-D18 cig1∷ura4+cig2∷ura4+puc1∷ura4+pas1∷ura4+leu1-32 |
| K571-2D | h−ura4-D18 pef1∷ura4+leu1-32 |
| K230-A6 | h−ura4-D18 cdc2-L7 ≪ cdc2HA+-ura4+leu1-32 |
| K566-11 | h−ura4-D18 pef1 ≪ pef1HA+-ura4+leu1-32 |
Libraries and Vectors
The S. pombe genomic libraries were constructed by inserting HindIII-digested wild-type (L972) genomic DNA into the HindIII-digested pALSK+ vector (HindIII library) and by inserting SpeI-digested wild-type genomic DNA into the SpeI-digested pALSK+ vector (SpeI library). The S. pombe cDNA library has been described (Okazaki et al., 1991; Sugiyama et al., 1994). The pALSK+ and pcL vectors were described previously (Tanaka et al., 2000). The pIK1 vector was constructed by inserting the kanMX6 marker into the pBluescriptII SK− vector. The pIU2HA vector was constructed by inserting a ura4+ gene, a sequence containing three copies of hemagglutinin (HA) epitope tag, and a polyadenylation signal of the nmt1+ gene into the pBluescriptII SK− vector. The pREP1 vector was described previously (Maundrell, 1993).
Isolation of the pas1+ Gene
The pas1+ gene was isolated as described previously (Okazaki et al., 1990; Miyamoto et al., 1994). The Δres1 (K156-D1) mutant cells were transfected with the HindIII library. The transfected cells were spread on MMA plates, incubated at 30°C for 17 h, and then selected at 21°C for 5–11 d. Plasmid DNA clones were recovered in Escherichia coli from candidates and analyzed by dot blotting with res1+, res2+, rep1+, rep2+, and cdc18+ genes as probes. The pas1+ cDNA clone and a genomic DNA clone containing its own promoter region were isolated by colony hybridization from the cDNA library and the SpeI library, respectively.
Gene Disruption
Gene disruption was performed by one-step gene replacement. The 1.1-kilobase (kb) XhoI–NdeI fragment of the pas1+ gene that contains ∼90% of the coding region was replaced with the 1.8-kb fragment of the ura4+ gene. Similarly, the 0.7-kb NdeI–BglII fragment of the pef1+ gene that contains ∼80% of the coding region was replaced with the ura4+ gene. The SphI fragment carrying the disrupted pas1 locus or the PstI–SpeI fragment carrying the disrupted pef1 locus was used to transform the ura4-D18 diploid strain, and stable ura+ transformants were isolated. The proper replacement of one wild-type allele with the disrupted constructs was confirmed by Southern blot analysis.
Assay for Conjugation
The mating frequencies were assayed at 27°C if temperature was not specified. Cells were grown to log phase in MM (+N/2%G), reinoculated in fresh medium, and grown to midlog phase (∼5 × 106 cells/ml). Cells were washed once with distilled water, inoculated in MM (+N/2%G), MM (+N/0.5%G), MM (−N/2%G), and MM (−N/0.5%G) at a density of 5 × 106 cells/ml, dispensed into test tubes to avoid repeated sonication, and incubated with gentry shaking. At the indicated time, cell suspensions were sonicated to disperse cell aggregates, and the number of zygotes formed was counted under the microscope. The percentage mating efficiencies were calculated by dividing the number of zygotes (one zygote counted as two cells) by the number of total cells.
Assay for Pheromone Sensitivity
Pheromone sensitivities of h− wild-type (L972) and h− Δpas1 (K193-A1) were assayed as follows. Cells were grown to log phase in MM (+N/2%G) at 30°C. Each culture was reinoculated in fresh medium and grown to midlog phase (∼5 × 106 cells/ml). Cells were washed once with MM (+N/0.5%G), inoculated in the same fresh medium at a density of 5 × 106 cells/ml, and divided into two parts. Chemically synthesized P-factor was then added to one of the two parts to a final concentration of 2 μg/ml. Cells were cultured at 30°C and harvested at the indicated time, and total RNA was prepared from each aliquot. Expression of sxa2+ and ura4+ was examined by Northern blot analysis.
Sensitivities to pheromone-induced cell cycle arrest were assayed as follows. The cells of h− Δsxa2 (K205-A1) and h− Δsxa2 Δpas1 (K209–22A) were grown to log phase in MM (+N/2%G) at 30°C. Each culture was reinoculated in fresh medium and grown to midlog phase. Cells were washed with MM (+N/0.1%G), inoculated in the same fresh medium at a density of 5 × 105 cells/ml, and divided into two parts, and both parts were incubated at 30°C for 4.5 h. Chemically synthesized P-factor was then added to one of the two parts to a final concentration of 2 μg/ml, and incubation was continued. The cell growth was examined by counting the cell number.
Assay for Acid Phosphatase Activity
Acid phosphatase activity was assayed as follows (modified from To-E et al., 1973). Cells of h− wild-type (L972) and h− Δpas1 (K193-A1) were grown to log phase in MM (+N/2%G) at 30°C. Each culture was reinoculated in EMMP (Moreno et al., 1991) lacking phosphate (−P) or EMMP plus phosphate (14.85 mM NaH2PO4, 0.65 mM Na2HPO4, pH 5.5) (+P) and grown for 12 h. Because up to 40% of acid phosphatase is secreted into the medium in fission yeast (Mitchison and Creanor, 1969; Creanor et al., 1983), acid phosphatase activity was assayed with the use of whole cell culture with medium. A total of 100 μl of cell culture was added to 400 μl of substrate solution (56.2 μg/ml p-nitrophenyl phosphate in 0.1 M sodium acetate, pH 4.1) and incubated for 1 h at 30°C. Reactions were stopped by the addition of 720 μl of saturated sodium carbonate. Cells were removed by centrifugation, and the absorbance at 420 nm was measured.
Protein Extracts, Immunoprecipitation, and Protein Kinase Assay
Full-length pas1+ and cdc13+ cDNAs were tagged at N termini with FLAG epitope by insertion into the pFLAG2 vector (Kodak, Rochester, NY), and the FLAG-tagged cDNAs were subcloned into the pREP1 vector. h− Δpas1 leu1 (K182-A7), h− cdc2HA+ leu1 (K230-A6), or h− pef1HA+ leu1 (K566−11) cells were transformed with these plasmids, and transformants were cultured to log phase in MM (+N/2%G) containing 10 μM thiamine at 30°C. Each culture was reinoculated into fresh medium containing 10 μM thiamine and grown to log phase again. To induce the FLAG-tagged genes, cells were collected, washed three times with MM (+N/2%G) without thiamine, inoculated into the same thiamine-minus medium at a density of 2 × 105 cells/ml, and cultured at 30°C for 16 h.
Total cell extracts were prepared as described previously (Booher et al., 1989; Moreno et al., 1989). About 5 × 108 cells were harvested, washed once with Stop buffer and once with H buffer (HB), and resuspended in 50 μl of HB. About 1 ml of chilled glass beads (∼500 μm) was added, and cells were broken by vigorous vortexing six times for 30 s each at 4°C. The beads were washed with 500 μl of ice-cold HB, and supernatant was removed from the glass beads. The lysate was centrifuged in a microfuge for 5 min at 15,000 rpm at 4°C. The soluble fraction was transferred to new tubes, kept on ice for 20 min, and centrifuged at 15,000 rpm for 20 min at 4°C again. The supernatant was recovered, and protein concentration was determined by the Bradford method (Bio-Rad [Richmond, CA] protein assay). NaCl was added to extracts to a final concentration of 150 mM before immunoprecipitation. For anti-FLAG immunoprecipitation, 4 mg of extracts was pretreated with mouse immunoglobulin G (IgG)-conjugated agarose (Jackson Immunoresearch, West Grove, PA) to reduce backgrounds and then incubated with 40 μl of anti-FLAG M2 affinity gel (20% suspension in HB) (Kodak) at 4°C. For p13suc1 bead precipitation, the extracts (4 mg of protein) were incubated with 40 μl of p13suc1 beads (20% suspension in HB) (Oncogene Research Products, Boston, MA) at 4°C. Precipitates were washed six times with ice-cold HB containing 150 mM NaCl, electrophoresed in SDS-polyacrylamide gels, and subjected to immunoblot with anti-FLAG (M2; Kodak), anti-PSTAIR (Yamashita et al., 1991), or anti-HA (12CA5; Boehringer Mannheim, Indianapolis, IN) mouse mAbs or an anti-FLAG (anti-OctA) (D-8; Santa Cruz Biotechnology, Santa Cruz, CA) rabbit polyclonal antibody.
Histone H1 kinase assay was performed as follows (modified from Moreno et al., 1989). Anti-FLAG immunoprecipitates were resuspended in 20 μl of KIN buffer (HB containing 1 mg/ml histone H1 [Boehringer Mannheim], 200 μM ATP, 0.5 μCi of [γ-32P]ATP, and 150 mM NaCl), and reaction mixtures were incubated for 20 min at 30°C. Reactions were stopped by the addition of 20 μl of 2× sample buffer, boiled for 5 min, and loaded on 12% SDS-polyacrylamide gels. Phosphorylated histone H1 was detected by autoradiography after an overnight exposure at −70°C.
Chromosomal Integration of the pas1+ Gene
A 6.8-kb SpeI fragment containing the pas1+ gene was subcloned into the pIK1 vector. This plasmid was digested at the unique BglII site that is located in the 5′-upstream region of the pas1+ ORF to promote integration via homologous recombination and transfected into the h− Δpas1 leu1 strain (K182-A7). The transfected cells were spread on YEA plates, incubated at 30°C for 18 h, replica plated onto YEA plates containing G418 (100 μg/ml), and incubated at 30°C for 2 d. Stable G418-resistant clones were selected, and proper integrants were identified by Southern blot analysis.
Chromosomal Integration of the cdc2HA+ and pef1HA+ Genes
A NotI restriction site was introduced just before the stop codon in the cdc2+ or the pef1+ gene. The SpeI–NotI fragment of cdc2+ and the EcoRV–NotI fragment of pef1+, which encode the C-terminal two-thirds of each kinase lacking the PSTAIR region, were subcloned into the pIU2HA vector that provides 3× HA tag. The vector plasmids with the inserts were linearized at the XbaI site in cdc2HA+ or at the XhoI site in pef1HA+ to promote integration via homologous recombination and transformed into the h− ura4-D18 cdc2-L7 leu1-32 (for the cdc2HA+) or the h− ura4-D18 leu1-32 (for the pef1HA+) strain. Stable ura+ transformants were isolated, and proper integrants were identified by Southern blot analysis.
Nucleotide Sequence Accession Number
The DDBJ-EMBL-GenBank accession numbers for pas1+ and pef1+ are AB 045126 and AB 045127, respectively.
RESULTS
Isolation of the pas1+ Gene
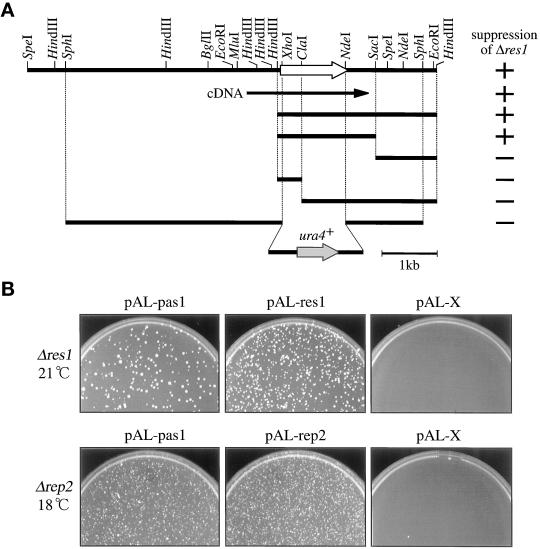
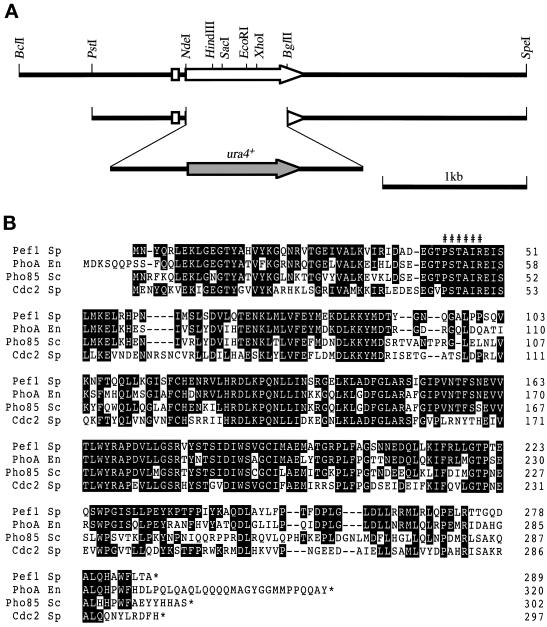
To understand the mechanism controlling the cell cycle start, we sought to identify novel factors functionally interacting with the Res-Cdc10p-Rep transcriptional regulator complexes. To this end, we searched for multicopy suppressors of the inability of the res1 null mutant (Δres1) to start the cell cycle at 21°C (Tanaka et al., 1992). To avoid repeated isolation of the known multicopy suppressors res1+, res2+, rep1+, rep2+, and cdc18+ (Tanaka et al., 1992; Kelly et al., 1993; Miyamoto et al., 1994; Sugiyama et al., 1994; Nakashima et al., 1995), we used an S. pombe HindIII-digested genomic DNA library, because all of the known suppressors except cdc18+ contain at least one HindIII site within their coding region and therefore they were expected to be eliminated from this library. After transfection and selection, 71 active plasmid clones were recovered in E. coli. As anticipated, most of the recovered clones were cdc18+. However, two active clones, H40 and H49, did not hybridize with any of the suppressor genes. Restriction mapping and hybridization analysis indicated that both clones contained a common 2.8-kb HindIII fragment with suppressor activity (Figure 1). Subcloning and suppression analysis revealed that the 1.8-kb HindIII–SacI fragment had activity. The gene in this fragment was named pas1+ (Pcl-like cyclin for activating start; see below) and characterized further. This initially isolated pas1+ gene was truncated at the promoter region; therefore, a genomic fragment spanning the entire pas1+ gene was isolated by colony hybridization (Figure 1A).
Figure 1.
Isolation of the pas1+ gene. (A) A restriction map of the pas1+ gene. The white and black arrows indicate the direction and extent of the pas1+ ORF and the pas1+ cDNA, respectively. The ability of the subclones to suppress the Δres1 mutant is shown as + or − in the right column. The structure of the 7-kb SphI fragment used for the disruption of the pas1+ gene is shown at the bottom. (B) Suppression of Δres1 and Δrep2 mutants by overexpression of the pas1+ gene. The Δres1 (K156-D1) and Δrep2 (N3-141S) mutants were transformed with the indicated plasmids, spread on MMA plates, and incubated at the indicated temperatures. pAL-pas1 has an insert of the initially isolated 2.8-kb HindIII fragment. pAL-X is the pALSK+ vector with no insert and is used as a negative control.
As noted above, the rep2 null mutant (Δrep2) cells are partially compromised in cell cycle start ability because inactive Res2p-Cdc10p complexes compete with active Res1p-Cdc10p for binding to MCB, thereby inhibiting the activation of MCB (Nakashima et al., 1995). The growth defect of the Δrep2 cells becomes evident at low temperatures, and at 18°C the mutant arrests in G1. This growth defect was also rescued by the expression of pas1+ (Figure 1B, lower panels).
The pas1+ Gene Encodes a Protein Homologous with Pho85-associated Cyclins
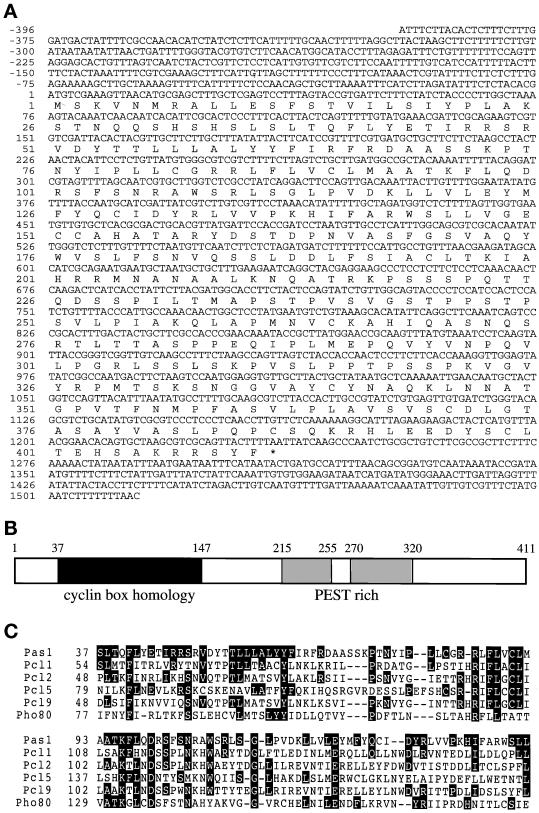
To elucidate the structure of pas1+-encoded protein, a cDNA clone spanning the entire coding region was isolated and sequenced. Its nucleotide sequence and the deduced amino acid sequence of the putative Pas1 protein are shown in Figure 2A. The putative Pas1 protein is composed of 411 amino acids with a calculated molecular mass of 45 kDa. An amino acid homology search revealed that Pas1p shares a limited but significant homology with the Pho85 kinase–associated cyclins (Pcls) of S. cerevisiae in the cyclin box (Figure 2C). Amino acid identity between Pas1p and Pcl1p in this region is 28%. In addition, Pas1p contains two of the typical PEST-rich sequences that are present in many G1 cyclins and considered to serve as a signal for proteolytic degradation (Figure 2B). The pas1+ mRNA was constitutively expressed during cell cycling and remained unchanged in the cells arrested at the execution points of cdc10+, res1+, and rep2+ (our unpublished results), indicating that pas1+ is not a target of transcriptional regulation by the Res-Cdc10p-Rep complexes.
Figure 2.
Primary structure of the Pas1 protein. (A) Nucleotide and predicted amino acid sequences of the pas1+ cDNA. The amino acid sequence is shown below the DNA sequence in a single-letter code. Nucleotide numbering starts at the initiating ATG of the predicted pas1+ translation product. (B) Scheme of the Pas1 protein. The cyclin box homologous region is shown in black, and two PEST-rich domains are shown in gray. (C) Amino acid homology in the cyclin box and its upstream region among Pas1p, Pcl1p (Hcs26p) (Ogas et al., 1991), Pcl2p (OrfDp) (Frohlich et al., 1991), Pcl5p, Pcl9p (Measday et al., 1997), and Pho80p (Toh-e and Shimauchi, 1986). Amino acids in which Pas1p is identical with the other five cyclins are shown in black.
pas1+ Is Required for Full cdc18+ mRNA Induction
To investigate the mechanism of action and the physiological role of pas1+, we constructed cells lacking the pas1+ gene by one-step gene replacement with the ura4+ gene (Figure 1A). After transfection, diploid cells deleted for one pas1+ allele were identified by Southern blot analysis and induced for meiosis to obtain haploid disruptant spores. Haploid cells lacking pas1+ (Δpas1) that germinated from the spores were viable and propagated well. To eliminate possible second mutations, they were back-crossed with the wild-type strain several times before extensive analysis. In proliferative abilities, Δpas1 cells were similar to wild-type cells under all of the nutritional conditions tested and at all temperatures between 18 and 36°C. The only noticeable difference was that the disruptant tended to arrest at a slightly lower cell density when grown to confluence.
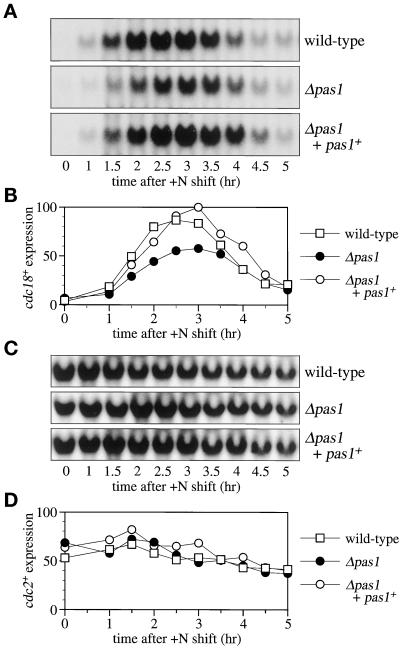
The aforementioned ability of pas1+ to suppress the G1 arrest phenotypes of Δres1 and Δrep2 mutants suggests that Pas1p activates either Res-Cdc10p-Rep complex(es) or factor(s) required for the G1-S transition but unrelated to the Res-Cdc10p-Rep transcriptional control system. First, to distinguish these two possibilities, we investigated whether deletion of pas1+ affected the level of the transcript of cdc18+, a major target gene regulated by Res-Cdc10p-Rep. The Δpas1 cells were synchronized to G1 by nitrogen starvation and then released to start the cell cycle in nitrogen-rich growth medium. The cells were harvested every 30 min, and the cdc18+ transcript was semiquantified by Northern hybridization. In the Δpas1 cells, the amount of the induced cdc18+ transcript was reduced to roughly 50% of the wild-type cell level. But it was restored to the wild-type cell level by chromosomal integration of a single copy of the pas1+ gene (Figure 3, A and B). In contrast, the transcript of the cdc2+ gene, which was not regulated by Res-Cdc10p-Rep and therefore was used as a negative control, was unchanged by the presence or absence of pas1+ (Figure 3, C and D). In addition, overexpression of pas1+ in the Δres1 cells suppressed not only arrest in G1 but also the reduction of the cdc18+ mRNA level at the restrictive temperature (our unpublished results). These results indicate that Pas1 cyclin activates the MCB-dependent transcription that is executed by Res-Cdc10-Rep.
Figure 3.
pas1+ is required for full cdc18+ mRNA induction. (A) The cdc18+ transcript is not fully induced during the G1-S transition in the Δpas1 mutant. Wild-type (EV3A), Δpas1 (K182-A7), and Δpas1+ pas1+(Δpas1 complemented by the pas1+ gene integration) (K182-A7-P1) cells were grown to midlog phase at 30°C in MM (+N/2%G) and then nitrogen-starved in MM (−N/2%G) for 48 h to be arrested in G1. The cells were then released by transfer into MM (+N/2%G) at 30°C. Cell aliquots were taken every 30 min, and cdc18+ expression was examined by Northern hybridization. (B) The cdc18+ mRNA levels shown in A were quantified with BAS2000 (Fuji Film, Tokyo, Japan). (C) cdc2+ transcription is not affected in the Δpas1 mutant. cdc2+ expression was examined by Northern hybridization with the use of the same filters used in A. (D) The cdc2+ mRNA levels shown in C were quantified with BAS2000 (Fuji Film).
The pas1+ Gene Promotes the Cell Cycle Start by Activating Res2p-Cdc10p
To identify the target(s) for the action of Pas1p, we carried out a series of genetic analyses. As mentioned above, Δpas1 cells grow at 30°C with no detectable defect. Δres1 cells also grow at this temperature, albeit poorly (Tanaka et al. 1992), owing to the presence of the Res2p-Cdc10p-Rep2p complex (Miyamoto et al., 1994; Nakashima et al., 1995). However, cells doubly deleted for res1+ and pas1+ were synthetically lethal at this temperature. Tetrad dissection of spores from >200 asci of the Δres1/res1+ Δpas1/pas1+ diploid cells yielded no viable haploid double mutant cells, which germinated but mostly arrested after one division with marked cell elongation (Figure 4A). The lethality of the double mutants was not caused by decreased expression of res2+, cdc10+, or rep2+ in the Δpas1 cells because the mRNA levels of the res2+, cdc10+, and rep2+ genes were unchanged regardless of the presence or absence of pas1+. This result indicates that in the absence of Pas1p the activity of the Res2p-Cdc10p-Rep2p transcriptional factor complex is not great enough to sustain the growth of the cells lacking Res1p. Unlike Pas1p, other G1 cyclins had no detectable genetic interaction with Res-Cdc10p-Rep. In proliferative ability, cells doubly deleted for res1+ and cig1+, cig2+, or puc1+ were indistinguishable from Δres1 single mutant cells; furthermore, when overexpressed, none of these cyclins could suppress the cold-sensitivity of Δres1 cells.
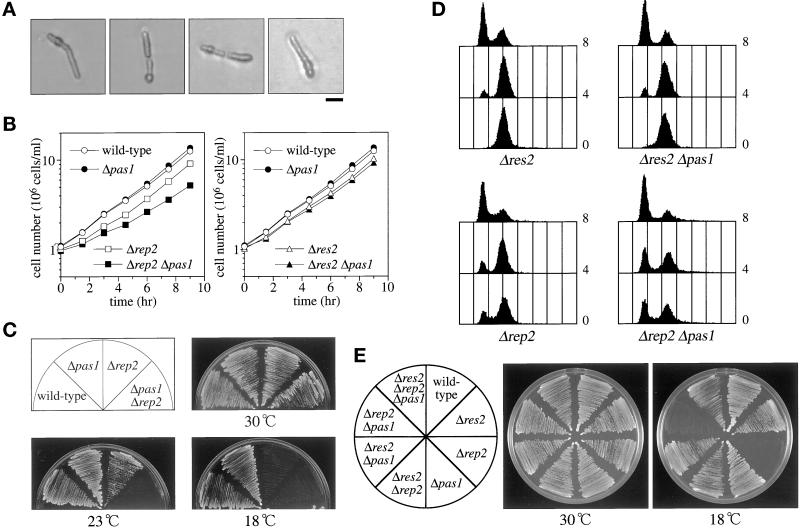
Figure 4.
Genetic interactions of pas1+ with res1+, res2+, and rep2+. (A) Terminal phenotype of Δres1 Δpas1 cells. Δres1/res1+ Δpas1/pas1+ diploid cells were tetrad dissected on YEA plates and incubated at 30°C. Cells that germinated from Δres1 Δpas1 double deletion spores (judged by genotypes of the other segregants) were photographed. Bar, 10 μm. (B) Deletion of pas1+ significantly reduces the growth rate of the Δrep2 mutant but not of the Δres2 mutant. The wild-type (L972), Δpas1 (K193-A1), Δrep2 (K166-A5), Δpas1 Δrep2 (K190-22B), Δres2 (K165-A12), and Δpas1 Δres2 (K189-3A) cells were cultured in MM (+N/2%G) at 30°C, and their growth was monitored by counting cell numbers. (C) Deletion of pas1+ greatly enhances the cold-sensitivity of the Δrep2 mutant. Cells were inoculated on MMA plates and incubated at the indicated temperatures. (D) Deletion of pas1+ enhances the slow G1 progression of the Δrep2 mutant but not that of the Δres2 mutant. Cells were grown to midlog phase at 30°C in MM (+N/2%G) and shifted to 18°C. Cells were sampled at 4 and 8 h after the temperature shift and analyzed by flow cytometry. (E) Deletion of the res2+ gene completely suppressed the cold-sensitivity of the Δpas1 Δrep2 double mutant. The wild-type (EV3A), Δres2 (M222), Δrep2 (N3-141S), Δpas1 (K182-A7), Δres2 Δrep2 (NP2-461), Δres2 Δpas1 (K189-23C), Δrep2 Δpas1 (K190-5D), and Δres2 Δrep2 Δpas1 (K539-1A) cells were inoculated on YEA plates and incubated at the indicated temperatures.
The suppression of the cold-sensitivity of Δrep2 cells by pas1+ (Figure 1B) indicates that Pas1p promotes the cell cycle start despite the absence of the Rep2 transcriptional activator subunit. This was confirmed by the synthetic effects displayed in the double mutants. Δrep2 cells are cold-sensitive but grow as rapidly as wild-type cells at 30°C. Deletion of the pas1+ gene, however, reduced the growth rate at 30°C (Figure 4B, left graph) and markedly enhanced cold-sensitivity (Figure 4, C and D). Whereas Δrep2 single mutant cells grow at temperatures as low as 18°C (Nakashima et al., 1995), cells doubly deleted for rep2+ and pas1+ were unable to grow even at 23°C (Figure 4C). All of these results imply that Pas1p activates either 1) only Res2p-Cdc10p independently of Rep2p or 2) both Res2p-Cdc10p and Res1p-Cdc10p.
To distinguish these possibilities, similar analysis was performed with Δres2 cells, in which only Res1p-Cdc10p complexes are active. Δres2 cells grow at temperatures for regular culture but slow in G1 progression or partially arrest at 18°C (Zhu et al., 1994). Deletion of pas1+ neither reduced the growth rate (Figure 4B, right graph) nor enhanced the cold-sensitivity (Figure 4E) of Δres2 cells. Confirming this, when shifted from 30 to 18°C, Δres2 single and Δres2 Δpas1 double mutant cells arrested in G1 with the same rate and extent, as shown by the flow cytometric patterns in Figure 4D. Thus, there was no detectable functional interaction between Pas1p and Res1p-Cdc10p. Together, these results led us to conclude that Pas1p promotes the cell cycle start via specifically activating the Res2p-Cdc10p complex without the Rep2 trans-activator subunit. The complete suppression of the cold-sensitivity of Δpas1 Δrep2 cells by deletion of res2+ (Figure 4E) is fully consistent with this conclusion.
The Rep2p-independent activation of Res2p-Cdc10p by Pas1p does not necessarily mean that Pas1p directly activates Res2p-Cdc10p without any trans-activator subunit. S. pombe contains Rep1p as a meiotic counterpart of Rep2p, which is highly induced during conjugation but still slightly expressed under nitrogen-starved conditions without mating partners (Sugiyama et al., 1994). Therefore, the possibility exists that slightly expressed Rep1p might be involved in the Rep2p-independent Res2p-Cdc10p activation by Pas1p. To examine this possibility, we compared the ability of pas1+ to rescue Δrep2 cells at low temperatures in the presence and absence of the rep1+ gene. If Pas1p requires Rep1p for the rescue of Δrep2 cells, cells doubly deleted for rep2+ and rep1+ would not be rescued by pas1+. The result shows that Pas1p rescued Δrep1 Δrep2 cells (NPP-12D) with an efficiency of 15.0%, which is comparable to the 17.8% efficiency for Δrep2 cells (N3-141S) (for suppression efficiencies, see MATERIALS AND METHODS). These results strongly suggest that Pas1p activates Res2p-Cdc10p independently of the known trans-activator subunits for this MCB-binding complex.
Cells Lacking pas1+ Are Proficient in Conjugation
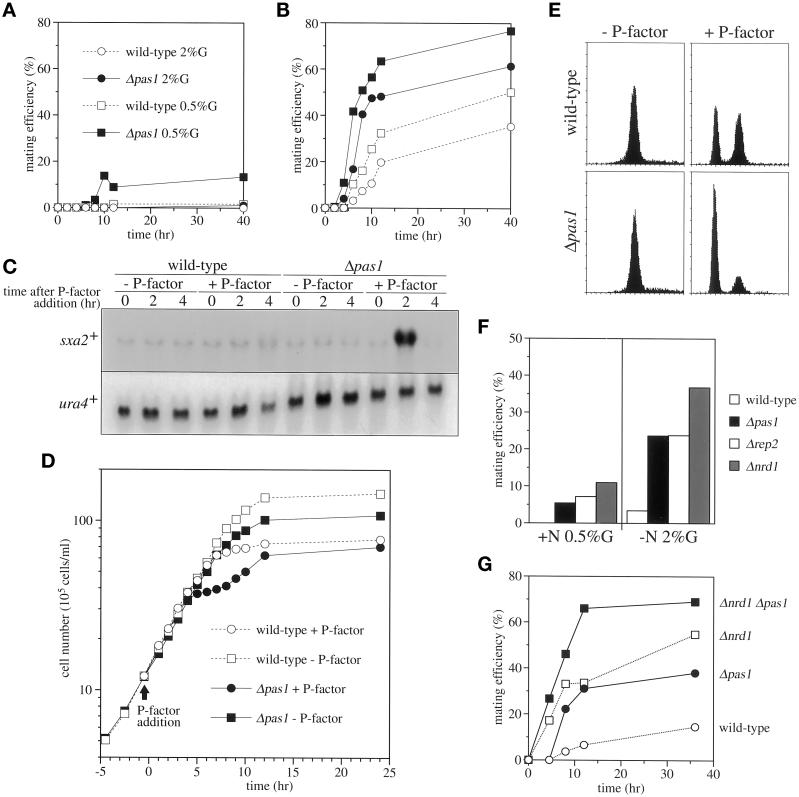
We previously found that Cig2 cyclin negatively regulated conjugation in addition to its role in the cell cycle start (Connolly and Beach, 1994; Obara-Ishihara and Okayama, 1994). To investigate the possibility that Pas1p might have a similar function, we examined the mating efficiencies of homothallic Δpas1 cells under various culture conditions. Both wild-type and Δpas1 cells did not conjugate in growth medium containing 2% glucose and 0.5% ammonium chloride as the sole carbon and nitrogen sources even at the stationary phase. However, when the glucose concentration was decreased to 0.5%, unlike wild-type cells, Δpas1 cells conjugated and performed meiosis with 15% efficiency (Figure 5A). Nitrogen starvation is not sufficient to induce efficient conjugation of wild-type S. pombe cells because the start of sexual differentiation is partially inhibited under high-glucose conditions. However, unlike wild-type cells, Δpas1 cells conjugated very efficiently in nitrogen-free glucose-rich medium (Figure 5B). Thus, cells lacking pas1+ were markedly enhanced in commitment to conjugation.
Figure 5.
Pas1 cyclin negatively regulates sexual differentiation. (A) Mating efficiencies of homothallic wild-type (L968) (open symbols) and Δpas1 mutant (K207-A1) (closed symbols) cells in nitrogen-rich MM. Cells were grown to log phase in MM (+N/2%G) and transferred to MM (+N/2%G) (circles) or MM (+N/0.5%G) (squares), and mating frequencies were assayed. (B) Mating efficiencies of homothallic wild-type (open symbols) and Δpas1 mutant (closed symbols) cells in MM without nitrogen. Cells were grown to log phase in MM (+N/2%G) and transferred to MM (−N/2%G) (circles) or MM (−N/0.5%G) (squares), and mating frequencies were assayed. (C) sxa2+ transcription is induced without nitrogen starvation in Δpas1 cells by P-factor. h− wild-type (L972) and h− Δpas1 (K193-A1) cells were inoculated into MM (+N/0.5%G) and cultured in the presence (+) or absence (−) of 2 μg/ml P-factor. Cells were harvested at the indicated times, and sxa2+ expression was examined by Northern hybridization. (D) Δpas1 cells are hypersensitive to mating pheromone and arrest the cell cycle without accompanying nutrient starvation. h− Δsxa2 (K205-A1) (referred to as wild-type; open symbols) and h− Δsxa2 Δpas1 (K209-22A) (referred to as Δpas1; closed symbols) cells were inoculated into MM (+N/0.1%G), divided into two parts, incubated at 30°C for 4.5 h, then cultured in the presence (+; circles) or absence (−; squares) of 2 μg/ml P-factor. Cell growth was monitored by counting cell numbers. (E) Δpas1 cells arrest in G1 phase upon exposure to P-factor. The same cell cultures used in D were harvested at 4 h after P-factor addition and analyzed by flow cytometry. (F) Δrep2 cells mate as efficiently as Δpas1 cells. Homothallic wild-type (L968), Δpas1 (K207-A1), Δrep2 (K166-A1), and Δnrd1 (14-1) cells were grown to log phase in MM (+N/2%G), transferred into MM (+N/0.5%G) or MM (−N/2%G), and cultured at 30°C. Mating efficiencies were determined by examining mated cells after 24 h of incubation. (G) Deletion of pas1+is not epistatic to deletion of nrd1+. Homothallic wild-type (L968), Δpas1 (K207-A1), Δnrd1 (14-1), and Δnrd1 Δpas1 (K228-29A) cells were grown to log phase in MM (+N/2%G) and transferred into MM (+N/0.5%G). Mating frequencies were determined as described above.
Cells lacking cig2+ are hyperfertile partly because they are facilitated to arrest in G1 under the nitrogen-starved condition (Obara-Ishihara and Okayama, 1994). In contrast, Δpas1 cells showed no detectable facilitation for G1 arrest in response to nitrogen or carbon starvation. Therefore, we investigated the sensitivity of the mutant to mating pheromones because the mating pheromone–mediated cell–cell communication is essential to elicit conjugation (for review, see Nielsen and Davey, 1995). P-factor, the mating pheromone secreted by h+ cells, is degraded by the serine carboxypeptidase encoded by the sxa2+ gene (Imai and Yamamoto, 1992). Interestingly, sxa2+ is activated upon exposure to P-factor via P-factor signaling to down-regulate the amount of P-factor, forming a negative feedback loop. However, the presence of P-factor is not sufficient for sxa2+ induction, and concurrent nutritional starvation is also needed because the P-factor signaling system becomes effective only when nutrient is exhausted. Accordingly, in wild-type h− cells cultured in nitrogen-rich medium containing 0.5% glucose and 2 μg/ml P-factor, no sxa2+ mRNA induction was observed (Figure 5C). In contrast, in the h− Δpas1 cells cultured in the same medium, sxa2+ mRNA was induced at 2 h transiently, showing that in these cells, the mating pheromone signal pathway was readily activated by P-factor without nutrient starvation.
Mating pheromones induce G1 arrest of the partner cells (Davey and Nielsen, 1994; Imai and Yamamoto, 1994) at least partly by inhibiting Cdc2p-Cdc13p/Cig2p (Stern and Nurse, 1997) via activation of the mating pheromone signal pathway. Therefore, given that the absence of Pas1p dispenses the requirement for nutrient starvation in P-factor–invoked activation of the mating pheromone signal pathway, it was predicted that P-factor might induce G1 arrest to Pas1p-lacking cells without nutrient starvation. To test this prediction, both h− wild-type cells and h− Δpas1 cells were cultured in P-factor–containing growth medium and their growth rates were compared. In this experiment, nitrogen-rich but low-glucose (0.1%) medium and strains with the Δsxa2 background were used to enhance the efficacy of P-factor. The addition of P-factor did not cause either growth delay (Figure 5D) or morphological changes to rapidly growing wild-type cells, although their progression through G1 phase was delayed, as indicated by the appearance of a G1 peak in flow cytometry (Figure 5E). On the other hand, upon P-factor exposure, Δpas1 cells transiently arrested even at midlog phase. Growth inhibition became apparent 4 h after P-factor addition and continued for ∼4 h (Figure 5D). During growth arrest, the cells extended conjugation tubes and increased their volume. Flow cytometry revealed that the majority of the cells 4 h after P-factor addition were in G1 (Figure 5E). After this transient growth arrest, cells began to repropagate. These results indicate that Pas1p plays a role in nutrient-controlled repression of the mating pheromone signaling pathway in addition to the activation of Res2p-Cdc10p.
The nutrient-controlled repression of the mating pheromone signaling pathway by Pas1 cyclin, however, could be an indirect effect of the activation of Res2p-Cdc10p because overexpression of res1+ inhibits conjugation (Tanaka et al., 1992) and cells lacking the res1+ gene have enhanced mating (Caliguiri and Beach, 1993; K.T. unpublished observation). To test this possibility, we compared the mating efficiencies of Δpas1 and Δrep2 cells. If Pas1p's effect on the repression of mating signaling is an indirect effect of the activation of Res2p-Cdc10p, the mating efficiency of Δrep2 cells would be much higher than that of Δpas1 cells because unlike Δpas1 cells, Δrep2 cells traverse G1 very slowly as a result of insufficient MCB activation and consequently are greatly facilitated to arrest in G1 (Nakashima et al.,1995). As shown in Figure 5F, Δrep2 cells were similar to Δpas1 cells in their ability to conjugate in nutrient-rich medium. This result strongly suggests that Pas1 cyclin controls mating pheromone signaling independently of the activation of Res2p-Cdc10p.
Nrd1p is an RNA-binding protein that blocks the commitment to conjugation until cells reach a critical level of nutrient starvation (Tsukahara et al., 1998). Cells lacking Nrd1p resemble those lacking Pas1p in their ability to conjugate without starvation (Figure 5, F and G). The phenotypic similarity led us to perform epistatic analysis of the two genes. Cells deleted for both pas1+ and nrd1+ were generated by crossing and compared with each single disruptant for mating proficiency. As shown in Figure 5G, the double disruptant had greater conjugation efficiency in MM (+N/0.5%G) than each single disruptant, suggesting that Pas1p controls mating pheromone signaling independently of Nrd1p.
Pas1p Is Not Involved in Phosphate Metabolism
The PHO80 gene of S. cerevisiae was identified as a negative regulator of the PHO5 acid phosphatase gene (Oshima, 1982). In the pho80 mutant, the PHO5 gene is constitutively expressed even in phosphate-rich medium. A similar regulation also takes place in fission yeast, and acid phosphatase activity is induced by phosphate starvation (Mitchison and Creanor, 1969; Dibenedetto, 1972). Because of the amino acid homology with Pho80p, it was not unreasonable to suspect that Pas1p might also be involved in phosphate metabolism. Therefore, we investigated the effect of the presence or absence of pas1+ on acid phosphatase activity that responded to phosphate availability. Upon growth in a high- or low-phosphate medium, acid phosphatase activity was repressed or induced to a similar extent in wild-type and Δpas1 cells (wild-type, 7.6-fold induction; Δpas1, 8.4-fold induction), showing that Pas1p is unlikely to participate in the regulation of phosphate metabolism. Thus, despite structural similarity, in its biological role Pas1 cyclin differs significantly from Pho80p of budding yeast.
Pas1 Cyclin Associates In Vivo with Cdc2 and Pef1 Kinases
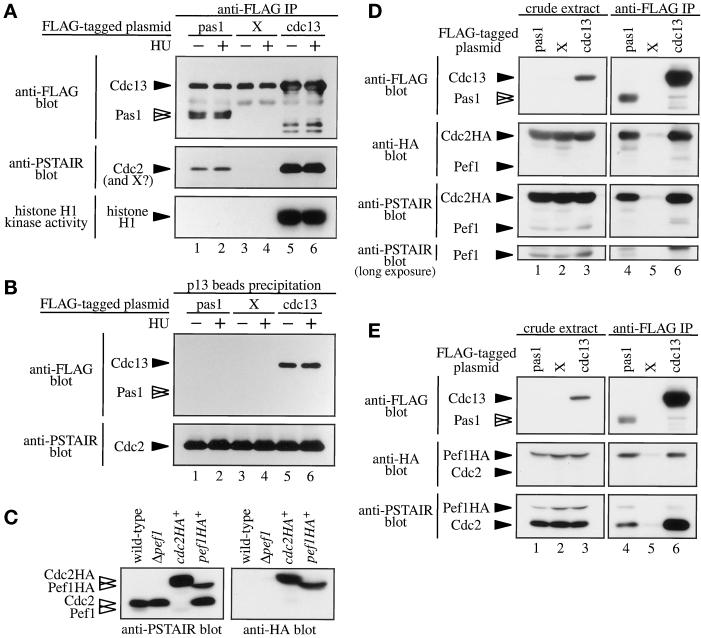
In S. cerevisiae, Pcl cyclins are associated with Pho85 kinase but not with Cdc28 kinase, the budding yeast counterpart of Cdc2p (Espinoza et al., 1994; Measday et al., 1994). To determine the associated protein kinase(s) and kinase activities, we expressed the FLAG-tagged Pas1p in S. pombe cells. The FLAG-tagged Cdc13 mitotic B-type cyclin was used as a positive control for association with Cdc2 kinase. When necessary, cells were arrested at the early S phase by culturing for 4 h in medium containing 12 mM hydroxyl urea before cell extraction. Res-Cdc10p-Rep is fully active at this arrest point (Baum et al., 1997). Cell lysates were then incubated with an anti-FLAG antibody or p13suc1 beads, and the precipitates were separated by SDS-PAGE followed by immunoblotting with anti-FLAG or anti-PSTAIR antibodies or assayed for histone H1 kinase activity. In the gel, FLAG-Cdc13p comigrated with IgG. As shown in Figure 6A, Pas1p was coprecipitated with the 34-kDa protein detectable with the anti-PSTAIR antibody. This PSTAIR protein was indistinguishable in size from the Cdc2 kinase associated with Cdc13p, although the amount was low. The amount and the mobility of the Pas1p-associated PSTAIR protein did not change between early S-phase cells and exponentially growing cells. This complex, however, phosphorylated histone H1 very poorly if at all compared with the Cdc2p-Cdc13p complex (Figure 6A). Myelin basic protein and casein were also poorly phosphorylated by the Pas1p-associated kinase.
Figure 6.
Pas1 cyclin associates in vivo with both Cdc2 and Pef1 kinases. (A) A Cdc2 kinase-like protein coprecipitates with Pas1 cyclin. Lysates were prepared from Δpas1 cells (K182-A7) expressing pREP1-FLAGpas1 (lanes 1 and 2), pREP1-X (lanes 3 and 4), or pREP1-FLAGcdc13 (lanes 5 and 6). Cells were cultured in MM (+N/2%G) in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 12 mM hydroxyl urea. Lysates were subjected to immunoprecipitation with the anti-FLAG M2 antibody. Immunoprecipitates were immunoblotted with the anti-FLAG M2 (top panel) or anti-PSTAIR (middle panel) antibodies, respectively, or assayed for histone H1 kinase activity (bottom panel). FLAG-Cdc13 protein comigrated with IgG (top panel; lanes 5 and 6). (B) The Pas1 cyclin-associated kinase does not bind Suc1p. The same lysates used in A were incubated with p13suc1 beads to pull down a Cdc2 kinase complex. Precipitates were immunoblotted with anti-FLAG M2 (upper panel) or anti-PSTAIR (lower panel) antibody. (C) Identification of Cdc2p and Pef1p molecules. Lysates were prepared from wild-type (EV3A), Δpef1 (K571-2D), cdc2HA+ (K230-A6), and pef1HA+ (K566-11) cells. The whole cell lysates were separated by SDS-PAGE and immunoblotted with anti-PSTAIR (left panel) or anti-HA (right panel) antibodies. (D and E) Pas1 cyclin associates in vivo with Cdc2 and Pef1 kinases. (D) Lysates were prepared from cdc2HA+ cells (K230-A6) expressing pREP1-FLAGpas1 (lanes 1 and 4), pREP1-X (lanes 2 and 5), or pREP1-FLAGcdc13 (lanes 3 and 6). The lysates were immunoprecipitated with the anti-FLAG M2 antibody. The whole cell lysate (left panels; lanes 1–3) and immunoprecipitates (right panels; lanes 4–6) were immunoblotted with the anti-FLAG D-8 (top panels), anti-HA (second panels), or anti-PSTAIR (third and bottom panels) antibody. In this experiment, an anti-FLAG D-8 rabbit polyclonal antibody was used to avoid an undesired reaction with mouse IgG (top panels). (E) Lysates were prepared from pef1HA+ cells (K566-11) expressing pREP1-FLAGpas1 (lanes 1 and 4), pREP1-X (lanes 2 and 5), or pREP1-FLAGcdc13 (lanes 3 and 6). The lysates were immunoprecipitated with the anti-FLAG M2 antibody. The whole cell lysate (left panels; lanes 1–3) and immunoprecipitates (right panels; lanes 4–6) were immunoblotted with the anti-FLAG D-8 (top panels), anti-HA (middle panels), or anti-PSTAIR (bottom panels) antibody. The FLAG-tagged proteins were detected by the anti-FLAG D-8 rabbit polyclonal antibody to avoid undesired reaction with mouse IgG (top panel).
Suc1p binds the Cdc2p that is associated with certain cyclins, including Cdc13p and Cig1p (Booher et al., 1989; Basi and Draetta, 1995). Consequently, Suc1p binding provides a convenient assay for characterizing the kinase complex. Proteins that bound p13suc1 beads were analyzed by immunoblotting with anti-FLAG and anti-PSTAIR antibodies. Unlike Cdc13p-associated Cdc2p, Pas1p-associated kinase did not bind to Suc1p, as indicated by the absence of FLAG-Pas1p in the p13Suc1-bound proteins (Figure 6B).
In the budding yeast, Pcl cyclins associate with Pho85 kinase but not with Cdc28 kinase. The S. pombe genome sequencing project recently identified an ORF (SPCC16C4.11) capable of encoding a protein highly homologous to Pho85 kinase of S. cerevisiae and PhoA kinase of Emericella nidulans (Figure 7). We named this putative gene pef1+ (pombe pho eighty-five), because as presented below Pef1 kinase encoded by this gene is a functional association partner of Pas1 cyclin. Both Cdc2p and Pef1p have a conserved PSTAIR motif, and their calculated molecular weights are similar (34,358 for Cdc2p and 32,736 for Pef1p). Because of this, it was difficult to identify the Pas1-associated PSTAIR kinase(s) without gene manipulation. Therefore, we constructed a Δpef1 mutant (see below for construction) and two epitope-tagged strains referred to as cdc2HA+ and pef1HA+. In the cdc2HA+ strain, the chromosomal cdc2+ was replaced with a cdc2+ gene having three copies of the influenza virus HA protein epitope at the C terminus. Likewise, the chromosomal pef1+ was replaced with a pef1+ gene having three copies of the HA protein in the pef1HA+ strain. With the use of these strains, Cdc2p and Pef1p could be identified by immunoblotting with anti-PSTAIR and anti-HA antibodies (Figure 6C). Anti-PSTAIR immunoblotting of wild-type cell extracts revealed two bands, a dense 34-kDa band and a faint 33-kDa band, the latter of which was absent in the Δpef1 cells. In addition, the original 34-kDa band shifted to 38 kDa (size increase caused by the HA tag) in the cdc2HA+ strain, and the original 33-kDa band shifted to 37 kDa in the pef1HA+ strain, with concomitant staining with the anti-HA antibody. Unexpectedly, HA tagging markedly increased the amount of the 33-kDa protein, perhaps as a result of stabilization of the protein, although its mechanism was unknown. Nonetheless, these results led us to assign the 34-kDa band to Cdc2p and the 33-kDa band to Pef1p. Given this information, we investigated the Pas1p-associated PSTAIR protein(s) in detail. We used epitope-tagged strains as the hosts for the same analysis mentioned above because the original Cdc2p and Pef1p migrated very closely in SDS-polyacrylamide gels (Figure 6C, wild-type lane). In the experiment with the cdc2HA+ strain, the major Pas1p-associated PSTAIR protein not only was recognized by the anti-HA antibody but also shifted to 38 kDa (Figure 6D, lane 4), verifying this Pas1p-associated protein to be Cdc2p. In addition, a tiny amount of a PSTAIR protein corresponding in size to Pef1 kinase also coprecipitated with Pas1p but not with Cdc13p (Figure 6D, bottom panels). To confirm this minor PSTAIR protein to be Pef1p, we performed the same analysis with the pef1HA+ strain (Figure 6E). In this analysis, the minor PSTAIR protein shifted to above Cdc2p with concomitant staining with the anti-HA antibody (Figure 6E, lane 4), demonstrating it to be Pef1p. Pef1p seemed to form a complex with Pas1p at a higher affinity than with Cdc13p. The amount of the Cdc13p-coprecipatated Pef1p was similar to that of Pas1p-coprecipitated Pef1p despite the fact that Cdc13p was 5- to 10-fold more abundant than Pas1p in these cells. On the other hand, Cdc2p seemed to form a complex with Pas1p and Cdc13p at a similar affinity, because the amounts of Pas1p-bound Cdc2p and Cdc13p-bound Cdc2p normalized for the amounts of the Pas1p and Cdc13p contained in the cell extracts appeared to be similar (Figure 6E).
Figure 7.
Structure of the Pef1 protein. (A) A restriction map of the pef1+ gene. The white arrow indicates the direction and extent of the pef1+ ORF. The structure of the PstI–SpeI fragment used for the disruption of the pef1+ gene is also shown. (B) Amino acid homology among S. pombe Pef1p (Pef1 Sp), E. nidulans PhoAp (M47) (PhoA En) (Bussink and Osmani, 1998), S. cerevisiae Pho85p (Pho85 Sc) (Uesono et al., 1987), and S. pombe Cdc2p (Cdc2 Sp) (Hindley and Phear, 1984). Amino acids in which Pef1p is identical to the other three kinases are shown in black. ###### indicates the conserved PSTAIR motif.
Pef1 Kinase Is Required for the Activation of Res-Cdc10p-Rep
To determine which of Pef1p or Cdc2p is a functional partner for Pas1 cyclin to activate Res2p-Cdc10p, we analyzed the properties of the Δpef1 cells. The pef1+ gene was isolated by colony hybridization, and cells lacking pef1+ were constructed by one-step gene replacement with the ura4+ gene (Figure 7A). Haploid Δpef1 cells that germinated from the spores of correctly gene-disrupted diploid cells were viable and propagated at all temperatures between 18 and 36°C, although their propagation was slower than that of wild-type cells. However, just like Δpas1 Δres1 cells, Δpef1 Δres1 cells were synthetically lethal. Tetrad dissection of >100 asci generated by crossing Δpef1 cells with Δres1 cells led to the production of no viable double mutant; instead, mutants germinated but mostly arrested as elongated cells. Moreover, the Δpef1 mutant was also found synthetically lethal with the Δrep2 mutation. In contrast, as expected, Δpef1 Δres2 cells were viable. Thus, in these genetic interactions, pef1+ behaved very similarly to pas1+, indicating that Pef1 kinase is a functional association partner for Pas1 cyclin that activates Res2p-Cdc10p. Detailed characterization of the pef1+ gene will be described elsewhere.
Pas1 Cyclin Genetically Interacts with Other Cyclin Genes Promoting the Cell Cycle Start
The last question we addressed concerns the functional relationship between Pas1p and other G1 cyclins. Fission yeast contains three types of G1 cyclins, Cig1/2p, Puc1p, and Pas1p, which are thought to have activity to regulate the G1-S transition. Either Cig1p or Cig2p is essential for the start of S phase in the absence of Cdc13 mitotic cyclin (Fisher and Nurse, 1996; Mondesert et al., 1996), and these cyclins act after the activation of Res-Cdc10p-Rep (Baum et al., 1997). Puc1p also acts in the G1-S transition, but this activity is detectable only in the Δcig1 Δcig2 background (Martin-Castellanos et al., 2000) (Figure 8C). However, this cyclin does not show any detectable interactions with Res-Cdc10p-Rep, despite its structural similarity to Cln cyclins of budding yeast.
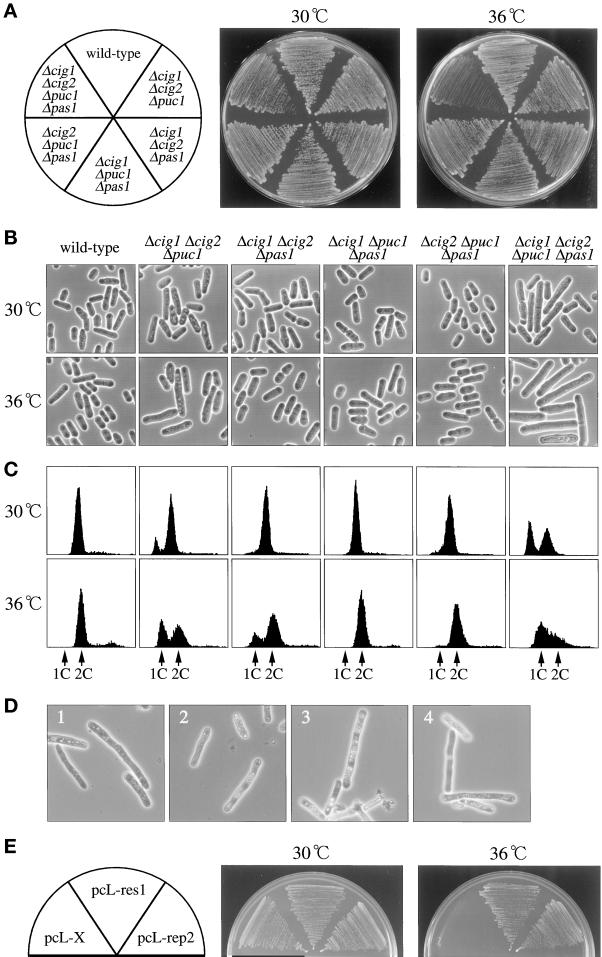
Figure 8.
Genetic interaction of pas1+ with other G1 cyclin genes. (A) Temperature-sensitive growth phenotype of the Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant. The wild-type (L972), triple null mutants (K224-4D, K224-51C, K224-59C, K224-35D), and quadruple null mutant (K224-42D) cells were inoculated on MMA plates and incubated at the indicated temperatures for 3 d. (B) Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant cells arrest with a cdc phenotype at 36°C. Cells were inoculated on MMA plates and incubated at the indicated temperatures for 18 h. (C) Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant cells arrest in G1 at 36°C. Cells were cultured in MM (+N/2%G) to midlog phase at 30°C (upper panels), shifted to 36°C, incubated for 3 h (lower panels), and analyzed by flow cytometry. (D) Terminal cell morphology of Δcig1 Δcig2 Δpuc1 Δrep2 quadruple mutant cells. Spores generated by crossing Δcig1 Δcig2 Δpuc1 cells with the Δrep2 haploid strain were tetrad dissected on YEA plates and incubated at 30°C. Cells that germinated from four independent Δcig1 Δcig2 Δpuc1 Δrep2 quadruple deletion spores (panels 1–4) (judged by genotypes of the other three segregants) were photographed under the microscope. (E) Suppression of the Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant by res1+ and rep2+ genes. The quadruple null mutant cells (K226-42A) were transformed with the indicated plasmids, streaked on MMA plates, and incubated at the indicated temperatures. pcL-X is the pcL vector with no insert and is used as a negative control.
Given these facts, we investigated the genetic interactions of pas1+ with other G1 cyclin genes by constructing triple and quadruple deletion mutants lacking cig1+, cig2+, puc1+, and/or pas1+. At 30°C, the Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant cells were viable (Figure 8A) and proliferated with slight cell elongation (Figure 8B) accompanied by a clear G1 peak, indicating slow G1 progression (Figure 8C). The mutant, however, could not proliferate at 36°C (Figure 8A). Upon a shift to this temperature, the mutant came to arrest in G1 with cell elongation, typical of a cdc phenotype (Figure 8, B and C). Thus, the four cyclins Cig1p, Cig2p, Puc1p, and Pas1p have a certain degree of functional redundancy, although the primary targets for the action of these cyclins differ significantly.
The genetic interaction of pas1+ with other cyclins is likely to be an indirect effect of the activation of Res2p-Cdc10p, because spores lacking cig1+, cig2+, puc1+, and rep2+ in place of pas1+ germinated and propagated a few times but ceased to proliferate at 30°C with cell elongation (Figure 8D). In addition, the temperature-sensitive proliferation of the Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant was efficiently suppressed by overexpression of res1+ or rep2+ (Figure 8E). These results indicate that Pas1 cyclin genetically interacts with other G1 cyclins via activation of the Rep2p-Cdc10p complex.
DISCUSSION
In S. pombe, the cell cycle start is controlled by two functionally redundant transcriptional activator complexes, Res1p-Cdc10p and Res2p-Cdc10p-Rep2p, the former of which functions predominantly for the start of the mitotic cycle (for review, see Woollard and Nurse, 1995; Okayama et al., 1996). Despite playing a relatively minor role in the onset of DNA synthesis, the Res2p-Cdc10p complex has an important regulatory role in the mitotic cell cycle. It acts as both activator and inhibitor of MCB-relying transcription, depending on the availability of at least the Rep2 trans-activator subunit. Lack of Rep2p prevents cells from growing at low temperatures and greatly facilitates starvation-induced G1 arrest (Nakashima et al., 1995). In contrast, lack of Res2p completely abrogates the periodicity of the MCB-dependent transcription, with a peak at G1-early S and a nadir at G2, resulting in constitutive expression (Baum et al., 1997). However, there may be a Rep2p-unrelated regulator for Res2p-Cdc10p activity because deletion of the C-terminal 41 amino acids from the Res2 protein completely abolishes the requirement for Rep2p in Res2p-Cdc10p activity (Sturm and Okayama, 1996).
Pas1 cyclin, described here, is likely to be one such regulator. All of the genetic and biochemical evidence obtained supports this claim. First, deletion of pas1+ reduced the expression of cdc18+ mRNA. Second, double deletion of pas1+ and res1+ led to synthetic lethality, whereas overexpression of pas1+ suppressed the cold-sensitive phenotype of the Δres1 mutant. Third, deletion of pas1+ greatly enhanced the cold-sensitivity of the Δrep2 mutant, whereas overexpression of pas1+ suppressed its cold-sensitive phenotype. Fourth, Δres2 Δpas1 cells are indistinguishable from Δres2 cells in doubling time during exponential growth and in the rate of temporal G1 arrest induced by exposure to a low temperature. These results indicate that pas1+ specifically activates Res2p-Cdc10p independently of Rep2p.
Fission yeast contains at least seven cyclins, Cdc13p, Cig1p, Cig2p, Puc1p, Pch1p, Mcs2p, and the Pas1p reported here. Cdc13p, Cig1p, and Cig2p are B-type cyclins that associate with Cdc2 kinase (for review, see Fisher and Nurse, 1995). The activity of Cdc13p- and Cig1p-associated Cdc2 kinase peaks during mitosis, whereas the activity of Cig2p-associated Cdc2 kinase peaks in G1-S phase (Booher et al., 1989; Moreno et al., 1989; Basi and Draetta, 1995; Martin-Castellanos et al., 1996; Mondesert et al., 1996). Only Cdc13p is essential for the G2-M transition, and neither Cig1p nor Cig2p substitute it. Cdc2p-Cig1p activity seems to be required for the inactivation of Rum1p, a Cdk inhibitor, which inhibits the activities of the Cdc2p-Cdc13p and Cdc2p-Cig2p complexes (Benito et al., 1998). A Cln-related cyclin, Puc1p, associates with Cdc2p (Forsburg and Nurse, 1994) and inactivates Rum1p at the end of G1 phase in the mitotic cell cycle (Martin-Castellanos et al., 2000). Pch1p is an essential fission yeast homologue of C-type cyclin and associates with Cdc2p, but its physiological role is unclear (Furnari et al., 1997). Mcs2p is a fission yeast counterpart of mammalian cyclin H and associates with Mop1 (also called Crk1) kinase, which acts as a Cdk-activating kinase (Buck et al., 1995; Damagnez et al., 1995). Of these cyclins, Cig2p is thought to play an important role in the cell cycle start, but this function is not specific to Cig2p and is shared by the other B-type cyclins. In fact, deletion of both cig1+ and cig2+ completely blocks the rereplication induced by the elimination of Cdc13p but imparts no visible effect on the cell containing Cdc13p (Fisher and Nurse, 1996; Mondesert et al., 1996). This situation appears to be similar to the requirement for one of the functionally redundant six Clb B-type cyclins in the onset of S phase in budding yeast (Schwob et al., 1994). Interestingly, the Δcig1 Δcig2 Δpuc1 Δpas1 quadruple mutant cells were also unable to start S phase at 36°C despite the presence of cdc13+.
Despite such functional redundancy, there is a clear distinction between Pas1p and other G1 cyclins. As mentioned above, Pas1p activates Res2p-Cdc10p, but Cdc13p, Cig1p, Cig2p, and Puc1p have no detectable ability to activate Res1p-Cdc10p or Res2p-Cdc10p-Rep2p. Overexpression or deletion of cig1+, cig2+, or puc1+ did not influence the proliferation properties of the Δres1 or Δrep2 mutant (K.T. unpublished observation). Furthermore, loss of pas1+ was mimicked by loss of rep2+ in the quadruple mutant (Figure 8D), and the four-cyclin quadruple mutant was rescued even by overexpression of res1+ or res2+ (Figure 8E). These results indicate that the seeming functional redundancy of Pas1p with other cyclins is an indirect effect of the activation of Res2p-Cdc10p. In this respect, Pas1p shares remarkable similarity with Cln3p of S. cerevisiae. Cln3 cyclin associated with Cdc28 kinase is considered to be a physiological activator of the Swi4p-Swi6p and Mbp1p-Swi6p transcriptional complexes (Tyers et al., 1993; Dirick et al., 1995; Stuart and Wittenberg, 1995; Levine et al., 1996). Moreover, like CLN3 (Wittenberg et al., 1990; Tyers et al., 1992), pas1+ is expressed throughout the cell cycle with little fluctuation in mRNA amount.
Despite the functional similarity of Pas1p and Cln3p and the fact that Pas1p can associate in vivo with Cdc2p as well as Pef1p (a fission yeast homologue of Pho85), our data indicate that the kinase partner for the cell cycle start action of Pas1 cyclin is Pef1p. Δpef1 cells were phenotypically similar to Δpas1 cells in genetic interactions with res1+, res2+, and rep2+. In contrast, cdc2+ has no indication for its involvement in activating these cell cycle start genes (Baum et al., 1997; Whitehall et al., 1999).
If the Pef1p-Pas1p complex activates Res2p-Cdc10p, then how does it activate Res2p-Cdc10p without Rep2p? The mechanism for this activation is unknown. In S. cerevisiae, the Pho85 kinase associated with Pho80 cyclin directly phosphorylates, and thereby regulates, Pho4p, a transcription factor required for the induction of the PHO5 acid phosphatase gene in response to phosphate starvation (for review, see Lenburg and O'Shea, 1996). Similarly, the Pef1p-Pas1p complex might directly phosphorylate Res2p. The phosphorylation of Res2p might induce its conformational changes in such a way that Res2p is allowed to interact with other unidentified coactivators or expose its intrinsic putative trans-activator domain overlapping with the N-terminal ankyrin repeats (Whitehall et al., 1999). Such possibilities may not be so remote, because deletion of the C-terminal 40 amino acids from Res2p totally negates the requirement for Rep2p in the activation of Res2p-Cdc10p (Sturm and Okayama, 1996).
Pas1 cyclin has an additional activity as a negative regulator of sexual differentiation. Cells deleted for pas1+ commit sexual development without critical nutrient starvation because of relaxed control of mating pheromone signaling (Figure 5). As shown in Figure 5F, this Pas1p activity is not an indirect effect of the stimulation of the cell cycle start by activating Res2p-Cdc10p. Consequently, Pas1p is likely to have the target that is involved in the control of mating pheromone signaling. This target, however, seems to be unrelated to any known mating-influencing factors, including the components of the cyclic AMP and stress MAPK signal pathways (for review, see Yamamoto, 1996) and Nrd1p, an RNA-binding protein that blocks commitment to mating by repressing Ste11p function until cells reach a critical level of starvation (Tsukahara et al., 1998). Unlike the cells deficient in these pathways, cells lacking Nrd1p phenotypically resembled cells lacking Pas1p. However, as shown in Figure 5G, we found no epistatic relation between nrd1+ and pas1+. The kinase partner for the Pas1p control of sexual differentiation is unknown. Detailed characterization of Δpef1 cells would help to solve this problem.
One important question that needs to be answered is how Pas1 cyclin is regulated. Because the expression of pas1+ is not cell cycle regulated, Pas1p activity must be regulated at a posttranscriptional level. In fact, Pas1p possesses two PEST-rich domains and is likely to be very unstable because it was difficult to detect in crude cell extract even when expressed from the strong nmt1+ promoter (Figure 6, D and E). Pas1p activity may be regulated by degradation by programmed proteolysis when cells traverse start or are starved for nutrient.
ACKNOWLEDGMENTS
We thank Masayuki Yamamoto and Yoshiyuki Imai for the sxa2+ gene and Δsxa2 mutant and Masakane Yamashita for anti-PSTAIR antibody. We also thank Chikashi Shimoda, Hiromi Maekawa, and Koei Okazaki for the pIU1 vector and S. pombe genomic libraries. This work was supported by grants from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Basi G, Draetta G. p13suc1 of Schizosaccharomyces pombe regulates two distinct forms of the mitotic cdc2 kinase. Mol Cell Biol. 1995;15:2028–2036. doi: 10.1128/mcb.15.4.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Nishitani H, Yanow S, Nurse P. Cdc18 transcription and proteolysis couple S phase to passage through mitosis. EMBO J. 1998;17:5689–5698. doi: 10.1093/emboj/17.19.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Wuarin J, Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4676–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Martin-Castellanos C, Moreno S. Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25rum1 CDK inhibitor. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach D. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Breeden L. Start-specific transcription in yeast. Curr Top Microbiol Immunol. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- Buck V, Russell P, Millar JBA. Identification of a cdk-activating kinase in fission yeast. EMBO J. 1995;14:6173–6183. doi: 10.1002/j.1460-2075.1995.tb00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussink HJ, Osmani SA. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 1998;17:3990–4003. doi: 10.1093/emboj/17.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliguiri M, Beach D. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell. 1993;72:607–619. doi: 10.1016/0092-8674(93)90079-6. [DOI] [PubMed] [Google Scholar]
- Connolly T, Beach D. Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol Cell Biol. 1994;14:768–776. doi: 10.1128/mcb.14.1.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Caligiuri M, Beach D. The Cdc2 protein kinase controls Cdc10/Sct1 complex formation. Mol Biol Cell. 1997;8:1105–1115. doi: 10.1091/mbc.8.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanor J, Elliott SG, Bisset YC, Mitchison JM. Absence of step changes in activity of certain enzymes during the cell cycle of budding and fission yeast. J Cell Sci. 1983;61:339–349. doi: 10.1242/jcs.61.1.339. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the START of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Damagnez V, Makela TP, Cottarel G. Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J. 1995;14:6164–6172. doi: 10.1002/j.1460-2075.1995.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J, Nielsen O. Mutations in cyr1 and pat1 reveal pheromone-induced G1 arrest in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1994;26:105–112. doi: 10.1007/BF00313796. [DOI] [PubMed] [Google Scholar]
- Dibenedetto G. Acid phosphatase in Saccharomyces pombe. I. Regulation and preliminary characterization. Biochim Biophys Acta. 1972;286:363–374. doi: 10.1016/0304-4165(72)90272-3. [DOI] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Espinoza FH, Ogas J, Herskowitz I, Morgan DO. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. Cyclins of the fission yeast Schizosaccharomyces pombe. Semin Cell Biol. 1995;6:73–78. doi: 10.1016/1043-4682(95)90003-9. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Nurse P. Identification of a G1-type cyclin puc1+ in the fission yeast Schizosaccharomyces pombe. Nature. 1991;351:245–248. doi: 10.1038/351245a0. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Nurse P. Analysis of the Schizosaccharomyces pombe cyclin puc1: evidence for a role in cell cycle exit. J Cell Sci. 1994;107:601–613. [PubMed] [Google Scholar]
- Frohlich KU, Fries HW, Rudiger M, Erdmann R, Botstein D, Mecke D. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari BA, Russell P, Leatherwood J. pch1+, a second essential C-type cyclin gene in Schizosaccharomyces pombe. J Biol Chem. 1997;272:12100–12106. doi: 10.1074/jbc.272.18.12100. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Hindley J, Phear GA. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe: patterns of splicing and homology to protein kinases. Gene. 1984;31:129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- Hofmann JFX, Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Yamamoto M. Schizosaccharomyces pombe sxa1+ and sxa2+ encode putative proteases involved in the mating response. Mol Cell Biol. 1992;12:1827–1834. doi: 10.1128/mcb.12.4.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Levine K, Huang K, Cross FR. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, McInerny CJ, Johnson AL, Fantes PA, Johnston LH. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco MA, de Prada JM, Moreno S. The puc1 cyclin regulates the G1 phase of the fission yeast cell cycle in response to cell size. Mol Biol Cell. 2000;11:543–554. doi: 10.1091/mbc.11.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM, Creanor J. Linear synthesis of sucrase and phosphatases during the cell cycle of Schizosaccharomyces pombe. J Cell Sci. 1969;5:373–391. doi: 10.1242/jcs.5.2.373. [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Tanaka K, Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1880. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Huang D, Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- Mondesert O, McGowan CH, Russell P. Cig2, a B-type cyclin, promotes the onset of S in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:181–191. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nakashima N, Tanaka K, Sturm S, Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O, Davey J. Pheromone communication in the fission yeast Schizosaccharomyces pombe. Semin Cell Biol. 1995;6:95–104. doi: 10.1016/1043-4682(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Phosphorylation of Sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol Biol Cell. 1998;9:2393–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara-Ishihara T, Okayama H. A B-type cyclin regulates conjugation via interacting with cell cycle ‘start’ genes in fission yeast. EMBO J. 1994;13:1863–1872. doi: 10.1002/j.1460-2075.1994.tb06455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Andrews BJ, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- Okayama H, Nagata A, Jinno S, Murakami H, Tanaka K, Nakashima N. Cell cycle control in fission yeast and mammals: identification of new regulatory mechanism. Adv Cancer Res. 1996;69:17–62. doi: 10.1016/s0065-230x(08)60859-3. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N, Okazaki K, Tanaka K, Okayama H. The ste4+ gene, essential for sexual differentiation of Schizosaccharomyces pombe, encodes a protein with a leucine zipper motif. Nucleic Acids Res. 1991;19:7043–7047. doi: 10.1093/nar/19.25.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y. Metabolism and gene expression. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring. Harbor, NY: Cold Spring. Harbor Laboratory; 1982. pp. 159–180. [Google Scholar]
- Reymond A, Marks J, Simanis V. The activity of S. pombe DSC-1-like factor is cell cycle regulated and dependent on the activity of p34cdc2. EMBO J. 1993;12:4325–4334. doi: 10.1002/j.1460-2075.1993.tb06117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Stern B, Nurse P. Fission yeast pheromone blocks S-phase by inhibiting the G1 cyclin B-p34cdc2 kinase. EMBO J. 1997;16:534–544. doi: 10.1093/emboj/16.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Sturm S, Okayama H. Domains determining the functional distinction of the fission yeast cell cycle “start” molecules Res1 and Res2. Mol Biol Cell. 1996;7:1967–1976. doi: 10.1091/mbc.7.12.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taba RM, Muroff I, Lydall D, Tebb G, Nasmyth K. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene products. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To-E A, Ueda Y, Kakimoto SI, Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A, Shimauchi T. Cloning and sequencing of the PHO80 gene and CEN15 of Saccharomyces cerevisiae. Yeast. 1986;2:129–139. doi: 10.1002/yea.320020209. [DOI] [PubMed] [Google Scholar]
- Tsukahara K, Yamamoto H, Okayama H. An RNA binding protein negatively controlling differentiation in fission yeast. Mol Cell Biol. 1998;18:4488–4498. doi: 10.1128/mcb.18.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Whitehall S, Stacey P, Dawson K, Jones N. Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol Biol Cell. 1999;10:3705–3715. doi: 10.1091/mbc.10.11.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- Woollard A, Nurse P. G1 regulation and checkpoints operating around START in fission yeast. BioEssays. 1995;17:481–490. doi: 10.1002/bies.950170604. [DOI] [PubMed] [Google Scholar]
- Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem Sci. 1996;21:18–22. [PubMed] [Google Scholar]
- Yamashita M, Yoshikuni M, Hirai T, Fukada S, Nagahama Y. A monoclonal-antibody against the PSTAIR sequence of p34cdc2, catalytic subunit of maturation-promoting factor, and key regulator of the cell cycle. Dev Growth Differ. 1991;33:617–624. doi: 10.1111/j.1440-169X.1991.00617.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Takeda T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]