Abstract
Timely diagnosis of respiratory syncytial virus (RSV) infection is critical for appropriate treatment of lower respiratory infection in young children. To facilitate diagnosis, we developed a rapid, specific, and sensitive TaqMan PCR method for detection of RSV A and RSV B. Two sets of primer-probe pairs were selected from the nucleotide sequences encoding the nucleocapsid protein—one targeting RSV A and the other targeting RSV B. The specificity of the TaqMan reverse transcription-PCR assay was evaluated by testing each primer-probe pair against various viruses derived from laboratory virus stocks, as well as clinical respiratory specimens. Fluorescent signals were observed only in the presence of RSV A and/or RSV B. The sensitivity of our quantitative PCR assay was determined on the basis of PFU and virus particle counts. The resulting assay sensitivity was found to be 0.023 PFU, or two copies of viral RNA, for RSV A and 0.018 PFU, or nine copies of viral RNA, for RSV B. This quantitative TaqMan PCR assay was utilized to diagnose 175 nasopharyngeal aspirates obtained from children in Hong Kong with respiratory symptoms during the winter of 2000 and 2001. Among these specimens, TaqMan PCR detected 36 RSV-positive samples, 10 of which were identified as RSV A and 26 of which were identified as RSV B, whereas culture confirmation identified 21 RSV-positive specimens and immunofluorescence identified 32 RSV-positive specimens, all of which were among those identified by PCR. The results confirmed the accuracy of our TaqMan PCR assay and demonstrated its improved sensitivity versus classical methods.
Respiratory syncytial virus (RSV) is one of the major respiratory pathogens causing lower respiratory tract diseases in children (10, 18, 23). Based on genetic and antigenic variations in the structural proteins, RSV is classified into two subgroups, A and B (3, 24). At present, there is no vaccine available for prevention of the viral infection, although antiviral therapy is known to be effective in some patients (1, 2, 11). For appropriate treatment of RSV infection, it is crucial to have an accurate and timely diagnostic method for detection of the virus.
A number of techniques are available for detection and identification of RSV, including cell culture, enzyme immunoassay (EIA), immunofluorescence (IF), and conventional reverse transcription (RT)-PCR (6, 7, 8, 9, 12, 16, 17, 21, 22, 25). The classical cell culture method requires prompt inoculation of the labile virus and is a time-consuming process. In addition, the test has low sensitivity, detecting only 50 to 60% of RSV infections (17). More rapid immunoassays for the detection of RSV, such as EIA and IF, also have limitations in sensitivity and specificity. The accuracy of EIA, for example, is in the range of 57 to 98%, and that of IF varies from 65 to 92% (12). This wide variability in assay sensitivity and specificity may lead to inaccurate diagnosis of RSV infection, and consequently, the assays may be of limited value in patient care. Furthermore, such rapid immunoassays are rarely capable of differentiating RSV subgroups A and B, which may be associated with different disease severities (28).
Conventional RT-PCR, which has recently been developed for the detection of RSV, is a more sensitive and specific diagnostic method that also allows for the subgrouping of the virus in a single reaction (6, 7, 8, 9, 17, 25). However, it still requires time-consuming post-PCR analysis. Real-time RT-PCR, on the other hand, eliminates post-PCR processing (13). This is achieved by combining conventional RT-PCR with advanced fluorescence detection technology, which allows the fluorescence signals to be analyzed and recorded during PCR cycling. The advanced method also performs automatic sample analysis with enhanced sensitivity and specificity. In this report, we describe the development and application of a TaqMan-based RT-PCR assay for simultaneous detection, subgrouping, and quantification of RSV A and B in clinical respiratory specimens.
MATERIALS AND METHODS
Virus stocks, plaque titration, and virus particle counts.
RSV A2 (subgroup A) and 2B (subgroup B) were propagated in Vero cells. About 100 ml each of subgroup A and B virus stocks were prepared, and the stocks were stored in 0.5-ml aliquots for future use. For plaque titration of RSV stocks, confluent Vero cells in 24-well plates were inoculated with 100 μl of a serial dilution of each virus at 37°C. After 60 min, 1 ml of 0.8% agarose was added to each well, and the plates were incubated in a CO2 incubator at 37°C for 1 week. The plaques were visualized by addition of agarose containing neutral red. For determination of virus particle counts, aliquots of RSV A and B virus stocks were enumerated by electron microscopy (EM) (Advanced Biotechnologies Inc., Columbia, Md.). The titers of our RSV A and B stocks were determined to be 4 × 106 and 3 × 106 PFU/ml, respectively. The particle counts of our RSV A and B stocks were 7.6 × 108 and 1.02 × 109/ml, respectively. The wild-type influenza viruses A/Sydney/5/97(H3N2), A/New Caledonia/20/99(H1N1), and B/Yamanashi/166/98 were propagated in embryonated eggs at 37°C.
Clinical specimens.
Nasopharyngeal aspirates were collected in 10 ml of Viral Transport Medium (Becton Dickinson, Sparks, Md.) from 175 children with acute respiratory tract infections at the Prince of Wales Hospital in Hong Kong during the winter season of 2000 and 2001. The specimens were immediately processed for virus isolation and IF at the virology laboratory of the Prince of Wales Hospital.
Aliquots of specimens, to which RNAlater (Ambion, Austin, Tex.) was added upon collection, were stored at −70°C and then shipped to the United States in the frozen state (−70°C). The culture methods for the detection of all viruses, including RSV, in clinical respiratory specimens were described previously (27). For detection of the viral antigen of RSV by IF, the cells from the clinical specimens were washed, concentrated, and spotted on slides. The slides were then stained using IMAGEN (Dako Diagnostics, Ely, United Kingdom) and analyzed as described previously (27).
Nasal specimens of asymptomatic subjects were collected from healthy adult volunteers in the United States between March and September 2001.
RNA extraction.
Viral RNA was extracted from 70 μl of clinical specimen, tissue culture supernatant, or allantoic fluid using the QIAamp viral RNA minikit (Qiagen, Chatsworth, Calif.) and following the manufacturer's instructions.
Design of primer-probe pairs.
The assay was designed to serve two purposes: (i) detection of a broad range of RSV strains and (ii) differentiation of RSV subgroups A and B. The N gene of RSV was chosen as the target because it is one of the most conserved genes in the RSV genome. The nucleotide sequences from this gene could possibly recognize a large variety of RSV strains in clinical specimens. In addition, 6 nucleotides in the N gene (Table 1) differ substantially between subgroups A and B (15). The probes were designed to contain these 6 nucleotides, thus providing subgroup specificity. The primers were also designed to be subgroup specific in order to enhance the assay specificity. After comparison of the sequences of the N genes from four strains of RSV A and four strains of RSV B (14, 15), two oligonucleotide primer-probe pairs were selected using Primer Express Software (Applied Biosystems, Foster City, Calif.). The nucleotide sequences of the primers (A21, A102, B17, and B120) and probes (APB48 and BPB45) are shown in Table 1. The primers were obtained from Invitrogen Life Technology (Carlsbad, Calif.), and the probes were synthesized by Applied Biosystems. Both probes contained oligonucleotides with the 5′ reporter dye 6-carboxyfluorescein and the 3′ quencher dye 6-carboxytetramethylrhodamine.
TABLE 1.
Oligonucleotide sequences for primers and probes
| RSV subgroup (target) | Primer or probea | Sequenceb | Nucleotide positions |
|---|---|---|---|
| A (N gene) | A21 | 5′ GCTCTTAGCAAAGTCAAGTTGAATGA | 19-45 |
| A102 | 5′ TGCTCCGTTGGATGGTGTATT | 81-101 | |
| APB48 | 5′ ACACTCAACAAAGATCAACTTCTGTCATCCAGC | 47-79 | |
| B (N gene) | B17 | 5′ GATGGCTCTTAGCAAAGTCAAGTTAA | 15-41 |
| B120 | 5′ TGTCAATATTATCTCCTGTACTACGTTGAA | 90-119 | |
| BPB45 | 5′ TGATACATTAAATAAGGATCAGCTGCTGTCATCCA | 43-77 |
Real-time PCR.
A 50-μl reaction mixture contained 5 μl of the purified RNA, 1.25 μl of multiscribe (Applied Biosystems) with RNase inhibitor, 25 μl of TaqMan one-step RT-PCR master mix including Rhodamine X as a passive reference, 0.5 μM primer, and 0.2 μM probe as suggested by the manufacturer. The real-time RT-PCRs were carried out in an ABI Prism 7700 sequence detection system (Applied Biosystems). RT was performed at 48°C for 30 min. This was followed by 10 min at 95°C, which activated the AmpliTaq DNA polymerase, and then by 40 cycles of 15 s at 95°C and 1 min at 60°C, which amplified the RT products.
During amplification, the ABI Prism sequence detector monitored fluorescence emissions at every thermal cycle. Rhodamine X was used as a passive reference to which the signal of the reporter dye was normalized during data analysis, which reduced non-PCR-related fluorescence fluctuation from well to well. The threshold cycle (CT) represents the cycle at which significantly increased fluorescence is first detected.
Standard precautions were taken throughout the PCR process to avoid cross-contamination. Negative controls and serial dilutions of positive controls were included in every PCR assay. All PCR procedures were performed in a well-designed PCR suite (5, 19).
RESULTS
Specificity.
The specificity of the TaqMan RT-PCR assay was evaluated by testing the method against viral RNAs derived from various sources. These included five laboratory virus stocks, which were prepared as described in Materials and Methods. Two of the laboratory stocks were RSV (A2 and 2B), and three were influenza virus [A/New Caledonia/20/99 (H1N1), A/Sydney/5/97(H3N2), and B/Yamanashi/166/98]. Additionally, 94 viruses derived from 175 well-characterized clinical respiratory specimens, described previously, were examined (27). They included adenovirus types 1, 3, 5, and 21; cytomegalovirus; coxsackie virus A9 and A16; enterovirus; herpesvirus type 1; influenza virus A and B; parainfluenza virus types 1, 2, and 3; poliovirus type 1; and RSV A and B. When the viruses were analyzed by TaqMan PCR, the results showed that all of the RSV strains and none of the other viruses were detected among the various laboratory stocks, as well as clinical respiratory specimens. In addition, no fluorescence signal was observed from RNA extracted from 12 nasopharyngeal aspirates obtained from asymptomatic subjects (data not shown).
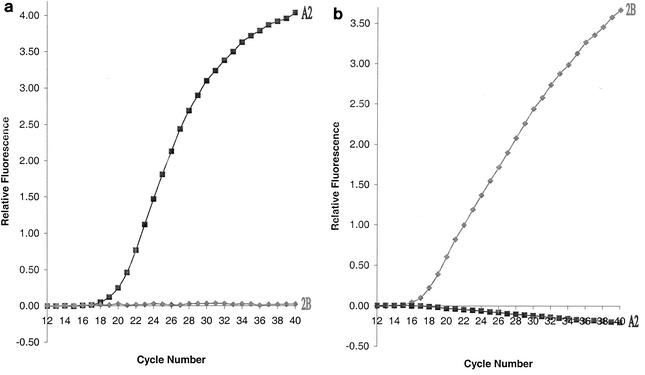
Fluorescent signal was observed only in the presence of RNA derived from the RSV A2 strain and the subgroup A-specific primer-probe pair (Fig. 1a). Similarly, the subgroup B-specific primer-probe pair detected only the RNA derived from the RSV 2B strain (Fig. 1b). Fluorescence was not observed in the negative controls, which contained no RNA (Fig. 2). To further confirm that the TaqMan primer-probe pairs are RSV and subgroup specific, nucleotide sequence analysis of the G gene (14) was performed for the RSV A and B virus stocks, as well as for 14 clinical isolates comprising 9 RSV A and 5 RSV B isolates. The results of sequence analysis confirmed that all the viruses corresponded to the subgroups predicted by the real-time RT-PCR assay (data not shown).
FIG. 1.
Specificity of RSV primer-probe pairs. RSV A primer pair A21-A102 and probe APB48 (a) and RSV B primer pair B17-B120 and probe BPB45 (b) were employed to detect RSV A2 (▪) and 2B (⧫) RNAs.
FIG. 2.
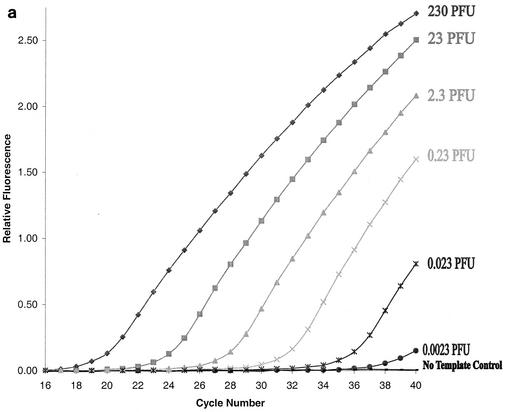
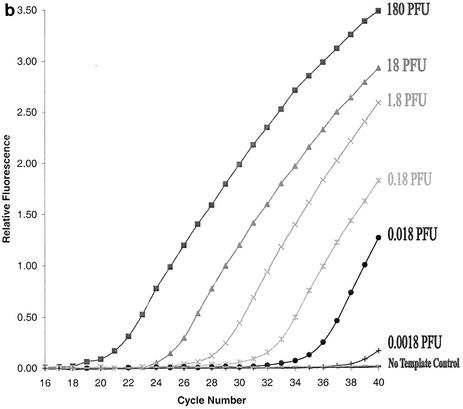
Sensitivity of the TaqMan RT-PCR assay. Tenfold serial dilutions of the RNA extracted from RSV A2 (0.0023 PFU to 230 PFU) (a) and RSV 2B (0.0018 PFU to 180 PFU) (b) were used in the PCR. Primer-probe pairs specific for RSV A (a) and B (b) were employed to detect the corresponding targets. The negative control contained RNase-free water instead of RNA template.
Quantification and sensitivity.
To measure the dynamic range of quantification and the sensitivity of our TaqMan PCR assay, two methods were used to quantify our RSV A and B virus stocks: (i) plaque assays and (ii) EM particle counts (see Materials and Methods). After virus quantification, viral RNA was extracted from 10-fold serial dilutions of each well-characterized virus stock and then amplified as described in Materials and Methods. The sensitivity of the TaqMan RT-PCR assay was determined for each RSV subgroup-specific primer-probe pair. Based on an 80% recovery of viral RNA in our routine extraction process (data not shown), detectable fluorescence signals were observed in RNA samples derived from 0.023 PFU of RSV A2 (Fig. 2a) or 0.018 PFU of RSV 2B (Fig. 2b), respectively. Based on EM particle counts, these PFU values correspond to two particles of RSV A2 and nine particles of RSV 2B. Thus, the sensitivity of our TaqMan PCR assay for RSV A and B was determined to be 0.023 PFU, or two copies of viral RNA, and 0.018 PFU, or nine copies of viral RNA, respectively.
In a quantitative TaqMan PCR assay, there is an inverse linear relationship between the cycle number (CT), at which a significant increment of fluorescence signal is first detected and the logarithm of the initial number of target molecules (13). For RSV A, the CT values were plotted against 10-fold serial dilutions of the virus stock (Fig. 2a). The results showed that the assay was linear over a wide range of virus concentrations—from 2.3 × 10−3 to 2.3 × 102 PFU, with a coefficient of correlation of >0.999. Similar results were also obtained for RSV B (data not shown). Thus, our assay provides quantitative results over 6 orders of magnitude for both RSV A and RSV B.
Comparison of culture confirmation, immunofluorescence, and TaqMan RT-PCR for detection of RSV in clinical specimens.
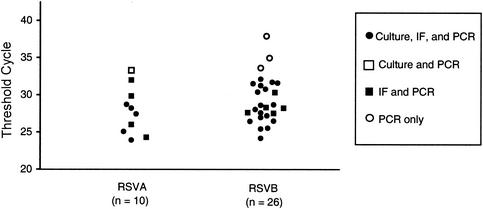
A total of 175 nasopharyngeal aspirates were collected from children with respiratory symptoms in Hong Kong during the winter of 2000 and 2001. For diagnosis of RSV infection, these specimens were tested independently by culture confirmation and IF at the virology laboratory of the Prince of Wales Hospital in Hong Kong and by TaqMan PCR at Wyeth, Pearl River, N.Y. Table 2 shows that, overall, RSV was identified in a total of 36 (21%) of the 175 specimens. Of these 36 RSV-positive specimens, 21 (58%) were detected by culture confirmation, 32 (89%) were detected by IF, and 36 (100%) were detected by PCR. The TaqMan RT-PCR assay identified all the positives that were identified by culture isolation or by IF. In addition, TaqMan RT-PCR was able to subgroup the 36 RSV-positive samples. Among them, 10 (28%) were identified as RSV A and 26 (72%) were identified as RSV B. The results indicated that all three methods were found suitable for analyzing clinical specimens containing moderate to high viral loads. IF and PCR were also found suitable for specimens containing moderate to low levels of RSV (Fig. 3). Overall, we found that TaqMan RT-PCR was about 40% more sensitive than culture and 10% more sensitive than IF in this study. Our assay not only detected RSV but also subgrouped the virus simultaneously. When the assay was utilized to analyze 175 clinical respiratory specimens, the results showed that about 2.5 times more RSV B than RSV A was found to be circulating among children in Hong Kong during the winter of 2000 and 2001 (Table 2).
TABLE 2.
Comparison of culture, IF, and real-time RT-PCR for detection of RSV A and B in 175 clinical specimens
| No. of samples | Resulta
|
|||
|---|---|---|---|---|
| Culture | IF | TaqMan PCR
|
||
| RSV A | RSV B | |||
| 5 | + | + | + | − |
| 15 | + | + | − | + |
| 1 | + | − | + | − |
| 4 | − | + | + | − |
| 8 | − | + | − | + |
| 3 | − | − | − | + |
| 139 | − | − | − | − |
+, positive; −, negative. The total numbers positive were as follows: culture, 21; IF, 32; TaqMan PCR, 10 (RSV A) and 26 (RSV B).
FIG. 3.
RSV-positive clinical specimens analyzed by TaqMan PCR. The samples were collected and analyzed as described in Materials and Methods. The threshold cycle (CT) represents the cycle number at which a significant increment of fluorescence signal is first detected. Each symbol represents the CT number for an individual specimen.
DISCUSSION
In this report, we describe the development and application of a rapid TaqMan-based RT-PCR assay not only for detection of RSV A and RSV B genomes but also for simultaneously subgrouping these viruses in clinical specimens. The assay is highly specific and sensitive and can provide results in ∼3 h. In contrast to conventional PCR, the TaqMan PCR assay generates fluorescence-labeled PCR products, which are measured and recorded during PCR cycling, thus eliminating time-consuming post-PCR processes, such as gel electrophoresis, blotting, and hybridization (26). Elimination of post-PCR processing increases the assay rapidity and reduces the chances of cross-contamination, thus eliminating false-positive results. Furthermore, the fluorescence signals of TaqMan PCR assays are derived from the total volume of a PCR mixture, whereas the signals of conventional RT-PCR are usually obtained from the gel electrophoresis of a fraction of a PCR product. Our TaqMan RT-PCR assay was able to detect RSV in the range of 1.8 to 2.3 PFU/ml. These detection limits are comparable to the sensitivity of 3.3 PFU/ml reported for the commercial Hexaplex PCR kit, which is based on conventional PCR methodology (17). In addition, the TaqMan PCR assay can exhibit enhanced specificity compared with conventional PCR because it employs one additional highly specific probe in the middle of the amplicon.
Our findings demonstrate that the TaqMan assay is more sensitive than culture confirmation. One major reason for lower recovery rates with culture detection is the lability of RSV that may be caused by improper handling of clinical specimens, as well as the variability inherent in specimen acquisition. TaqMan RT-PCR can overcome these problems by measuring the total number of viral genomes irrespective of virus viability. A combination of PCR and culture confirmation methods, however, provides a useful means of assessing the quality of clinical specimens.
To overcome the issue of false-positive results in PCR assays, we implemented the following precautions: (i) negative controls (∼5% of the total number of samples undergoing analysis), containing no RNA template, were included in every assay; (ii) nasal specimens from 12 asymptomatic subjects were collected outside the winter season and assessed by our TaqMan PCR assay. False positives were never detected among the negative controls or among specimens obtained from asymptomatic subjects.
Immunofluorescence is widely used for the quick detection of RSV in nasal aspirates (7, 20). In this study, the IF sensitivity is closer to that of PCR (Table 2), but it requires a skillful person to perform and analyze the results. These requirements hamper automation of the IF assay for use in high-throughput analyses of large numbers of clinical specimens. In addition, clinical respiratory specimens are often viscous and may not contain enough epithelial cells for examination, which is problematic for IF (20). In the case of TaqMan PCR, the addition of RNAlater to the clinical specimens appears not only to preserve the RNA in the clinical specimens but also to reduce sample viscosity and consequently ease sample manipulation. Furthermore, this method does not depend on the recovery of epithelial cells. Overall, TaqMan PCR offers automatic data analysis, which eliminates operator subjectivity and provides accurate subgrouping information.
In conclusion, we established a rapid, sensitive, and specific quantitative TaqMan-based real-time RT-PCR assay for simultaneous detection and subgrouping of RSV A and B viruses. The assay has the potential to operate in a high-throughput format when combined with the use of an automatic RNA isolation workstation, which allows results to be obtained within 6 h after the collection of specimens. Despite the suitability of traditional assays for the diagnosis of respiratory viral infections, the real-time RT-PCR assay provides a rapid and highly sensitive alternative for investigating RSV in clinical specimens obtained in surveillance, diagnostic, prophylactic, and therapeutic settings. Furthermore, this new option permits a more extensive examination of the relationships of viral load and RSV subgroup with disease progression or severity (4, 28).
Acknowledgments
We thank Giuseppe Palladino for providing wild-type influenza viruses and Fenglan Li and Ping Zhao for technical assistance and sample management.
REFERENCES
- 1.American Academy of Pediatrics, Committee on Infectious Diseases. 1996. Reassessment of the indications for ribavirin therapy in respiratory syncytial virus infections. Pediatrics 97:137-140. [PubMed] [Google Scholar]
- 2.American Academy of Pediatrics, Committee on Infectious Diseases and Committee on Fetus and Newborn. 1998. Prevention of respiratory syncytial virus infections: indications for the use of palivizumab and update on the use of RSV-IGIV. Pediatrics 102:1211-1216. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, L. J., J. C. Hierholzer, C. Tsou, R. M. Hendry, B. F. Fernie, Y. Stone, and K. McIntosh. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 151:626-633. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham, S. C., A. J. Bush, and J. P. Devincenzo. 2000. Nasal quantity of respiratory syncytial virus correlates with disease severity in hospitalized infants. Pediatr. Infect. Dis. J. 19:113-117. [DOI] [PubMed] [Google Scholar]
- 5.Cha, T., K. Kao, J. Zhao, P. E. Fast, P. M. Mendelman, and A. Arvin. 2000. Genotypic stability of cold-adapted influenza virus vaccine in an efficacy clinical trial. J. Clin. Microbiol. 38:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubie, H. A., J. M. Inglis, E. E. Leslie, A. T. Edmunds, and B. Totapally. 1992. Detection of respiratory syncytial virus in acute bronchiolitis in infants. J. Med. Virol. 38:283-287. [DOI] [PubMed] [Google Scholar]
- 7.Eugene-Ruellan, G., F. Freymuth, C. Bahloul, H. Badrane, A. Vabret, and N. Tordo. 1998. Detection of respiratory syncytial virus A and B and parainfluenza virus 3 sequences in respiratory tracts of infants by a single PCR with primers targeted to the l-polymerase gene and differential hybridization. J. Clin. Microbiol. 36:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falsey, A. R., M. A. Formica, and E. E. Walsh. 2002. Diagnosis of respiratory syncytial virus infection: comparison of reverse transcription-PCR to viral culture and serology in adults with respiratory illness. J. Clin. Microbiol. 40:817-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freymuth, F., G. Eugene, A. Vabret, J. Petitjean, E. Gennetay, J. Brouard, J. F. Duhamel, and B. Guillois. 1995. Detection of respiratory syncytial virus by reverse transcription-PCR and hybridization with a DNA enzyme immunoassay. J. Clin. Microbiol. 33:3352-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glezen, P., and F. W. Denny. 1973. Epidemiology of acute lower respiratory disease in children. N. Engl. J. Med. 288:498-505. [DOI] [PubMed] [Google Scholar]
- 11.Hall, C. B., J. T. McBride, C. L. Gala, S. W. Hildreth, and K. C. Schnabel. 1985. Ribavirin treatment of respiratory syncytial viral infection in infants with underlying cardiopulmonary disease. JAMA 254:3047-3051. [PubMed] [Google Scholar]
- 12.Halstead, D. C., S. Todd, and G. Fritch. 1990. Evaluation of five methods for respiratory syncytial virus detection. J. Clin. Microbiol. 28:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, P. R., and P. L. Collins. 1989. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J. Gen. Virol. 70:1539-1547. [DOI] [PubMed] [Google Scholar]
- 16.Johnston, S. L., and C. S. Siegel. 1990. Evaluation of direct immunofluorescence, enzyme immunoassay, centrifugation culture, and conventional culture for the detection of respiratory syncytial virus. J. Clin. Microbiol. 28:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehl, S. C., K. J. Henrickson, W. Hua, and J. Fan. 2001. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol. 39:1696-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. W., J. O. Arrobio, C. D. Brandt, B. C. Jeffries, G. Pyles, J. L. Reid, R. M. Chanock, and R. H. Parrott. 1973. Epidemiology of respiratory syncytial virus infection in Washington, DC. I. Importance of the virus in different respiratory tract disease syndromes and temporal distribution of infection. Am. J. Epidemiol. 98:216-225. [DOI] [PubMed] [Google Scholar]
- 19.Kitchin, P. A., and J. S. Bootman. 1993. Quality control of the polymerase chain reaction. Rev. Med. Virol. 3:107-114. [Google Scholar]
- 20.Landry, M. L., and D. Ferguson. 2000. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J. Clin. Microbiol. 38:708-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters, H. B., K. O. Weber, J. R. Groothuis, C. G. Wren, and B. A. Lauer. 1987. Comparison of nasopharyngeal washings and swab specimens for diagnosis of respiratory syncytial virus by EIA, FAT, and cell culture. Diagn. Microbiol. Infect. Dis. 8:101-105. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza, J., A. Rojas, J. M. Navarro, C. Plata, and M. de la Rosa. 1992. Evaluation of three rapid enzyme immunoassays and cell culture for detection of respiratory syncytial virus. Eur. J. Clin. Microbiol. Infect. Dis. 11:452-454. [DOI] [PubMed] [Google Scholar]
- 23.Mufson, M. A., H. D. Levine, R. E. Wasil, H. E. Mocega-Gonzalez, and H. E. Krause. 1973. Epidemiology of respiratory syncytial virus infection among infants and children in Chicago. Am. J. Epidemiol. 98:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mufson, M. A., C. Orvell, B. Rafnar, and E. Norrby. 1985. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 66:2111-2124. [DOI] [PubMed] [Google Scholar]
- 25.Paton, A. W., J. C. Paton, A. J. Lawrence, P. N. Goldwater, and R. J. Harris. 1992. Rapid detection of respiratory syncytial virus in nasopharyngeal aspirates by reverse transcription and polymerase chain reaction amplification. J. Clin. Microbiol. 30:901-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweiger, B., I. Zadow, R. Heckler, H. Timm, and G. Pauli. 2000. Application of afluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J. Clin. Microbiol. 38:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung, R. Y., J. Yin, S. J. Oppenheimer, J. S. Tam, and J. Lau. 1993. Treatment of respiratory syncytial virus infection with recombinant interferon alfa-2a. Arch. Dis. Child. 69:440-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh, E. E., K. M. McConnochie, C. E. Long, and C. B. Hall. 1997. Severity of respiratory syncytial virus infection is related to virus strain. J. Infect. Dis. 175:814-820. [DOI] [PubMed] [Google Scholar]