Abstract
The largest reported outbreak of waterborne Escherichia coli O157:H7 in the United States occurred in upstate New York following a county fair in August 1999. Culture methods were used to isolate E. coli O157:H7 from specimens from 128 of 775 patients with suspected infections. Campylobacter jejuni was also isolated from stools of 44 persons who developed diarrheal illness after attending this fair. There was one case of a confirmed coinfection with E. coli O157:H7 and C. jejuni. Molecular detection of stx1 and stx2 Shiga toxin genes, immunomagnetic separation (IMS), and selective culture enrichment were utilized to detect and isolate E. coli O157:H7 from an unchlorinated well and its distribution points, a dry well, and a nearby septic tank. PCR for stx1 and stx2 was shown to provide a useful screen for toxin-producing E. coli O157:H7, and IMS subculture improved recovery. Pulsed-field gel electrophoresis (PFGE) was used to compare patient and environmental E. coli O157:H7 isolates. Among patient isolates, 117 of 128 (91.5%) were type 1 or 1a (three or fewer bands different). Among the water distribution system isolates, 13 of 19 (68%) were type 1 or 1a. Additionally, PFGE of C. jejuni isolates revealed that 29 of 35 (83%) had indistinguishable PFGE patterns. The PFGE results implicated the water distribution system as the main source of the E. coli O157:H7 outbreak. This investigation demonstrates the potential for outbreaks involving more than one pathogen and the importance of analyzing isolates from multiple patients and environmental samples to develop a better understanding of bacterial transmission during an outbreak.
Escherichia coli O157:H7 is a primary cause of severe and bloody diarrhea. Complications, particularly hemolytic-uremic syndrome (HUS) (39, 52), have made infections with this organism a public health priority. Ground beef and other bovine products have often been implicated as sources (19, 32), along with other food products (1, 6, 22, 30, 43, 51) and person-to-person transmission (5, 34). Occasional outbreaks have also been associated with public drinking water (42) and swimming in contaminated water (16).
Campylobacter spp. are the most commonly reported bacterial cause of gastrointestinal illness in the United States (25, 29). Although outbreaks have been reported, most cases occur sporadically (35). Cases have been associated most frequently with consumption of contaminated poultry (44), and less frequently with contaminated milk (31) and water (36). When pulsed-field gel electrophoresis (PFGE) is performed on outbreak isolates, multiple types are often identified, implying a polyclonal origin of the outbreak strains (20, 49). Only a few outbreaks involving both E. coli O157:H7 and Campylobacter jejuni have been reported (26; M. Cosgriff, www.promedmail.org).
The PCR is a rapid and reliable tool for the molecular-based diagnosis of a variety of infectious diseases (14). PCR analysis for screening drinking water and environmental samples has been reported (46-48) and has been utilized to identify E. coli in primary water specimens (15, 27), stool specimens (33, 56), and outbreaks (23, 24). In contrast, isolation of E. coli O157:H7 from water and other environmental samples is laborious. Culture is problematic due to large numbers of other flora that either overgrow or mimic the non-sorbitol-fermenting E. coli O157:H7 (12). Recently, immunomagnetic separation (IMS) has helped improve recovery by providing an antibody-based concentration procedure that uses magnetic beads coated with antibody against E. coli O157. Although many report the usefulness of IMS for testing artificially contaminated samples, few reports have documented the use of IMS with naturally occurring, epidemiologically linked specimens (11, 54). Furthermore, there are no reports documenting the use of IMS in support of a waterborne outbreak investigation.
PFGE is useful for subtyping E. coli O157:H7 isolates during outbreak investigations (1, 4, 5). PFGE is reproducible and has sufficient discriminatory power to allow detection of minor genetic variations among isolates (18, 53). Although the use of PFGE for the subtyping of Campylobacter spp. has not been as well described, it has been shown to be useful for strain discrimination under some circumstances (40, 55).
This study describes the laboratory analysis of patient and environmental isolates from the largest outbreak of waterborne E. coli O157:H7 and C. jejuni in the United States (8). This analysis illustrates the utility of screening diverse sample types using a PCR assay for stx1 and stx2. Moreover, it demonstrates the improved recovery of E. coli O157:H7 using IMS protocols which enabled a definitive match of PFGE patterns between isolates obtained from contaminated water and from ill patients.
MATERIALS AND METHODS
Primary specimens.
The Washington County Fair (WCF) was held from 23 August through 29 August 1999 in New York State. There were approximately 111,000 admissions to this large agricultural fair, and exhibitors displayed hundreds of farm animals. The fairground that is the site of this fair does not have a public water supply and its 50 acres is situated on porous soil that is composed of sand and gravel. Additionally, drought conditions existed during this time period, causing a decrease in the water table, which necessitated the use of additional shallow wells for a water supply. Also, a heavy rainfall occurred on one day during the week of the fair.
Once a link between diarrheal illness and this fair was suspected, water was obtained from six shallow wells at the fairgrounds, four of which were unchlorinated in 1999. Additional samples were collected from distal points in the distribution system, a septic tank adjacent to the pump house for well 6, cow manure from a pile close to well 6, soil from a nearby dry well, and ice from a vendor at the fair. Stool samples were also received from several patients who attended the fair and were ill with HUS but whose routine cultures were negative for E. coli O157:H7. A comprehensive epidemiological analysis has been performed (J. Ackelsberg, B. Wallace, D. Schoonmaker-Bopp, D. Dziewulski, S. Sivapalasingam, R. Limberger, P. Smith, M. Burke, B. Sauders, S. Kondracki, S. Olson, D. Morse, D. L. Swerdlow, and the WCF Outbreak Team, unpublished data).
Primary isolation and IMS.
Aliquots containing 100, 250, or 500 ml of liquid specimens (water, septic tank contents, or melted ice) were filtered through 0.45-μm-pore-size filters, and the filters were then placed in 225 ml of E. coli broth supplemented with novobiocin (ECN) (20 μg/ml). Twenty-five-gram aliquots of solid and semisolid specimens (cow manure, soil, or stool) were homogenized with 225 ml of ECN in a stomacher (model 400; Seward Ltd., London, United Kingdom) at normal speed for 2 min. Specimens were incubated in ECN with shaking for 12 to 18 h at 35 to 37°C (primary enrichment), and this was followed by plating onto sorbitol-MacConkey agar (SMAC) and onto SMAC supplemented with cefixime (0.05 mg/liter) and potassium tellurite (2.5 mg/liter) (CTSMAC). Colorless colonies suspected to be E. coli O157:H7 were subcultured and identified as described below. Primary enrichment cultures from most specimens were also subjected to IMS using beads coated with adsorbed and affinity-purified antibodies against all strains of E. coli O157 (Dynabeads anti-E. coli O157; Dynal, Wirral, United Kingdom) according to the manufacturer's directions, and this was followed by direct plating of beads onto SMAC and CTSMAC. Some cultures underwent a secondary overnight ECN enrichment of the beads at 42°C and plating of the broth and bead-bacteria complex onto SMAC and CTSMAC to further select for E. coli O157:H7 (7, 21). This method was adapted from the technique of Wells et al. (J. G. Wells, K. D. Greene, C. A. Bopp, M. E., Proctor, L. M. Slutsker, and P. Mead, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. P-21, 1999). For selected specimens the IMS enrichment was repeated on the secondary enrichment overnight and an aliquot was transferred for a third enrichment at 42°C.
stx1 and stx2 PCR.
The primer sequences used for PCR are shown in Table 1. Primers VT1a, VT1b, and VT2b were identical to those described by Pollard et al. (37), whereas VT2aa was designed at the Wadsworth Center and is capable of identifying stx2 as well as the additional antigenically distinct Shiga toxins stx2c and stx2e. Multiplex PCR amplification of stx1 and stx2 sequences was performed on primary and secondary enrichment broths from the samples. Crude template DNA was prepared as follows. A 200-μl aliquot of each broth sample was centrifuged at 13,000 × g for 2 min, and the pellets were resuspended in 200 μl of phosphate-buffered saline. The tubes were centrifuged and washed with phosphate-buffered saline again. After a third centrifugation, the pellets were resuspended in 1× GeneAmp PCR buffer II (100 mM Tris-HCl [pH 8.3], 500 mM KCl; Perkin-Elmer Cetus). The tubes were placed in a 95°C heat block for 15 min. DNA was amplified in a final reaction volume of 50 μl consisting of 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 1.25 U of Amplitaq Gold DNA polymerase (Perkin-Elmer Cetus), a 1 μM concentration (each) of the VT1-VT2 primer pair, and 10 μl of crude template DNA. Thermocycling conditions in a GeneAmp 9600 thermocycler (Perkin-Elmer Cetus) were as follows: initial cycle of 9 min at 95°C, followed by 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C for 30 cycles, with a 7-min extension at 72°C. PCR products were visualized on a 2% agarose gel stained with ethidium bromide. Sample types such as cow manure, soil, and stools were tested for PCR inhibition by 1:10 dilution of the template DNA and/or by spiking of a duplicate PCR with a lysate of an E. coli O157:H7 strain known to produce Shiga toxin 1 and Shiga toxin 2.
TABLE 1.
Primers used for amplification of stx1, stx2, eaeA, and hlyA genes by PCR
| Primer | Gene | Oligonucleotide sequence (5′→3′) | Amplicon product (bp) | Reference or source |
|---|---|---|---|---|
| VT1a | stx1 | GAAGAGTCCGTGGGATTACG | 130 | 37 |
| VT1b | stx1 | AGCGATGCAGCTATTAATAA | 37 | |
| VT2aa | stx2 | CGACCCCTCTTGAACATATATCTC | 397 | Wadsworth Center (1996) |
| VT2b | stx2 | GCTCTGGATGCATCTCTGGT | 37 | |
| AE19 | eaeA | CAGGTCGTCGTGTCTGCTAAA | 1,087 | 17 |
| AE20 | eaeA | TCAGCGTGGTTGGATCAACCT | 17 | |
| MFS1F | hlyA | ACGATGTGGTTTATTCTGGA | 166 | 13 |
| MFS1R | hlyA | CTTCACGTCACCATACATAT | 13 |
eaeA gene PCR.
The primer sequences (AE19 and AE20) used for PCR are shown in Table 1. PCR assays were performed according to the methods in published studies (13) with slight modifications. Briefly, DNA was amplified in a final reaction volume of 100 μl consisting of 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of each dNTP, 2.5 U of Amplitaq Gold DNA polymerase, a 1 μM concentration (each) of the AE19-AE20 primer pair, and 10 μl of crude template DNA. Thermocycling conditions were as follows: initial cycle of 9 min at 95°C, followed by 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C for 35 cycles.
60-MDa plasmid (hlyA gene) PCR.
The primer sequences (MFS1F and MFS1R) used for PCR are shown in Table 1. PCR assays were performed according to the methods in published studies (15) with slight modifications. Briefly, DNA was amplified in a final reaction volume of 100 μl consisting of 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of each dNTP, 2.5 U of Amplitaq Gold DNA polymerase, a 1 μM concentration (each) of the MFS1F-MFS1R primer pair, and 10 μl of crude template DNA. Thermocycling conditions were as follows: initial cycle of 9 min at 95°C, followed by 30 s at 94°C, 30 s at 62°C, and 30 s at 72°C for 35 cycles.
Bacterial isolates.
Patient isolates of E. coli O157:H7 were obtained from clinical laboratories, which processed stool samples by conventional methods. Briefly, stools were plated onto SMAC followed by identification of sorbitol-negative isolates as E. coli O157 using biochemical systems and O157 serological kits. At the Wadsworth Center, patient and environmental isolates of E. coli O157:H7 were confirmed using triple sugar iron, urea, citrate, indole, a latex agglutination kit for serogroup O157 (Oxoid, Hampshire, England), and tube agglutination for H7 (Difco, Detroit, Mich.).
Campylobacter isolates from clinical laboratories were plated on Mueller-Hinton II agar with 5% sheep blood (MHSBA) and incubated at 35 to 37°C microaerophilically in a tri-gas incubator with 85% N2, 10% CO2, and 5% O2. Identification was confirmed by aerobic (37°C), anaerobic (37°C), and microaerophilic (25, 37, and 42°C) growth, as well as assays for oxidase; indoxyl acetate; hippurate hydrolysis; catalase; MacConkey agar growth; and susceptibility to nalidixic acid, cephalothin, and polymyxin B.
PFGE.
DNA was prepared from E. coli O157:H7 cells grown for 24 h on brain heart infusion agar at 37°C according to established protocols (9). Briefly, bacteria were embedded in agarose, lysed, and treated with protease. DNA was digested with restriction endonuclease XbaI and in some cases AvrII (New England Biolabs, Beverly, Mass.), and the fragments were separated in 1.0% agarose gels on a clamped homogenous electric field apparatus (CHEF Mapper; Bio-Rad Laboratories, Richmond, Calif.). The initial pulse time of 2.2 s was increased linearly to 54.2 s over 22 h. Gels were stained with ethidium bromide, destained in water, and visualized with a Gel Doc 1000 gel analysis system (Bio-Rad Laboratories, Hercules, Calif.).
DNA was prepared from Campylobacter sp. cells that had been grown on MHSBA and incubated at 35 to 37°C for 24 h under microaerophilic conditions. The DNA was prepared as for the E. coli O157:H7 isolates with slight modifications (38). The DNA was digested with SmaI (New England Biolabs). The initial pulse time of 5 s was increased linearly to 35 s over 22 h.
We considered PFGE patterns to be identical if all bands greater than 50 kb in size were the same. Related subtypes were defined by three or fewer band differences, and unrelated types were defined by four or more band differences (45). Patterns were submitted to PulseNet (41) central for comparison to the national E. coli O157:H7 PFGE pattern database.
RESULTS
Epidemiological investigation.
The outbreak was recognized when the Bureau of Communicable Disease Control of the New York State Department of Health became aware of an increased number of persons being evaluated for diarrheal illness at local hospitals. Some hospitalized children showed worsening kidney function, and E. coli O157:H7 was isolated from the stool of one patient. The next day, area hospitals were alerted to consider E. coli O157:H7 infection in patients who presented with diarrheal illness.
An epidemiological investigation revealed that many of these patients had attended the WCF, one of the largest agricultural fairs in New York State. The WCF 50-acre site does not utilize a public water supply, and in 1999 at the time of this outbreak, its water came from shallow wells. Water from the fairgrounds was obtained from six wells (four of which were unchlorinated). Analysis of samples from each of the wells showed that coliform bacteria were present in two of the wells and that one of these (well 6), an unchlorinated well that supplied drinking water, contained E. coli. Additionally, two potential environmental sources of E. coli contamination near well 6 were identified: a cow manure storage site (80 ft away) and a dormitory septic tank with a seepage pit only 36 ft from well 6. Environmental samples were collected from well 6, distal points in the distribution system of well 6, the septic tank for the dormitory, cow manure from the nearby storage site, soil from a dry well, and ice from a vendor. Details of the epidemiologic investigation will be published elsewhere (Ackelsberg et al., unpublished data).
Molecular screening for Shiga toxin genes stx1 and stx2.
A summary of results obtained by screening specimens from this outbreak for Shiga toxin genes by PCR is shown in Table 2. Notably, stx1 and stx2 genes were detected in the well water from well 6 and distal points in the water distribution system (Fig. 1). Additional positive samples include primary specimens from the septic tank and cow manure sites as well as soil samples from the dry well and two submitted stool samples (Table 2). Ice specimens were negative by PCR upon initial screening and remained negative after the second and third enrichments (Table 2; Fig. 1). All PCR-positive enrichment broths were E. coli O157:H7 culture positive except for the cow manure specimens, but no PCR-negative enrichment broths were culture positive. The cow manure samples may have contained Shiga toxin genes from non-E. coli O157:H7 strains. The three water samples that were positive for stx1 and stx2 were also positive by PCR for the virulence factor genes eaeA and hlyA (data not shown). A reduction in the amount of amplified PCR product obtained from the spiked positive control indicated that a partial inhibition to the PCR occurred in some of the primary cow manure, stool, and soil samples. In previous validation tests, we noted that samples showing a partial inhibition did not detract from our ability to detect E. coli O157:H7 by PCR, since overnight growth amplification provided sufficient template for amplification. In addition, no inhibition occurred in the double-enriched and triple-enriched broths (not shown).
TABLE 2.
Enriched specimens screened for stx1 and stx2 genes by PCR
| Type of specimen tested | No. of specimens tested (no. positive) for stx1 and stx2
|
||
|---|---|---|---|
| Primary enriched | Double enriched | Triple enriched | |
| Water | 34 (3) | 0 | 0 |
| Cow manure | 6 (3) | 3 (0) | 3 (0) |
| Soil from dry well | 2 (0) | 2 (2) | 2 (2) |
| Ice | 10 (0) | 6 (0) | 2 (0) |
| Stool | 6 (0) | 5 (2) | 0 |
| Septic tank | 5 (5) | 0 | 0 |
| Total (n = 86) | 63 (11) | 16 (4) | 7 (2) |
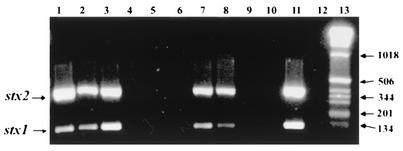
FIG. 1.
Multiplex PCR amplification of Shiga toxin genes (stx1 and stx2) from selected environmental specimens. The expected amplified products were 397 bp for stx2 and 138 bp for stx1. Lanes 1 and 2, primary enriched water samples from well 6; lane 3, primary enriched sample from the distribution system of well 6; lanes 4 to 6, secondary enrichments of cow manure samples; lanes 7 and 8, secondary enrichments of samples from a dry well; lanes 9 and 10, secondary enrichments of ice samples from a vendor who utilized well 6; lane 11, positive control; lane 12, negative template control; lane 13, 1-kb DNA ladder (Gibco BRL) (base pairs indicated at right).
Selective enrichment and IMS treatment of environmental samples and clinical specimens.
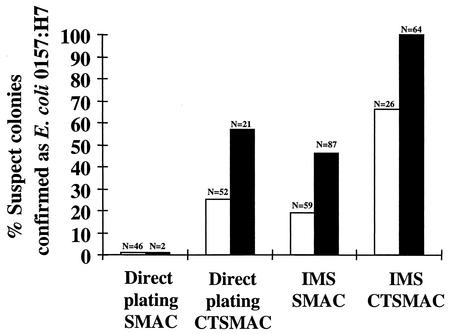
As shown in Fig. 2, no E. coli O157:H7 was recovered by direct plating on SMAC of primary or secondary enrichment broths from environmental samples. Direct plating of primary and secondary broths on CTSMAC, however, resulted in successful recovery of E. coli O157:H7 (Fig. 2). The implementation of IMS further increased the percentage of recovery from both the SMAC and CTSMAC cultures (Fig. 2). Additionally, two stool samples that were previously culture negative, from patients with HUS, were received from a clinical laboratory. They were also selectively enriched and subjected to IMS, which resulted in the successful isolation of E. coli O157:H7 from these specimens (data not shown).
FIG. 2.
Recovery of E. coli O157:H7 after primary enrichment (open bars) and secondary enrichment (solid bars), IMS treatment, and plating on various culture media. The n values indicate the total number of colonies isolated from these plates that were suspected to be E. coli O157:H7. The percentages were determined by dividing the number of E. coli O157:H7 confirmed identifications by the number of isolates examined. The first two columns, which encompass direct plating on SMAC, represent a value of 0%.
PFGE of E. coli O157:H7 isolates from patients and the environment.
During this outbreak, 128 isolates of E. coli O157:H7 from 117 patients were examined by PFGE. From the group of patient isolates identified as E. coli O157:H7, it was observed that the majority had enzyme digestion restriction patterns designated as types 1 and 1a (91.5%). Related subtypes and types unrelated to the outbreak pattern comprised the remaining 8.5% (Table 3; Fig. 3). Multiple isolates from nine patients were analyzed (20 isolates), and each patient was found to harbor strains representing a single PFGE pattern (data not shown).
TABLE 3.
PFGE patterns of E. coli O157:H7 isolates according to source
| PFGE pattern | No. (%) of isolates
|
|||
|---|---|---|---|---|
| Patient | Water | Soil from dry well | Septic tank | |
| 1 | 76 (65.0) | 12 (63.1) | 0 | 1 (25) |
| 1a | 31 (26.5) | 1 (5.3) | 0 | 3 (75) |
| 1b | 2 (1.7) | 0 | 0 | 0 |
| 1c | 1 (0.9) | 0 | 0 | 0 |
| 1e | 0 | 6 (31.6) | 0 | 0 |
| 1f | 0 | 0 | 10 (100) | 0 |
| 1g | 2 (1.7) | 0 | 0 | 0 |
| 1h | 1 (0.9) | 0 | 0 | 0 |
| 2 | 1 (0.9) | 0 | 0 | 0 |
| 4 | 3 (2.6) | 0 | 0 | 0 |
| Total | 117 | 19 | 10 | 4 |
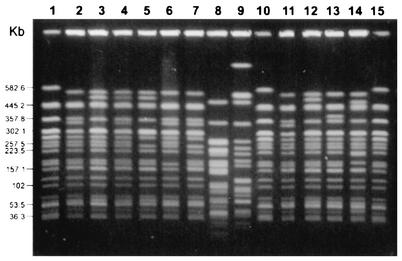
FIG. 3.
PFGE analysis of selected E. coli O157:H7 isolates after digestion with XbaI. Lanes 1, 10, and 15 contain control E. coli strain G5244, with numbers at left indicating the band size in kilobase pairs. Lanes 1 to 9 contain clinical isolates from outbreak-associated patients, and lanes 11 to 14 contain environmental isolates associated with the outbreak. Lane 2, PFGE type 1; lanes 3 to 7, related subtypes (three or fewer band differences) 1a, 1b, 1c, 1g, and 1 h, respectively; lanes 8 and 9, PFGE types 2 and 4; lane 11, PFGE type 1; lanes 12 to 14, subtypes (three or fewer band differences) 1a, 1e, and 1f, respectively.
Among water distribution system isolates, 12 (63.1%) were type 1, 1 (5.3%) was subtype 1a, and 6 (31.6%) were subtype 1e (which was not found in patient isolates). Among the dormitory septic tank isolates, one (25%) was type 1, and three (75%) were subtype 1a. All of the soil isolates were subtype 1f. Type 1 and type 1a, the predominant patient patterns in this outbreak, were not previously seen in the 165 E. coli O157:H7 isolates fingerprinted in our laboratory during the year prior to the outbreak. Additionally, since the outbreak, only 1 out of 300 isolates typed by our laboratory has matched the type 1 pattern, indicating that this pattern is rare. This predominant PFGE pattern, pattern 1, was also noted by four other states as part of the PulseNet database, and at least one of these cases was epidemiologically linked to this outbreak.
PFGE of C. jejuni isolates from patients.
Clinical laboratories confirmed C. jejuni in specimens from 44 patients believed to be associated with the WCF outbreak, one of whom was coinfected with E. coli O157:H7 type 4. Thirty-five of the outbreak C. jejuni isolates were submitted to our laboratory for species confirmation and PFGE. Although multiple PFGE patterns were detected, the majority 29 (83%) were type 1 (Fig. 4), together with one isolate each of types 2, 3, 3a, 4, 5, and 6. C. jejuni was not isolated from well water or from the septic tank.
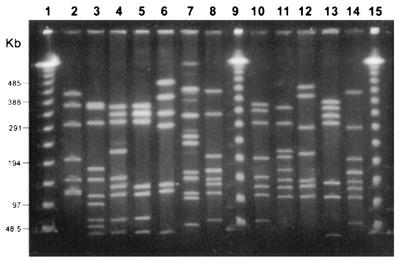
FIG. 4.
PFGE analysis of selected C. jejuni isolates after digestion with SmaI. Numbers at left indicate band size in kilobase pairs. Lanes 1, 9, and 15 contain lambda ladder (48.5 kb) for size estimation. Lanes 2 to 8 contain clinical isolates from outbreak-associated patients. Lane 2, PFGE type 1, which comprised 83% of the patient isolates; lanes 3 to 8, unrelated types (more than four band differences) and subtypes (three or fewer band differences) 2, 3, 3a, 4, 5, and 6, respectively. Lanes 10 to 14 contain patient isolates not associated with the outbreak.
DISCUSSION
This outbreak, which included 775 patients with suspected cases of infection, was the largest outbreak of diarrhea associated with waterborne E. coli O157:H7 ever reported in the United States. Since water is a commonly consumed beverage and is used in food preparation and for hand washing, it was important to establish the presence of the pathogen in the water and to evaluate the genetic relatedness of the patient and water isolates from this investigation.
Given the long turnaround time associated with traditional culture methods, PCR was used as a primary screen for the detection of stx1 and stx2 toxin genes. PCR-positive results implicated well 6 and its distribution system as the source. This led to the laboratory's effort to isolate and fingerprint the organism, thus successfully confirming the suspected epidemiological link. We found no evidence that culturing of PCR-negative specimens would improve sensitivity. Also, positive results from the specific PCR tests for eaeA and hlyA genes, which are associated with virulence, helped to characterize the organisms isolated from water as pathogenic and probable E. coli O157:H7.
This investigation also demonstrated that IMS subculture improved the yield of recovery for E. coli O157:H7. Direct plating onto SMAC did not allow isolation of E. coli O157:H7, perhaps due to overgrowth of competing gram-negative organisms. Direct plating of primary and secondary broths to CTSMAC plates did, however, result in successful recovery of E. coli O157:H7. Use of CTSMAC increased selection of E. coli O157:H7 and decreased growth of non-O157 organisms. In addition, IMS further improved the success of isolation. When IMS was combined with secondary enrichment and plating on CTSMAC medium, a 100% confirmation rate was achieved for suspect colonies. The submitted stool samples that were previously culture-negative when processed by conventional methods and then found to be positive by our protocol demonstrated the usefulness of this protocol. We conclude from these experiments that the use of CTSMAC is necessary for successful recovery of E. coli O157:H7 from environmental samples. We also conclude, as have others (Wells et al., 99th Gen. Meet. Am. Soc. Microbiol.), that the implementation of the secondary IMS procedure combined with plating on CTSMAC medium is useful for both environmental and clinical samples in which the organism may be present in low numbers, and for which conventional isolation procedures are unsuccessful.
PFGE was used to genetically type patient isolates as well as multiple isolates from environmental samples. The two predominant PFGE types, 1 and 1a, comprised 91.5% of the E. coli isolates examined from patients with culture-confirmed infection. These two types were also present in isolates obtained from well 6, distal parts of the water distribution system, and the dormitory septic tank. The presence of isolates with this rare PFGE type in the vast majority of patients and in water from well 6 and parts of the distribution system provided evidence that the water was most likely the source of exposure for the illness. Analysis of the septic system from the nearby dormitory building using dye seepage tests demonstrated a hydraulic connection between the septic system and the water in well 6. Patient interviews and multivariate analysis also suggested that water could be a potential source of exposure (Ackelsberg et al., unpublished data). Thus, the laboratory and epidemiological data convincingly implicated water as the primary source of E. coli O157:H7.
The vast majority of the E. coli O157:H7 isolates from patients were of one related type, suggesting primary contamination of water and subsequent patient infection from a single unknown source. It is unclear whether the source was of bovine or human origin, since both cattle and humans can excrete multiple subtypes (2, 3, 10, 28). One hypothesis is that cow manure contaminated with E. coli O157:H7 was carried into the dormitory on muddy boots, was washed into the septic system, and subsequently seeped into well 6 (Ackelsberg et al., unpublished data). Unfortunately, we were unable to detect E. coli O157:H7 in a limited sample of six cow manure specimens and are unable to confirm this hypothesis. Laboratory data also cannot rule out the possibility of a primary contamination from a human source in the septic tank. Moreover the presence of multiple PFGE types and subtypes, although few, further complicates the analysis. Only PFGE patterns 1, 1a, and 1e were detected in the water, although patient samples contained additional types and subtypes. There are several explanations that can account for these results. One possibility is that not all of the types and subtypes were able to be cultured from the water. Alternatively, an exogenous source of E. coli O157:H7 contamination could have contributed to the number of other types noted in isolates from ill patients. For example, some patients could have become infected through direct contact with the farm animals or even outside of the fair, which could explain the few patient isolates for which types different from the predominant pattern found in the water were found. In addition, the public announcement of an outbreak likely led to an increase in laboratory testing by concerned patients and physicians, which may have resulted in isolation of additional diverse E. coli O157:H7 strains from patients not involved in the outbreak. Conceivably, a combination of the above factors could explain the presence of the few other PFGE types found in the patient isolates.
There are some caveats to the genetic analysis of these isolates. Isolates from patients were obtained from a local laboratory (the isolates were presumably from a single colony) and sent to the Wadsworth Center on agar slants. Unlike the environmental samples, from which independently isolated colonies were typed by PFGE, the patient isolates were only typed from one original colony. Thus, there may have been genetic variation in additional patient isolates tested that we were unable to detect because only single colonies had been sent for analysis. Of the 117 E. coli O157:H7 isolates cultured from patient specimens, 96.6% were related to the major type 1 pattern (45). Testing with another enzyme confirmed that various subtypes of pattern 1 were genetically related (data not shown). Furthermore, the predominant PFGE patterns, 1 and 1a, have rarely been detected by our laboratory, suggesting that this outbreak originated primarily from a single genetically related strain. Studies have demonstrated that multiple subtypes can be shed from both bovine and human infections (2, 3, 10, 28). Although this is often explained by a loss of either the 60-MDa plasmid or the bacteriophage harboring the Shiga toxin genes, the exact mechanisms involved are unclear. Unfortunately, outbreak analysis often focuses on isolation of E. coli O157:H7 from a patient sample, followed by PFGE of a single colony. This complex outbreak may have benefited from PFGE typing of additional patient isolates much in the same way the environmental isolate typing resulted in demonstration of multiple subtypes from the same source.
The occurrence of 29 concurrent Campylobacter cases with isolates having a single PFGE type also suggests primary contamination from one source, with some secondary contamination from other sources. The presence of a predominant PFGE type in 83% of the C. jejuni isolates that were available for PFGE testing places this outbreak as one of the few outbreaks in which a predominant C. jejuni clone has been demonstrated. Our laboratory and others have demonstrated broad heterogeneity among PFGE types from Campylobacter isolates, which is believed to be related to genetic instability of the organism (20, 49, 50). Therefore, the finding of essentially one PFGE type suggests a single source of contamination, with the other types resulting from rare exogenous sources or genetic instability. Because we were unable to culture C. jejuni from the water samples, we were unable to definitively link the C. jejuni infections to the water. However, the predominance of a single type suggests a massive contamination from a single source.
In summary, since Shiga toxins are important virulence factors of E. coli O157:H7, the initial PCR screening for stx1 and stx2 was an effective method for identifying samples to be processed for culture isolation and fingerprinting. Successful isolation was accomplished using IMS subculture and selective culture medium. Furthermore, we demonstrated the relatedness of isolates from patients and water by PFGE fingerprinting, thereby implicating the water distribution system as the main source of exposure in this large outbreak of E. coli O157:H7 infection. The presence of multiple PFGE types among patient and environment isolates demonstrates the importance of testing multiple isolates and of using a discriminating fingerprinting technique during an outbreak investigation.
Acknowledgments
We gratefully acknowledge the New York State Department of Health, the regional laboratory partners, and the Centers for Disease Control and Prevention and the contributions of numerous personnel from these institutions who were involved in the activities associated with this outbreak.
REFERENCES
- 1.Ackers, M. L., B. E. Mahon, E. Leahy, B. Goode, T. Damrow, P. S. Hayes, W. F. Bibb, D. H. Rice, T. J. Barrett, L. Hutwagner, P. M. Griffin, and L. Slutsker. 1998. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J. Infect. Dis. 177:1588-1593. [DOI] [PubMed] [Google Scholar]
- 2.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 3.Akiba, M., T. Sameshima, and M. Nakazawa. 1999. The shift of genetic subtypes of Escherichia coli O157:H7 isolates from cattle. Epidemiol. Infect. 122:343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ammon, A., L. R. Petersen, and H. Karch. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H7. J. Infect. Dis. 179:1274-1277. [DOI] [PubMed] [Google Scholar]
- 5.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 6.Besser, R. E., S. M. Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA 269:2217-2220. [PubMed] [Google Scholar]
- 7.Blais, B. W., R. A. Booth, L. M. Phillippe, and H. Yamazaki. 1997. Effect of temperature and agitation on enrichment of Escherichia coli O157:H7 in ground beef using modified EC broth with novobiocin. Int. J. Food Microbiol. 36:221-225. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1999. Outbreak of Escherichia coli O157:H7 and Campylobacter among attendees of the Washington County Fair—New York, 1999. Morb. Mortal. Wkly. Rep. 48:803-805. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2000. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: training manual. Centers for Disease Control and Prevention, Atlanta, Ga.
- 10.Cobbold, R., and P. Desmarchelier. 2001. Characterization and clonal relationships of Shiga-toxigenic Escherichia coli (STEC) isolated from Australian dairy cattle. Vet. Microbiol. 79:323-335. [DOI] [PubMed] [Google Scholar]
- 11.Cubbon, M. D., J. E. Coia, M. F. Hanson, and F. M. Thomson-Carter. 1996. A comparison of immunomagnetic separation, direct culture and polymerase chain reaction for the detection of verocytotoxin-producing Escherichia coli O157 in human faeces. J. Med. Microbiol. 44:219-222. [DOI] [PubMed] [Google Scholar]
- 12.de Boer, E., and A. E. Heuvelink. 2000. Methods for the detection and isolation of Shiga toxin-producing Escherichia coli. J. Appl. Microbiol. 88:133S-143S. [DOI] [PubMed]
- 13.Fratamico, P. M., S. K. Sackitey, M. Wiedmann, and M. Y. Deng. 1995. Detection of Escherichia coli O157:H7 by multiplex PCR. J. Clin. Microbiol. 33:2188-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredricks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-486. [DOI] [PubMed] [Google Scholar]
- 15.Fricker, E. J., and C. R. Fricker. 1994. Application of the polymerase chain reaction to the identification of Escherichia coli and coliforms in water. Lett. Appl. Microbiol. 19:44-46. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, M. S., T. Roels, J. E. Koehler, L. Feldman, W. F. Bibb, and P. Blake. 1999. Escherichia coli O157:H7 outbreak associated with an improperly chlorinated swimming pool. Clin. Infect. Dis. 29:298-303. [DOI] [PubMed] [Google Scholar]
- 17.Gannon, V. P., M. Rashed, R. K. King, and E. J. Thomas. 1993. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J. Clin. Microbiol. 31:1268-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouveia, S., M. E. Proctor, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1998. Genomic comparisons and Shiga toxin production among Escherichia coli O157:H7 isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J. Clin. Microbiol. 36:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 20.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hepburn, N. F., M. MacRae, M. Johnston, J. Mooney, and I. D. Ogden. 2002. Optimizing enrichment conditions for the isolation of Escherichia coli O157 in soils by immunomagnetic separation. Lett. Appl. Microbiol. 34:365-369. [DOI] [PubMed] [Google Scholar]
- 22.Hilborn, E. D., J. H. Mermin, P. A. Mshar, J. L. Hadler, A. Voetsch, C. Wojtkunski, M. Swartz, R. Mshar, M. A. Lambert-Fair, J. A. Farrar, M. K. Glynn, and L. Slutsker. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159:1758-1764. [DOI] [PubMed] [Google Scholar]
- 23.Hilborn, E. D., P. A. Mshar, T. R. Fiorentino, Z. F. Dembek, T. J. Barrett, R. T. Howard, and M. L. Cartter. 2000. An outbreak of Escherichia coli O157:H7 infections and haemolytic uraemic syndrome associated with consumption of unpasteurized apple cider. Epidemiol. Infect. 124:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huerta, M., I. Grotto, M. Gdalevich, D. Mimouni, B. Gavrieli, M. Yavzori, D. Cohen, and O. Shpilberg. 2000. A waterborne outbreak of gastroenteritis in the Golan Heights due to enterotoxigenic Escherichia coli. Infection 28:267-271. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, K. E., and C. M. Nolan. 1985. Community-wide surveillance of Campylobacter jejuni infection. Evaluation of a laboratory-based method. Diagn. Microbiol. Infect. Dis. 3:389-396. [DOI] [PubMed] [Google Scholar]
- 26.Jones, I. G., and M. Roworth. 1996. An outbreak of Escherichia coli 0157 and campylobacteriosis associated with contamination of a drinking water supply. Public Health 110:277-282. [DOI] [PubMed] [Google Scholar]
- 27.Juck, D., J. Ingram, M. Prevost, J. Coallier, and C. Greer. 1996. Nested PCR protocol for the rapid detection of Escherichia coli in potable water. Can. J. Microbiol. 42:862-866. [DOI] [PubMed] [Google Scholar]
- 28.Karch, H., H. Russmann, H. Schmidt, A. Schwarzkopf, and J. Heesemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michino, H., K. Araki, S. Minami, S. Takaya, N. Sakai, M. Miyazaki, A. Ono, and H. Yanagawa. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787-796. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, D., C. Gunneberg, D. Gunnell, T. D. Healing, S. Lamerton, N. Soltanpoor, D. A. Lewis, and D. G. White. 1994. An outbreak of Campylobacter infection associated with the consumption of unpasteurised milk at a large festival in England. Eur. J. Epidemiol. 10:581-585. [DOI] [PubMed] [Google Scholar]
- 32.Ostroff, S. M., P. M. Griffin, R. V. Tauxe, L. D. Shipman, K. D. Greene, J. G. Wells, J. H. Lewis, P. A. Blake, and J. M. Kobayashi. 1990. A statewide outbreak of Escherichia coli 0157:H7 infections in Washington State. Am. J. Epidemiol. 132:239-247. [DOI] [PubMed] [Google Scholar]
- 33.Paton, A. W., J. C. Paton, P. N. Goldwater, and P. A. Manning. 1993. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J. Clin. Microbiol. 31:3063-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavia, A. T., C. R. Nichols, D. P. Green, R. V. Tauxe, S. Mottice, K. D. Greene, J. G. Wells, R. L. Siegler, E. D. Brewer, D. Hannon, and P. A. Blake. 1990. Hemolytic-uremic syndrome during an outbreak of Escherichia coli O157:H7 infections in institutions for mentally retarded persons: clinical and epidemiologic observations. J. Pediatr. 116:544-551. [DOI] [PubMed] [Google Scholar]
- 35.Pebody, R. G., M. J. Ryan, and P. G. Wall. 1997. Outbreaks of campylobacter infection: rare events for a common pathogen. Commun. Dis. Rep. CDR Rev. 7:R33-R37. [PubMed] [Google Scholar]
- 36.Penner, J. L., A. D. Pearson, and J. N. Hennessy. 1983. Investigation of a waterborne outbreak of Campylobacter jejuni enteritis with a serotyping scheme based on thermostable antigens. J. Clin. Microbiol. 18:1362-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard, D. R., W. M. Johnson, H. Lior, S. D. Tyler, and K. R. Rozee. 1990. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J. Clin. Microbiol. 28:540-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 40.Steele, M., B. McNab, L. Fruhner, S. DeGrandis, D. Woodward, and J. A. Odumeru. 1998. Epidemiological typing of Campylobacter isolates from meat processing plants by pulsed-field gel electrophoresis, fatty acid profile typing, serotyping, and biotyping. Appl. Environ. Microbiol. 64:2346-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swerdlow, D. L., B. A. Woodruff, R. C. Brady, P. M. Griffin, S. Tippen, H. D. Donnell, Jr., E. Geldreich, B. J. Payne, A. Meyer, Jr., J. G. Wells, K. D. Greene, M. Bright, N. H. Bean, and P. A. Blake. 1992. A waterborne outbreak in Missouri of Escherichia coli O157:H7 associated with bloody diarrhea and death. Ann. Intern. Med. 117:812-819. [DOI] [PubMed] [Google Scholar]
- 43.Tamblyn, S., J. deGrosbois, D. Taylor, and J. Stratton. 1999. An outbreak of Escherichia coli O157:H7 infection associated with unpasteurized non-commercial, custom-pressed apple cider—Ontario, 1998. Can. Commun. Dis. Rep. 25:113-120. [PubMed] [Google Scholar]
- 44.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. American Society for Microbiology, Washington, D. C.
- 45.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsen, H. Y., and L. Z. Jian. 1998. Development and use of a multiplex PCR system for the rapid screening of heat labile toxin I, heat stable toxin II and shiga-like toxin I and II genes of Escherichia coli in water. J. Appl. Microbiol. 84:585-592. [DOI] [PubMed] [Google Scholar]
- 47.Waage, A. S., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of low numbers of Salmonella in environmental water, sewage and food samples by a nested polymerase chain reaction assay. J. Appl. Microbiol. 87:418-428. [DOI] [PubMed] [Google Scholar]
- 48.Waage, A. S., T. Vardund, V. Lund, and G. Kapperud. 1999. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 65:1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe, H., A. Wada, Y. Inagaki, K. Itoh, and K. Tamura. 1996. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet 348:831-832. [DOI] [PubMed] [Google Scholar]
- 52.Wells, J. G., B. R. Davis, I. K. Wachsmuth, L. W. Riley, R. S. Remis, R. Sokolow, and G. K. Morris. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J. Clin. Microbiol. 18:512-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willshaw, G. A., H. R. Smith, T. Cheasty, P. G. Wall, and B. Rowe. 1997. Vero cytotoxin-producing Escherichia coli O157 outbreaks in England and Wales, 1995: phenotypic methods and genotypic subtyping. Emerg. Infect. Dis. 3:561-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright, D. J., P. A. Chapman, and C. A. Siddons. 1994. Immunomagnetic separation as a sensitive method for isolating Escherichia coli O157 from food samples. Epidemiol. Infect. 113:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]
- 56.Yavzori, M., N. Porath, O. Ochana, R. Dagan, R. Orni-Wasserlauf, and D. Cohen. 1998. Detection of enterotoxigenic Escherichia coli in stool specimens by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 31:503-509. [DOI] [PubMed] [Google Scholar]