Abstract
In order to implement a new and reliable method for characterizing different species of Legionella, a genetic fingerprinting study with an automated ribotyping system (RiboPrinter) was completed with members of this genus which were deposited at the American Type Culture Collection. The RiboPrinter examined the different patterns of EcoRI digestion fragments from the rRNA operons of 110 strains, representing 48 of the 49 described Legionella species as well as 70 serogroups of those species. Distinctive and consistent patterns were obtained for the type strains of the 48 species investigated. Legionella pneumophila subsp. fraseri and L. pneumophila subsp. pascullei each generated a specific pattern, whereas L. pneumophila subsp. pneumophila produced six different fingerprint patterns. No correlation seemed to exist between the ribotypes obtained and the 15 serotypes of L. pneumophila. For the other species, those with two known serogroups presented two distinctive patterns with the RiboPrinter with the exception of L. hackeliae and L. quinlivanii, which yielded only one pattern. We also encountered ribotypes for strains which were not identified to the species level. The ribotypes generated for these strains with the RiboPrinter did not match those generated for known type strains, suggesting the putative description of new serogroups or species. Although the automated system did not have sufficient discriminatory ability to serve as an epidemiological tool in a clinical setting, it appeared to be a powerful tool for general genomic analysis of the Legionella isolates (e.g., determination of new species) and assessment of the interrelationship among Legionella strains through the RiboPrinter database connection.
Since the first isolation of Legionella pneumophila, the causative agent of Legionnaires' disease, nearly 30 years ago (37), members of this genus have been isolated from a wide range of environments and geographical locations (17, 20). To date, members of the family Legionellaceae comprise 49 described species (Deutsche Sammlung von Mikroorganismen und Zellkulturen website [ftp://ftp.dsmz.de/pub/DSMZ/bactnom/bactname.pdf]), with 15 serogroups (Sgs) described for L. pneumophila; 2 Sgs each described for L. bozemanii, L. longbeachae, L. feeleii, L. hackeliae, L. sainthelensi, L. spiritensis, L. erythra, and L. quinlivanii; and a single Sg each described for the remaining species (3). Over the years, several species of Legionella that were initially isolated from environmental sources but that were not implicated as etiological agents have later been shown to be human pathogens (9, 22, 25, 35). Approximately 70 to 90% of Legionella infections are caused by L. pneumophila Sgs 1 and 6, and others species are responsible for between 5 and 30% of the cases of infection (18, 42). Nineteen species have been recognized to be pathogenic for humans (14), causing pneumonia (Legionnaires' disease) (11, 19), a mild febrile disease (Pontiac fever) (31, 55), and most recently, soft tissue abscesses (25). Pneumonia caused by Legionella is becoming a public health problem, since this organism has the potential to cause large outbreaks (1, 40) and to infect young immunocompetent adults (51) or newborns after water birth (21). Legionnaires' disease, if left untreated, leads to an average mortality rate of 15% (16).
Various methods for typing of members of the family Legionellaceae have been developed in the past. These include antibiotic susceptibility testing (54), plasmid analysis (47), fatty acid profiling (29), multilocus enzyme electrophoresis (45), pulsed-field gel electrophoresis (39), and various DNA fingerprinting protocols by PCR (14, 34, 41, 53). Most of them have focused specifically on the subtyping of L. pneumophila Sgs 1 and 6 because these Sgs are responsible for the majority of legionellosis cases.
At present, the majority of the Legionella isolates are detected and typed by serological designation (3, 30, 36, 49). This has been satisfactory for the most commonly occurring species and serovars. However, antisera are not available commercially for many of the less well known species. Immunological cross-reactions among some species have also been reported to be troublesome (3, 6, 33, 49). With the increasing number of described Legionella species, methods based on serology will become more difficult and cumbersome to use in environmental and clinical studies.
Ribotyping, a molecular method based on the analysis of the restriction fragment length polymorphisms (RFLPs) of rRNA genes (23), was also used to characterize Legionella strains (2, 52). The feasibility of ribotyping for the differentiation of Legionella species was initially tested by Grimont et al. (24) with 28 members of the Legionellaceae family.
To further investigate this approach, we tested a larger number of Legionella species using an automated ribotyping system. A total of 110 strains representing 48 species and 3 subspecies, as well as a newly reported Sg for L. londiniensis (Sg 2; F. Lo Presti [Centre National de Référence des Légionelles, Lyon, France], personal communication to the American Type Culture Collection [ATCC]) were included in the study. L. lytica (28) is a symbiont of amoeba and requires cocultivation with the host, so it was not feasible to include a culture of this isolate for testing. The results of that study are presented in this paper.
MATERIALS AND METHODS
Cultures, media, and growth conditions.
A total of 110 Legionella strains comprising 48 species, including 3 subspecies and 70 Sgs deposited at ATCC, were used for this study (Table 1). Each culture was grown in an atmosphere of 5% CO2 at 37°C for 24 to 48 h on ATCC 1099 Charcoal Yeast Extract buffered medium (the medium formulation is described at the website http://www.atcc.org/SearchCatalogs/MediaFormulations.cfm).
TABLE 1.
Legionella strains included in the studya
| Species | Subspecies | Sg | Other | Source | Original designation | ATCC no. |
|---|---|---|---|---|---|---|
| L. adelaidensis | 1 | TS | Cooling tower water, Adelaide, Australia | 1762-AUS-E | 49625 | |
| L. anisa | 1 | TS | Hot water, sink, Los Angeles, Calif. | WA-316-C3 | 35292 | |
| L. anisa | Sink faucet, Chicago, Il. | CH-47-C3 | 35291 | |||
| L. anisa | Sink faucet, Chicago, Il. | CH-47-C1 | 35290 | |||
| L. beliardensis | 1 | TS | Water from a calorifier in reanimation unit, Montbeliard, France | Montbeliard A1 | 700512 | |
| L. birminghamensis | 1 | TS | Lung biopsy, cardiac transplant recipient, Alabama | 1407-AL-H | 43702 | |
| L. birminghamensis | Water near Clermont-Ferrand, France | CF VII no. 3A | 700709 | |||
| L. bozemanii | 1 | TS | Lung tissue, pneumonia, Key West, Fla. | WIGA | 33217 | |
| L. bozemanii | 2 | RS | Human lung aspirate, Toronto, Ontario, Canada | Toronto 3 | 35545 | |
| L. brunensis | 1 | TS | Cooling tower water, Brno, Czechoslovakia | 444-1 | 43878 | |
| L. cherrii | 1 | TS | Thermally altered water, Michigan | ORW | 35252 | |
| L. cincinnatiensis | 1 | TS | Lung tissue, pneumonia, Cincinnati, Ohio | 72-OH-H | 43753 | |
| L. drozanskii | 1 | TS | Tank of well water, Leeds, United Kingdom | LLAP-1 | 700990 | |
| L. dumoffii | 1 | TS | Water in cooling tower, New York, N.Y. | NY 23 | 33279 | |
| L. dumoffii | Human lung, Los Angeles, Calif. | Wadsworth 81-782A | 35850 | |||
| L. dumoffii | Thermal spa water | A3a3F | 700714 | |||
| L. erythra | 1 | TS | Water in cooling tower, Seattle, Wash. | SE-32A-C8 | 35303 | |
| L. erythra | 2 | RS | Strain isolated in Paris, France | LC217 | BAA-536 | |
| L. fairfieldensis | 1 | TS | Cooling tower water, Fairfield, Victoria, Australia | 1725-AUS-E | 49588 | |
| L. fallonii | 1 | TS | Ship air-conditioning system, United Kingdom | LLAP-10 | 700992 | |
| L. feeleii | 1 | TS | Grinding machine coolant fluid, Windsor, Ontario, Canada | WO-44C-C3 | 35072 | |
| L. feeleii | 2 | RS | Human lung tissue, Wisconsin, Wis. | 691-WI-H | 35849 | |
| L. feeleii | 1 | Bronchoalveolar lavage, pneumonia, Savoy, France | Ly126.92b | 700513 | ||
| L. feeleii | 1 | Bronchoalveolar lavage, pneumonia and HIV, Lyon, France | Ly166.96 | 700514 | ||
| L. geestiana | 1 | TS | Hot-water tap, Geest Office building, London, United Kingdom | 1308 | 49504 | |
| L. genomospecies 1 | Cooling-water tower, Adelaide, Australia | 2055-AUS-E | 51913 | |||
| L. gormanii | 1 | TS | Soil from a creek bank, Atlanta, Ga. | LS-13 | 33297 | |
| L. gormanii | Bronchial brush, Pneumonia, Calif. | 86A5796 | 43769 | |||
| L. gratiana | 1 | TS | Thermal spa water, Savoy region, France | Lyon-8420412 | 49413 | |
| L. gresilensis | 1 | TS | Water from shower in a thermal spa. Gréoux, France | Gréoux 11 D13 | 700509 | |
| L. gresilensis | Water sample from a potash mine near Mulhouse, France | Mulhouse 12 A23 | 700759 | |||
| L. hackeliae | 1 | TS | Human bronchial biopsy specimen, Ann. Arbor., Mich. | Lansing 2 | 35250 | |
| L. hackeliae | 2 | RS | Human lung aspirate, Pittsburgh, Pa. | 798-PA-H | 35999 | |
| L. israelensis | 1 | TS | Water, Israel | Bercovier 4 | 43119 | |
| L. jamestowniensis | 1 | TS | Wet soil, Janestown, N.Y. | JA-26-G16-E2 | 35298 | |
| L. jordanis | 1 | TS | Jordan River, Bloomington, Ind. | BL-540 | 33623 | |
| L. jordanis | Patient with pneumonia, France | Ly95.90 | 700762 | |||
| L. lansingensis | 1 | TS | Bronchial aspirate pneumonia and leukemia, Lansing, Mich. | 1677-MI-H | 49751 | |
| L. londiniensis | 1 | TS | Office building cooling tower, London, United Kingdom | 1477 | 49505 | |
| L. londiniensis | 2 | RS | Water, Mulhouse, France | Mulhouse B26 | 700510 | |
| L. longbeachae | 1 | TS | Human lung, pneumonia, Long Beach, Calif. | Long Beach 4 | 33462 | |
| L. longbeachae | 2 | RS | Human lung, Tucker, Ga. | Tucker 1 | 33484 | |
| L. maceachernii | 1 | TS | Water in home evaporator cooler, Phoeniz, Ariz. | PX-1-G2-E2 | 35300 | |
| L. micdadei | 1 | TS | Human blood, pneumonia, Fort Bragg, Calif. | TATLOCK | 33218 | |
| L. micdadei | Lung tissue, pneumonia, Pittsburgh, Pa. | EK | 33204 | |||
| L. micdadei | Transtracheal aspirate, Pittsburgh, Pa. | VAMC-MCC | 33344 | |||
| L. micdadei | Showerhead, Pittsburgh, Pa. | VAM-7W | 33345 | |||
| L. micdadei | Ultrasonic nebulizer, Pittsburgh, Pa. | VAM-PGH-12 | 33346 | |||
| L. moravica | 1 | TS | Cooling-tower water, Jihlava, Czechoslovakia | 316-36 | 43877 | |
| L. nautarum | 1 | TS | Domestic hot-water tap, Greenwich, London, United Kingdom | 1224 | 49506 | |
| L. oakridgensis | 1 | TS | Industrial cooling-tower water, Pennsylvania | Oak Ridge 10 | 33761 | |
| L. oakridgensis | Bronchoalveolar lavage, pneumonia, Nantes, France | Nantes-930101868 | 700515 | |||
| L. oakridgensis | Bronchoalveolar lavage, pneumonia, Nantes, France | Nantes-930101937 | 700516 | |||
| L. parisiensis | 1 | TS | Water in cooling tower, Paris, France | PF-209C-C2 | 35299 | |
| L. parisiensis | Tracheal aspirate, liver transplant, France | FLP2 | 700174 | |||
| L. pneumophila | fraseri | 4 | TS | Human lung, pneumonia, Los Angeles, Calif. | Los Angeles-1 | 33156 |
| L. pneumophila | fraseri | 5 | RS | Cooling tower, Dallas, Tex. | Dallas 1E | 33216 |
| L. pneumophila | fraseri | 15 | RS | Human lung, fatal pneumonia, Royal Oak, Mich. | Lansing 3 | 35251 |
| L. pneumophila | pascullei | TS | Water from showerhead, Pittsburgh, Pa. | U8W | 33737 | |
| L. pneumophila | pascullei | 13 | RS | Water from showerhead, Pittsburgh, Pa. | U7W | 33736 |
| L. pneumophila | pascullei | Tap water, Pittsburgh, Pa. | MICU B | 33735 | ||
| L. pneumophila | pneumophila | 1 | TS | Human lung, pneumonia, Philadelphia, Pa. | Philadelphia-1 | 33152 |
| L. pneumophila | pneumophila | 1 | Tap water, Pittsburgh, Pa. | 684 | 33733 | |
| L. pneumophila | pneumophila | 1 | Tap water, Pittsburgh, Pa. | 687 | 33734 | |
| L. pneumophila | pneumophila | 2 | RS | Human lung, Togus, Maine | Togus-1 | 33154 |
| L. pneumophila | pneumophila | 3 | RS | Creek water, Bloomington, Ind. | Bloomington-2 | 33155 |
| L. pneumophila | pneumophila | 6 | RS | Human lung biopsy specimen, Chicago, Ill. | Chicago 2 | 33215 |
| L. pneumophila | pneumophila | 8 | RS | Human lung, Concord, Calif. | Concord 3 | 35096 |
| L. pneumophila | pneumophila | 9 | RS | Tap water, Leden, Holland | IN-23-G1-C2 | 35289 |
| L. pneumophila | pneumophila | 11 | RS | Human endotracheal tube, Pittsburgh, Pa. | 797-PA-H | 43130 |
| L. pneumophila | pneumophila | 10 | RS | Respiratory tract secretions, Holland | Leiden 1 | 43283 |
| L. pneumophila | pneumophila | 12 | RS | Human lung, pneumonia, Denver, Col. | 570-CO-H | 43290 |
| L. pneumophila | pneumophila | 14 | RS | Bronchial aspirate, pneumonia, Minn. | 1169-MN-H | 43703 |
| L. pneumophila | pneumophila | 13 | RS | Lung aspirate, pneumonia, Calif. | B2A3105 | 43736 |
| L. pneumophila | 1 | Human lung, Knoxville, Tenn. | Knoxville-1 | 33153 | ||
| L. pneumophila | 7 | RS | Showerhead, Illinois | Chicago 8 | 33823 | |
| L. pneumophila | 1 | CDC-Quebec | Allentown 1 | 43106 | ||
| L. pneumophila | 1 | CDC-Quebec | Heysham 1 | 43107 | ||
| L. pneumophila | 1 | CDC-Quebec | Benidorm 030 E | 43108 | ||
| L. pneumophila | 1 | CDC-Quebec | OLDA | 43109 | ||
| L. pneumophila | 1 | CDC-Quebec | France 5811 | 43112 | ||
| L. pneumophila | 1 | CDC-Quebec | Camperdown 1 | 43113 | ||
| L. pneumophila | Clinical sample, State Health Department California | RIO | 43660 | |||
| L. pneumophila | Hospital faucet, Pittsburgh Veterans Affairs Medical Center, Pittsburgh, Pa. | 11EJ | 43661 | |||
| L. pneumophila | Hospital faucet, Calif. | FAUC 19 | 43662 | |||
| L. pneumophila | 1 | Bronchoalveolar lavage fluid, pneumonia, France | CA1 | 700711 | ||
| L. pneumophila | 1 | Transtracheal aspirate, 1978 | F1724 | BAA-74 | ||
| L. quateirensis | 1 | TS | Shower in hotel bathroom, Quarteira, Portugal | 1335 | 49507 | |
| L. quinlivanii | 1 | TS | Water in bus air conditioner, Australia | 1442-AUS-E | 43830 | |
| L. quinlivanii | 2 | RS | Cooling-tower pond, London, United Kingdom | LC870 | BAA-538 | |
| L. rowbothamii | 1 | TS | Water and sludge from an industrial liquifier tower, United Kingdom | LLAP-6 | 700991 | |
| L. rubrilucens | 1 | TS | Tap water, Los Angeles, Calif. | WA-270A-C2 | 35304 | |
| L. sainthelensi | 1 | TS | Spring water, Mt. St. Helens, Wash. | MSH-4 | 35248 | |
| L. sainthelensi | 2 | RS | Human bronchial washings, pneumonia, Calif. | 1489-CA-H | 49322 | |
| L. santicrucis | 1 | TS | Tap water, St. Croix, U.S. Virgin Islands | SC-63-C7 | 35301 | |
| L. shakespearei | 1 | TS | Cooling-tower water, Stratford upon Avon, United Kingdom | 214 | 49655 | |
| L. species | Water from a well, Montpellier, France | IB V no 3 | 700511 | |||
| L. species | Water, Bourbonne-les-Bains, France | Nancy II no. 1 | 700703 | |||
| L. species | Water, La Rochelle, France | La Rochelle A2.1 | 700705 | |||
| L. species | Water, Bourbonne-les-Bains, France | Nancy II no. 3 | 700706 | |||
| L. species | Environmental isolate, Venissieux, France | IBV no. 2 | 700761 | |||
| L. species | Clinical isolate, France | ParisB1 | 700833 | |||
| L. spiritensis | Sg1 | TS | Spirit Lake, Mt. St. Helens, Wash. | MSH-9 | 35249 | |
| L. spiritensis | Sg2 | RS | Cooling tower, United Kingdom | ML 76 | BAA-537 | |
| L. steigerwalii | TS | Tap water, St. Croix, U.S. Virgin Islands | SC-18-C9 | 35302 | ||
| L. taurinensis | Sg1 | TS | Water from a hospital oxygen bubble humidifier, Turin, Italy | Turin no 1 | 700508 | |
| L. tucsonensis | TS | Pleural fluid, renal transplant, Tucson, Ariz. | 1087-AZ-H | 49180 | ||
| L. wadsworthii | Sg1 | TS | Human sputum, pneumonia, Los Angeles, Calif. | Wadsworth 81-716A | 33877 | |
| L. waltersii | Sg1 | TS | Drinking water distribution system, Adelaide, Australia | 2074-AUS-E | 51914 | |
| L. worsleiensis | Sg1 | TS | Industrial cooling tower, Worsley, United Kingdom | 1347 | 49508 |
TS, type strain; RS, reference strain; CDC, Centers for Disease Control and Prevention HIV, human immunodeficiency virus.
Sample preparation and processing.
All Legionella strains were characterized by use of the RiboPrinter system (Qualicon Inc., Wilmington, Del.) as described previously (13) with respect to the procedures and conditions recommended by the manufacturer (12, 46). EcoRI was used as the restriction enzyme. At the end of the process, a densitometric scan depicting the distributions and molecular weights of the restriction fragments was obtained for each sample analyzed. This output was saved in the RiboPrinter's computer. Ribotype groups (ribogroups) were defined by the RiboPrinter's proprietary algorithm, which compared the pattern of each isolate to those of others in the database and assigned groups by the differences in band number, position, and signal intensity. A given ribogroup was defined as a group of ribotypes with similarity values >0.93. Strains used for repeatability testing were analyzed by using the RiboPrinter's proprietary algorithm, which was a modified version of the coefficient of simple correlation (38). Similarity values obtained are reported in Table 2.
TABLE 2.
Strains used for repeatability testing
| Strain | ATCC no. | No. of repeated tests | Similarity valuesa |
|---|---|---|---|
| L. dumoffii | 33279 | 2 | 1.0, 0.99 |
| L. longbeachae | 43462 | 3 | 1.0, 0.99, 0.97 |
| L. lansingensis | 49751 | 3 | 1.0, 0.98, 0.97 |
| L. birminghamensis | 43702 | 4 | 1.0, 0.98, 0.98, 0.98 |
| L. feeleii | 35849 | 2 | 1.0, 0.95 |
| L. anisa | 35292 | 2 | 1.0, 0.95 |
| L. pneumophila subsp. fraseri | 33156 | 2 | 1.0, 0.96 |
| L. pnemophila subsp. pascullei | 33737 | 5 | 1.0, 0.99, 0.98, 0.96, 0.98 |
| L. cincinnatiensis | 43753 | 2 | 1.0, 0.98 |
| L. fallonii | 700992 | 2 | 1.0, 0.95 |
| L. gratiana | 49413 | 2 | 1.0, 0.98 |
| L. parisiensis | 700174 | 2 | 1.0, 0.96 |
| L. dumoffii | 700714 | 2 | 1.0, 0.95 |
| L. pneumophila subsp. pneumophila | 33152 | 3 | 1.0, 0.96, 0.95 |
| L. pneumophila subsp. pneumophila | 43283 | 2 | 1.0, 0.99 |
| L. pneumophila subsp. pneumophila | 43130 | 3 | 1.0, 0.99, 0.99 |
| L. pneumophila | BAA-74 | 3 | 1.0, 0.99, 0.98 |
| L. pneumophila subsp. pneumophila | 35289 | 6 | 1.0, 0.95, 0.97, 0.98, 0.97, 0.97 |
| L. cherrii | 33252 | 2 | 1.0, 0.98 |
| L. moravica | 43877 | 4 | 1.0, 0.99, 0.97, 0.97 |
| L. jordanis | 33623 | 2 | 1.0, 0.97 |
| L. quinlivanii | 43830 | 3 | 1.0, 1.0 |
| L. drosanskii | 700990 | 3 | 1.0, 0.99 |
| L. hackeliae | 35250 | 2 | 1.0, 0.99 |
| L. micdadei | 33218 | 2 | 1.0, 0.98 |
| L. maceachernii | 35300 | 6 | 1.0, 0.97, 0.98, 0.98, 0.97, 0.98 |
| L. bozemanii | 33217 | 7 | 1.0, 0.98, 0.98, 0.98, 0.99, 0.99, 0.96 |
| L. londinensis | 49505 | 4 | 1.0, 0.98, 0.97, 0.96 |
| L. nautarum | 49506 | 2 | 1.0, 0.98 |
| L. steigerwaltii | 35302 | 2 | 1.0, 0.99 |
Similarity values were generated by comparing all runs to the first one with the RiboPrinters proprietary algorithm (see Materials and Methods).
Ribotype analysis.
For each batch of eight samples, ribotypes were normalized to the positions of the molecular weight standards with Qualicon software. Computerized ribotypes were exported for analysis in .txt files and imported into BioNumerics software (version 2.5; Applied Maths, Sint-Martens-Latem, Belgium) by using the Qualicon macro. Clustering analysis was performed by the unweighted pair group method with arithmetic averages (UPGMA) method based on the Dice (15) coefficient for band matching, with a position tolerance setting of 1.0% (default values are 1% of position tolerance and 0.5% of optimization). Bands for analysis with the Dice coefficient were assigned manually, according to densitometric curves and the accompanying hard-copy photograph.
RESULTS
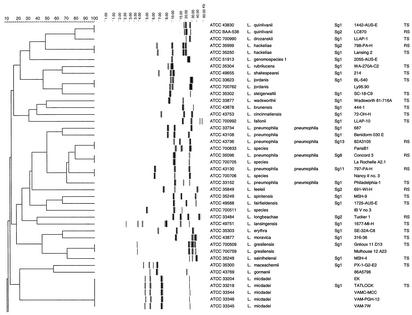
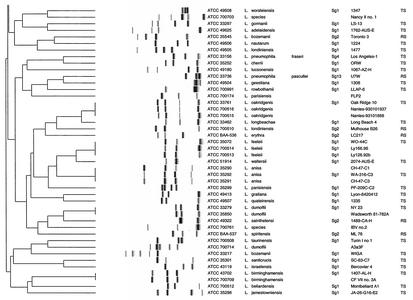
All 110 Legionella strains in this study could be processed with the RiboPrinter, resulting in 100% typeability. Two dendrograms were derived by using the BioNumerics software (version 2.5). Figure 1 presents the results for all L. pneumophila strains included in this study, while Fig. 2 presents the results for all other strains. The second dendrogram also included eight L. pneumophila strains (ATCC 43108, ATCC 33734, ATCC 43736, ATCC 35096, ATCC 33152, ATCC 43130, ATCC 33736, and ATCC 33156), which corresponded to the eight ribogroups displayed in Fig. 1.
FIG. 1.
Comparative analysis of the EcoRI ribotypes obtained with the RiboPrinter for the collection of L. pneumophila strains. Clustering was performed by the UPGMA method, and similarity analysis was based on the use of the Dice coefficient (see Materials and Methods). In the dendrogram scale, correlation levels were converted to percent homology levels. TS, type strain; RS, reference strain.
FIG. 2.
Comparative analysis of the EcoRI ribotypes obtained with the RiboPrinter for the ATCC collection of Legionella strains. Clustering was performed by the UPGMA method, and similarity analysis was based on the use of the Dice coefficient (see Materials and Methods). In the dendrogram scale, correlation levels were converted to percent homology levels. TS, type strain; RS, reference strain.
Pattern reproducibility was investigated by ribotyping of 30 isolates more than twice, which resulted in mean similarity values ranging from 0.95 to 1.00 (Table 2). The patterns obtained after EcoRI cleavage and probe hybridization contained two to six fragments (mainly three to four) in the range of 3 to 60 kb. Sixty-seven different patterns were generated for the 110 strains tested, and distinctive patterns were obtained for the type strains of 48 species.
Examination of 32 L. pneumophila strains allowed us to distinguish three separate clusters and eight different ribogroups within these clusters (Fig. 1). Two ribogroups, which clustered separately from each other and away from the major cluster of L. pneumophila, were formed by three strains each of L. pneumophila subsp. fraseri and L. pneumophila subsp. pascullei. We could distinguish six ribogroups within the major cluster of L. pneumophila strains. The first ribogroup was made up of ribotypes from three strains of L. pneumophila Sg 1 (ATCC 43106, ATCC 43108, ATCC BAA-74). The second ribogroup consisted of two strains of L. pneumophila subsp. pneumophila Sg 1 (ATCC 33733, ATCC 33734). The third ribogroup included identical ribotypes from nine strains (ATCC 35289, ATCC 43107, ATCC 43109, ATCC 43113, ATCC 43660, ATCC 43662, ATCC 43661, ATCC 43736, ATCC 700711), corresponding to three different Sgs (L. pneumophila Sg 1, L. pneumophila subsp. pneumophila Sg 9 and Sg 13). The fourth ribogroup comprised identical ribotypes from seven strains (ATCC 33155, ATCC 33153, ATCC 33823, ATCC 35096, ATCC 43112, ATCC 43283, ATCC 43703), corresponding to six Sgs (L. pneumophila Sg 1 and Sg 7, L. pneumophila subsp. pneumophila Sg 3, Sg 8, Sg 10, and Sg 14). Only four L. pneumophila subsp. pneumophila strains (ATCC 33152, ATCC 33154, ATCC 33215, ATCC 43290) made up of the fifth ribogroup, each with a different Sg (Sg 1, Sg 2, Sg 6, and Sg 12). The sixth ribogroup contained one strain of L. pneumophila subsp. pneumophila Sg 11 (ATCC 43130).
For 10 other species examined (Fig. 2) we were able to distinguish two ribogroups (L. feeleii, L. dumoffii) or ribotypes (L. bozemanii, L. erythra, L. londiniensis, L. longbeachae, L. parisiensis, L. sainthelensi, L. spiritensis, and L. gormanii). For the remaining 37 species, which included Legionella genomospecies 1 (ATCC 51913), a unique and distinctive ribogroup (L. micdadei, L. gresilensis, L. oakridgensis, L. anisa, L. hackeliae, L. quinlivanii, L. jordanis, and L. birminghamensis) or ribotype was observed for each species (Fig. 2). Finally, we also noted that three Legionella sp. strains (ATCC 700511, ATCC 700703, ATCC 700761) showed different fingerprint patterns which were not observed among other Legionella members.
DISCUSSION
The previous studies on the ribotyping of the members of the Legionellaceae family were done by traditional, time-consuming manual techniques and focused on a limited number of strains. In this study, we used an automated microbial genotyping system, the RiboPrinter (Qualicon), to investigate a large panel of Legionella species. We used the EcoRI restriction enzyme, which generated a number of fragments similar to the number observed by manual ribotyping of Legionella strains with various restriction enzymes (EcoRV, HindIII, and PstI) (2, 24). There was evidence that in Escherichia coli automated riboprints correlated well with the fingerprinting patterns generated by traditional methods (13). Furthermore, the EcoRI ribotypes obtained manually by Schoonmaker et al. (44) for L. pneumophila strains (ATCC 33153, ATCC 33152, ATCC 33216) were identical to the corresponding ribotypes obtained in the present study. The reproducibility of our data is demonstrated in Table 2. These results indicate that the RiboPrinter is a powerful device with excellent reproducibility, in addition to a high throughput capacity, which allows the analysis of 32 isolates per day.
The main dendrogram that was derived from our study indicated that each of the 48 type strains produced a distinctive and consistent fingerprint pattern (Fig. 2). This suggested that the patterns for Legionella strains obtained with the RiboPrinter could be used to identify new isolates by comparison to the patterns generated for known species. However, they may not be suitable for phylogenetic purposes, as our dendrogram did not agree with the phylogenetic tree that was generated by 16S rRNA analysis (28). By examination of Fig. 1, two separate clusters for L. pneumophila subsp. fraseri and L. pneumophila subsp. pascullei were clearly observed, while the patterns for all L. pneumophila subsp. pneumophila strains clustered in a common group. This is in agreement with results based on DNA hybridization (10) as well as those of a previous study showing that L. pneumophila subsp. fraseri could be separated from other subspecies of L. pneumophila by four restriction enzymes (3). We also noticed that there did not seem to be any correlation between the 15 serotypes of L. pneumophila and their patterns obtained with the RiboPrinter. This is in accordance with previous reports indicating that the separation of L. pneumophila into different Sgs has no apparent relation to the underlying genetic structure of the microorganism (27, 45). Furthermore, strains of Sg 1 were present in five of the six ribogroups of L. pneumophila. These results are in agreement with previous findings which indicated that L. pneumophila Sg 1 is a fairly heterogeneous group (3). In addition, we noticed three strains (ATCC 700706, ATCC 700705, ATCC 700833) deposited without species names clearly clustered with three different L. pneumophila ribogroups (Fig. 2), which strongly suggests that these strains belong to this species. These results should be further confirmed by other molecular tests such as 16S rRNA gene sequencing or DNA-DNA hybridization studies.
A few Legionella species (L. micdadei, L. gresilensis, L. oakridgensis, L. anisa, L. jordanis, L. birminghamensis) from various origins seemed to display genetic homogeneity within the species, exhibiting one ribogroup per species, as seen previously with the subspecies of L. pneumophila. L. hackeliae (ATCC 35250, ATCC 33216) and L. quinlivanii (ATCC 43830, ATCC BAA-538), with two serotypes each, also displayed this feature (one ribogroup per species), which is in agreement with earlier studies based on manual ribotyping analysis (8, 24).
On the other hand, L. bozemanii, L. erythra, L. londiniensis, L. longbeachae, L. parisiensis, L. sainthelensi, L. spiritensis, L. gormanii, L. dumoffii, and L. feeleii displayed remarkable genetic diversity. Homologies of less than 16% were consistently observed between ribogroups (L. feeleii, L. dumoffii) or ribotypes (L. bozemanii, L. erythra, L. londiniensis, L. longbeachae, L. parisiensis, L. sainthelensi, L. spiritensis, L. gormanii) within a given species. Interestingly, for seven of these species (L. bozemanii, L. erythra, L. londiniensis, L. longbeachae, L. sainthelensi, L. spiritensis, and L. feeleii), two serogroups have previously been described or reported (5, 7, 26, 43, 48, 50). For these species, each Sg seemed to be associated with a given ribotype or ribogroup (L. feeleii Sg 1). This observation is in agreement with previous findings based on manual ribotyping (24) and randomly amplified polymorphic DNA analysis (34). L. dumoffii, L. parisiensis, and L. gormanii each has a single Sg; however, two different fingerprint patterns were observed for each species. Regarding L. parisiensis ATCC 700174 (35), L. gormanii ATCC 43769 (22), and L. dumoffii ATCC 700714 (M. Molmeret [Centre National de Référence des Légionelles, Lyon, France], personal communication to ATCC), our results suggest that these strains may correspond to new putative serotypes, having less than 16% of homology within their own ribogroup. This, however, needs to be confirmed by further serological studies.
For all the other species which were represented in the study by only one strain, each type strain produced a distinctive and consistent identifying pattern.
Automated ribotyping may represent an alternative tool for determination of putative new species before labor-intensive techniques are needed. For example, ATCC 700509 and ATCC 700761 were deposited at ATCC as Legionella spp. and clearly showed two unequivocal and distinctive patterns in our study. Recently, these two strains were described as two novel species with the names L. gresilensis sp. nov. (type strain, ATCC 700509) and L. beliardensis sp. nov. (type strain, ATCC 700761) (32). In the same way, the RiboPrinter displayed three distinctive patterns for three Legionella strains (ATCC 700511, ATCC 700703, ATCC 700761) in our dendrogram (Fig. 2), and thus, these strains may possibly represent new species of Legionella or new Sgs of known species. Interestingly, ATCC 700511 has been reported to produce a specific randomly amplified polymorphic DNA pattern (34), and our results could be considered new data to support this strain as a new species. Nevertheless, further investigation by 16S rRNA gene sequencing and DNA homology studies are necessary before a conclusion should be made.
In the same way, ATCC 51913, reported as Legionella genomospecies 1, displayed a distinctive pattern with the RiboPrinter. This strain was related to L. quinlivanii Sg 2 serologically and to L. quinlivanii Sg 1 and Sg 2 genetically (4). However, the pattern obtained with the RiboPrinter clearly differed from those associated with L. quinlivanii Sg 1 and Sg 2. Thus, this strain could possibly be considered a new Sg of L. quinlivanii or a novel species.
The automated ribotyping system with EcoRI restriction digestion has been shown to be a powerful tool for general genomic analysis of Legionella isolates (e.g., determination of new species or serotypes within a given species). However, this method lacked the discriminatory power required for routine analysis of nosocomial etiological agents, as epidemiologically unrelated strains within a given species may present identical ribotypes. This could be illustrated by examination of the major cluster of L. pneumophila strains in which six ribogroups have been identified. Four ribogroups (ribogroups 1, 3, 4, and 5) contained epidemiologically unrelated strains, as shown by their different serotypes, as well as strains with identical or unknown serotypes which were most unlikely related due to their very different geographical origins or sources (Table 1). For example, the nine isolates of ribogroup 3 came from Holland, the Centers for Disease Control and Prevention-Quebec, California, and Pennsylvania. Strains within ribogroups 4 and 5 were isolated from different parts of the United States as well as other countries.
When our study was initiated, EcoRI was the only enzyme available for use with the instrument. In the past few years additional restriction enzymes have been added, which may improve the discriminatory ability of the system. Enzymes such as ClaI, NciI, PstI, and HindIII have already been tested for use in the ribotyping of the Legionellaceae family manually (2, 24, 44), but a combination of enzymes was needed for good differentiation among the species (2).
Conclusion.
Automated ribotyping can serve as a rapid and reproducible method for characterization of the members of the Legionellaceae family. Increased awareness of the diseases caused by Legionella has resulted in closer monitoring and investigation of potential sources of infection. This will no doubt increase the number of Legionella species being isolated and examined from environmental and clinical studies. Given its worldwide distribution and interconnection, the RiboPrinter system will enable immediate comparisons of ribotypes through connection of databases of ribotypes and assessment of interrelationships within the Legionellaceae family. Despite the limitations with the use of the RiboPrinter as a tool for epidemiological analysis of nosocomial members of the Legionellaceae family, this automated system holds promise as a very useful addition to the ever expanding molecular typing repertoire.
Acknowledgments
We are grateful to Maryse De-Ré and Armelle Marecat, Institut Pasteur de Lille, for skillful technical assistance. We also thank Monique Reyrolle for providing strain LC217 and Pierre Farge (Centre National de Référence des Légionelles, Lyon, France) for helpful information.
REFERENCES
- 1.Aavitsland, P. 30August2001, posting date. Outbreak of legionellosis in Stavanger, Norway. Eurosurv. Wkly. 5(35). [Online.] http://www.eurosurveillance.org/.
- 2.Bangsborg, J., P. Gerner-Smidt, H. Colding, N. Fiehn, B. Bruun, and N. Hoiby. 1995. Restriction fragment length polymorphism of rRNA genes for molecular typing of members of the family Legionellaceae. J. Clin. Microbiol. 33:402-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 4.Benson, R. F., W. L. Thacker, M. I. Daneshvar, and D. J. Brenner. 1996. Legionella waltersii sp. nov. and an unnamed Legionella genomospecies isolated from water in Australia. Int. J. Syst. Bacteriol. 46:631-634. [DOI] [PubMed] [Google Scholar]
- 5.Benson, R. F., W. L. Thacker, F. C. Fang, B. Kanter, W. R. Mayberry, and D. J. Brenner. 1990. Legionella sainthelensi serogroup 2 isolated from patients with pneumonia. Res. Microbiol. 141:453-463. [DOI] [PubMed] [Google Scholar]
- 6.Benson, R. F., W. L. Thacker, B. B. Plikaytis, and H. W. Wilkinson. 1987. Cross-reactions in Legionella antisera with Bordetella pertussis strains. J. Clin. Microbiol. 25:594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibb, W. F., R. J. Sorg, B. M. Thomason, M. D. Hicklin, A. G. Steigerwalt, D. J. Brenner, and M. R. Wulf. 1981. Recognition of a second serogroup of Legionella longbeachae. J. Clin. Microbiol. 14:674-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birtles, R. J., N. Doshi, N. A. Saunders, and T. G. Harrison. 1991. Second serogroup of Legionella quinlivanii isolated from two unrelated sources in the United Kingdom. J. Appl. Bacteriol. 71:402-406. [DOI] [PubMed] [Google Scholar]
- 9.Bornstein, N., A. Mercatello, D. Marmet, M. Surgot, Y. Deveaux, and J. Fleurette. 1989. Pleural infection caused by Legionella anisa. J. Clin. Microbiol. 27:2100-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner, D. J., A. G. Steigerwalt, P. Epple, W. F. Bibb, R. M. McKinney, R. W. Starnes, J. M. Colville, R. K. Selander, P. H. Edelstein, and C. W. Moss. 1988. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J. Clin. Microbiol. 26:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner, D. J., A. G. Steigerwalt, and J. E. McDade. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 90:656-658. [DOI] [PubMed] [Google Scholar]
- 12.Bruce, J. L. 1996. Automated system rapidly identifies and characterizes micro organisms in food. Food Technol. 50:77-81. [Google Scholar]
- 13.Clermont, O., C. Cordevant, S. Bonacorsi, A. Marecat, M. Lange, and E. Bingen. 2001. Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J. Clin. Microbiol. 39:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gheldre, Y., N. Maes, F. Lo Presti, J. Etienne, and M. Struelens. 2001. Rapid identification of clinically relevant Legionella spp. by analysis of transfer DNA intergenic spacer length polymorphism. J. Clin. Microbiol. 39:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dice, L. R. 1945. Measures of the amount of ecological associations between species. J. Ecol. 26:297-302. [Google Scholar]
- 16.Edelstein, P. H. 1995. Antimicrobial chemotherapy for Legionnaires' disease: a review. Clin. Infect. Dis. 21:265-276. [DOI] [PubMed] [Google Scholar]
- 17.Edelstein, P. H., and N. P. Cianciotto. 2002 1999. Legionella species and Legionnaires' disease. In The prokaryotes. An evolving electronic database for the microbiological community, 3rd ed. [Online.] Springer-Verlag, Berlin, Germany. http://link.springer.de/link/service/books/10125/bibs/1003001/10030331.htm. Accessed 16 June 2002.
- 18.Edelstein, P. H., and R. D. Meyer. 1994. Legionella pneumonia, p. 455-484. In J. E. Pennington (ed.), Respiratory infections: diagnosis and management. Raven Press, Ltd., New York, N.Y.
- 19.File, T. M., Jr., J. S. Tan, and J. F. Plouffe. 1998. The role of atypical pathogens: Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella pneumophila in respiratory infection. Infect. Dis. Clin. N. Am. 12:569-592. [DOI] [PubMed] [Google Scholar]
- 20.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzin, L., C. Scolfaro, D. Cabodi, M. Valera, and P. A. Tovo. 2001. Legionella pneumophila pneumonia in a newborn after water birth: a new mode of transmission. Clin. Infect. Dis. 33:103-104. [DOI] [PubMed] [Google Scholar]
- 22.Griffith, M. E., D. S. Lindquist, R. F. Benson, W. L. Thacker, D. J. Brenner, and H. W. Wilkinson. 1988. First isolation of Legionella gormanii from human disease. J. Clin. Microbiol. 26:380-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimont, F., and P. A. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. 137B:165-175. [DOI] [PubMed] [Google Scholar]
- 24.Grimont, F., M. Lefevre, E. Ageron, and P. A. Grimont. 1989. rRNA gene restriction patterns of Legionella species: a molecular identification system. Res. Microbiol. 140:615-626. [DOI] [PubMed] [Google Scholar]
- 25.Gubler, J. G., M. Schorr, V. Gaia, R. Zbinden, and M. Altwegg. 2001. Recurrent soft tissue abscesses caused by Legionella cincinnatiensis. J. Clin. Microbiol. 39:4568-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison, T. G., N. A. Saunders, N. Doshi, R. Wait, and A. G. Taylor. 1988. Serological diversity within the species Legionella spiritensis. J. Appl. Bacteriol. 65:425-431. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, T. G., N. A. Saunders, A. Haththotuwa, N. Doshi, and A. G. Taylor. 1992. Further evidence that genotypically closely related strains of Legionella pneumophila can express different serogroup specific antigens. J. Med. Microbiol. 37:155-161. [DOI] [PubMed] [Google Scholar]
- 28.Hookey, J. V., N. A. Saunders, N. K. Fry, R. J. Birtles, and T. G. Harrison. 1996. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int. J. Syst. E vol. Microbiol. 46:526-531. [Google Scholar]
- 29.Jantzen, E., A. Sonesson, T. Tangen, and J. Eng. 1993. Hydroxy-fatty acid profiles of Legionella species: diagnostic usefulness assessed by principal component analysis. J. Clin. Microbiol. 31:1413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joly, J. R., R. M. McKinney, J. O. Tobin, W. F. Bibb, I. D. Watkins, and D. Ramsay. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23:768-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann, A. F., J. E. McDade, C. M. Patton, J. V. Bennett, P. Skaliy, J. C. Feeley, D. C. Anderson, M. E. Potter, V. F. Newhouse, M. B. Gregg, and P. S. Brachman. 1981. Pontiac fever: isolation of the etiologic agent (Legionella pneumophila) and demonstration of its mode of transmission. Am. J. Epidemiol. 114:337-347. [DOI] [PubMed] [Google Scholar]
- 32.Lo Presti, F., S. Riffard, H. Meugnier, M. Reyrolle, Y. Lasne, P. A. Grimont, F. Grimont, R. F. Benson, D. J. Brenner, A. G. Steigerwalt, J. Etienne, and J. Freney. 2001. Legionella gresilensis sp. nov. and Legionella beliardensis sp. nov., isolated from water in France. Int. J. Syst. E vol. Microbiol. 51:1949-1957. [DOI] [PubMed] [Google Scholar]
- 33.Lo Presti, F., S. Riffard, H. Meugnier, M. Reyrolle, Y. Lasne, P. A. Grimont, F. Grimont, F. Vandenesch, J. Etienne, J. Fleurette, and J. Freney. 1999. Legionella taurinensis sp. nov., a new species antigenically similar to Legionella spiritensis. Int. J. Syst. Bacteriol. 49:397-403. [DOI] [PubMed] [Google Scholar]
- 34.Lo Presti, F., S. Riffard, F. Vandenesch, and J. Etienne. 1998. Identification of Legionella species by random amplified polymorphic DNA profiles. J. Clin. Microbiol. 36:3193-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo Presti, F., S. Riffard, F. Vandenesch, M. Reyrolle, E. Ronco, P. Ichai, and J. Etienne. 1997. The first clinical isolate of Legionella parisiensis, from a liver transplant patient with pneumonia. J. Clin. Microbiol. 35:1706-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luck, P. C., L. Bender, M. Ott, J. H. Helbig, and J. Hacker. 1991. Analysis of Legionella pneumophila serogroup 6 strains isolated from a hospital warm water supply over a three-year period by using genomic long-range mapping techniques and monoclonal antibodies. Appl. Environ. Microbiol. 57:3226-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDade, J., C. Shepard, D. Fraser, T. Tsai, M. Redus, and W. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 38.Neter, J., W. Wasseman, and M. H. Kutner. 1990. The coefficient of simple correlation, p. 101-102. In Applied linear stastistical models, 3rd ed. Irwin, Boston, Mass.
- 39.Ott, M., L. Bender, R. Marre, and J. Hacker. 1991. Pulsed-field electrophoresis of genomic restriction fragments for the detection of nosocomial Legionella pneumophila in hospital water supplies. J. Clin. Microbiol. 29:813-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portero, R. C., and C. A. Joseph. 12July2001, posting date. Community outbreak of Legionnaires' disease in Murcia, Spain. Eurosurv. Wkly. 5(28). [Online.] http://www.eurosuveillance.org/.
- 41.Riffard, S., F. L. Presti, F. Vandenesch, F. Forey, M. Reyrolle, and J. Etienne. 1998. Comparative analysis of infrequent-restriction-site PCR and pulsed-field gel electrophoresis for epidemiological typing of Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 36:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers, F. G., and A. W. Pasculle. 1991. Legionella, p. 442-453. In A. Balows, W. J. Hausler, Jr., K. L. Herman, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 43.Saunders, N. A., N. Doshi, and T. G. Harrison. 1992. A second serogroup of Legionella erythra serologically indistinguishable from Legionella rubrilucens. J. Appl. Bacteriol. 72:262-265. [DOI] [PubMed] [Google Scholar]
- 44.Schoonmaker, D., T. Heimberger, and G. Birkhead. 1992. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J. Clin. Microbiol. 30:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selander, R. K., R. M. McKinney, T. S. Whittam, W. F. Bibb, D. J. Brenner, F. S. Nolte, and P. E. Pattison. 1985. Genetic structure of populations of Legionella pneumophila. J. Bacteriol. 163:1021-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sethi, M. R. 1997. Fully automated microbial characterization and identification for industrial microbiologists. Am. Lab. 5:31-35. [Google Scholar]
- 47.Stout, J. E., J. Joly, M. Para, J. Plouffe, C. Ciesielski, M. J. Blaser, and V. L. Yu. 1988. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila. J. Infect. Dis. 157:486-495. [DOI] [PubMed] [Google Scholar]
- 48.Tang, P. W., S. Toma, C. W. Moss, A. G. Steigerwalt, T. G. Cooligan, and D. J. Brenner. 1984. Legionella bozemanii serogroup 2: a new etiological agent. J. Clin. Microbiol. 19:30-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thacker, W. L., B. B. Plikaytis, and H. W. Wilkinson. 1985. Identification of 22 Legionella species and 33 serogroups with the slide agglutination test. J. Clin. Microbiol. 21:779-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thacker, W. L., H. W. Wilkinson, B. B. Plikaytis, A. G. Steigerwalt, W. R. Mayberry, C. W. Moss, and D. J. Brenner. 1985. Second serogroup of Legionella feeleii strains isolated from humans. J. Clin. Microbiol. 22:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai, H. C., S. S. Lee, W. R. Lin, C. K. Huang, Y. S. Chen, S. R. Wann, H. H. Lin, M. Y. Yen, and Y. C. Liu. 2001. Legionnaires' disease in an immunocompetent young adult. Gaoxiong Yi Xue Ke Xue Za Zhi 17:331-335. [PubMed] [Google Scholar]
- 52.van Belkum, A. 2002. Molecular tools for epidemiological investigations into Legionella pneumophila infections, p. 227-236. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Lück (ed.), Legionella. American Society for Microbiology, Washington, D.C.
- 53.van Belkum, A., M. Struelens, and W. Quint. 1993. Typing of Legionella pneumophila strains by polymerase chain reaction-mediated DNA fingerprinting. J. Clin. Microbiol. 31:2198-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vickers, R. M., J. E. Stout, L. S. Tompkins, N. J. Troup, and V. L. Yu. 1992. Cefamandole-susceptible strains of Legionella pneumophila serogroup 1: implications for diagnosis and utility as an epidemiological marker. J. Clin. Microbiol. 30:537-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn, W. C., Jr. 1988. Legionnaires' disease: historical perspective. Clin. Microbiol. Rev. 1:60-81. [DOI] [PMC free article] [PubMed] [Google Scholar]