Abstract
Escherichia coli causes the vast majority of urinary tract infections (UTI) in both ambulatory and hospital patients. Several uropathogenic virulence factors have been identified, but half of all E. coli isolates that cause UTI have none or only one of the known virulence factors. Thus, it is reasonable to presume that other bacterial factors may be important in UTI pathogenesis. In order to find additional uropathogenic E. coli genes, we used genomic subtraction to identify DNA regions present in a uropathogenic strain of E. coli (1128-11). Genomic subtraction yielded 40 tester-specific fragments, including a novel heat-resistant agglutinin (hra) gene fragment. hra occurred in 55% of 486 UTI strains compared to 28% of 165 rectal strains (P = 0.001). The hra gene in 1128-11 was cloned, sequenced, and found to have 91% homology to the hra gene from E. coli meningitis strain RS218. The genetic organization of genes flanking hra in 1128-11 is distinct from the hra found in E. coli strains J96 and RS218. In our UTI and rectal specimen collections, hra was positively associated with a number of known virulence genes, including pathogenicity island genes hly and cnf, which are absent in 1128-11. The presence of hra in 1128-11 independent of hly/cnf suggests multiple mechanisms by which hra can be acquired by pathogenic E. coli strains. The flanking genes suggest that in 1128-11, hra may be part of a novel variant of a pathogenicity island V.
Uropathogenic Escherichia coli (UPEC) strains account for 90% of all urinary tract infections (UTI) among ambulatory patients and up to 50% of all nosocomial UTI (11). UPEC strains have a number of gene virulence factors, including adhesins, siderophores, toxins, capsule, and protease, that are implicated in UTI pathogenesis (1, 4), but as many as half of all E. coli strains that cause UTI have none or only one of the known virulence factors. Thus, it is reasonable to presume that other bacterial factors may be important in UTI pathogenesis.
We used genomic subtraction to search for new virulence gene candidates for UTI pathogenesis. Genomic subtraction is a PCR-based method to subtract gene sequences that are common between a “tester” and “driver” strain and yield sequences that are unique to the tester strain. This procedure is based on suppressive subtractive hybridization (6). Strain pairs were selected from our UPEC and rectal E. coli specimen collections (reviewed in reference 13) based on molecular epidemiologic information in order to maximize the potential of identifying new virulence gene candidates. For this subtraction, we selected a first-UTI-causing E. coli strain, 1128-11, as the tester and UTI strains 366-11 (used in a previous subtraction and described in reference 17) and CFT073 as the combined drivers. The resulting gene fragments were screened against our pathogenic and nonpathogenic E. coli collections in order to determine their potential significance in UPEC.
We describe our successful use of this strategy to identify a heat-resistant agglutinin gene (hra) associated with UTI. The identified hra gene is 91% homologous to a gene found on RS218, the sequenced E. coli meningitis strain.
MATERIALS AND METHODS
Selection of strains for subtraction. (i) Tester.
Strain 1128-11 has the virulence genes fim, prf, aer, kpsMT, ompT, and papGAD (9). This combination of virulence genes occurs three times more frequently in UTI isolates than in rectal E. coli isolates. 1128-11 does not contain the known uropathogenic factors hly, sfa, drb, or cnf that are implicated in UTI virulence, thus increasing our probability of finding new UTI genes. Strain 1128-11 was selected as the tester strain for this subtraction from among three other strains with this combination of virulence genes because it had a pulsed-field gel band pattern that was most representative of the patterns in that group.
(ii) Driver.
The choice of driver strain was determined by our aim to find new genetic regions that had not been discovered through our initial subtraction (17). Therefore, we chose the tester strain of the previous subtraction, 366-11, as the driver. To avoid finding genes that have already been discovered and sequenced, we also included the newly sequenced pyelonephritis-causing E. coli strain CFT073 in our driver.
Genomic subtraction.
A commercial kit (Clontech PCR-Select bacterial genome subtraction kit) was used to identify gene fragments specific to the tester strain through differential cloning. The genomic DNA of drivers (366-11 and CFT073) was subtracted from that of the tester (1128-11) following manufacturer's protocols to obtain tester-specific DNA. A high-copy-number plasmid specific to the 1128-11 tester strain was added to the driver DNA 366-11 to suppress its overrepresentation in the final tester specific fragments.
Briefly, genomic DNA was isolated from tester (1128-11) and driver (366-11 and CFT-073) strains, purified using phenol-chloroform extraction, and digested with RsaI. Following purification of the digested DNA, the tester DNA was ligated with the adapter provided with the kit. The tester-specific DNA (sPCR) fragments were cloned into PCR2.1 plasmid vector using the TOPO TA cloning kit (Invitrogen) and transformed. The transformants were tested for inserts by PCR using M13R and T7 primers, which flank the cloned insert regions, followed by a nested PCR using nested primers supplied with the genomic subtraction kit. The sPCR products were spotted on nylon membranes and probed with fluorescence-labeled tester genomic DNA and driver genomic DNA using a commercially available kit (ECF random prime labeling and detection kit; Amersham). sPCR fragments that bound to both tester and driver DNA were removed from further analysis. Duplicate sPCR fragments were removed by cross-hybridization among all the tester specific sPCR fragments.
Genome walking.
The regions flanking the hra gene in 1128-11 were obtained by PCR using a commercial Genome Walker kit (Clontech Inc.). Briefly, the method involves ligation of adapters to purified uncloned libraries of genomic DNA digested with different restriction enzymes. The adapter primers provided in the kit and two sets of primers specific to hra were used to PCR regions upstream and downstream of the hra gene according to manufacturer's specifications. The primers for the primary PCR were HRAP1 (5′-CCAGAGCGATATCCGGGGTTACGTCATA-3′) and HRAP2 (5′-TATGACGTAACCCCGGATATC-GCTCTGG-3′). The conditions for PCR in the PE Biosystems 9600 thermal cycler were as follows: seven cycles of 2 s at 94°C and 3 to 6 min at 67°C and 30 cycles of 2 s at 94°C, 3 to 6 min at 72°C, and 3 to 6 min at 72°C. The nested PCRs were carried out with primers that were designed to confirm the primary PCR products. The primers for the nested PCR were NHRAP1 (5′-GAAGTTGTCAGCAGAGCCTGAACGTGAC-3′) and NHRAP2 (5′-GTCACGTTCAGGCTCTGCTGACAACTTC-3′). The conditions used were five cycles of 2 s at 94°C and 3 to 6 min at 67°C and 22 cycles of 2 s at 94°C, 3 to 6 min at 72°C, and 3 to 6 min at 72°C. The PCR products were cloned into a TOPO4 vector (Invitrogen), transformed into DH5α cells, and sequenced at the DNA Sequencing core (University of Michigan). Blast searches (http://www.ncbi.nlm.nih.gov/BLAST) on the sequences revealed identities based on their homology to known genes and translated gene products.
Fluorescein labeling of genomic DNA and sPCR fragments.
Restriction enzyme-digested genomic DNA or sPCR fragments were labeled with a fluorophore (ECF labeling kit, Amersham Pharmaceuticals) following manufacturer's instructions. Labeled probes were stored at −20°C.
E. coli collections.
We screened hra against a total of 486 UTI, 165 rectal, 79 periurethral, and 155 vaginal isolates from various collections (reviewed in reference 13). First-UTI isolates included E. coli isolates collected from the student health services of the University of Michigan and University of Texas at Austin from women 18 to 39 years old. The UTI 40-65 group consisted of E. coli isolates from women in the age group 40 to 65 years with a UTI in western Michigan and Israel. Recurring UTI isolates are E. coli isolates from women at the University of Michigan student health services who presented with three or more UTI within the previous 12 months. Pyelonephritis isolates are from children 18 to 24 months old from five hospitals in Finland. Vaginal isolates were collected from women 18 to 39 years old with and without UTI, and rectal and periurethral isolates were from women 18 to 39 years old without UTI. All isolates were previously screened for the presence or absence of adhesins, P-pili (papGAD, papGJ96, and prsGJ96), S-fimbrial adhesin (sfa), aerobactin (aer), group II capsule (kpsMT), cytotoxic necrotizing factor (cnf), Dr family of adhesins (drb), hemolysin (hly), outer membrane protease T (ompT), Irg homolog adhesin (iha), uropathogenic specific protein (usp), and catechole siderophore receptor gene (iroNEcoli) as described previously (2, 13).
A subset of strains from the above E. coli collection was used to study the distribution of hra among the UTI- and non-UTI-causing E. coli. This subset was prepared by selecting a minimum of 88 strains randomly from each different collection of UTI- and non-UTI-causing E. coli isolates from the various epidemiologic studies. Due to the smaller size of the recurring-UTI and periurethral strains collections, only 61 and 79 strains, respectively, were included for screening from these two collections. The total subset contains 885 strains.
Nylon membrane hybridizations.
The presence of hra in E. coli strains was determined using dot blot hybridization with fluorescence-labeled probes as described previously (18). Briefly, bacterial DNA was prepared by growing strains overnight in Luria-Bertani medium in a 96-well deep-well plate (volume per well, 1-ml; Corning Inc.). Bacterial cells were pelleted by centrifuging at 3,000 rpm in a Beckman desktop centrifuge and lysed with 800 μl of 0.4 N NaOH-10 mM EDTA at 70°C for 30 min. The bacterial lysate was arrayed on nylon membrane (Hybond H+; Amersham Pharmacia) using a BIO-dot Microfiltration apparatus (Bio-Rad Laboratories). Nylon membranes were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dried, and fixed by UV light.
Hybridization of probes to membranes.
Fluorescently labeled sPCR fragments were hybridized to Nylon membranes and detected using the fluorescein-based detection kit (Amersham) as described previously (18). Hybridization intensities were detected using a Storm 860 PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant software, version 5.0. The signal intensity of each spot was normalized to the intensity of 1128-11 (positive control). All strains were tested for the presence or absence of hra with a minimum of two independent membranes. Ambiguous results were retested on duplicate membranes and confirmed by Southern hybridization using previously described protocols (18). Sequencing of sPCR fragment DNA was performed at the University of Michigan Molecular Biology Core Facility using an Applied Biosystems model 373A automated sequencer.
Data analysis.
The magnitude of the association between hra and known virulence factors was estimated using the odds ratios and 95% confidence intervals, and the significance was tested using the chi-square test. All analyses were done using SAS (version 8.0). Excel (Microsoft) software was used for data entry. Software packages from DNAStar (Madison, Wis.) were used for primer design, DNA sequence comparison, and analysis.
RESULTS
Genomic subtraction.
Genomic subtraction with uropathogenic E. coli strain 1128-11 as the tester and 366-11 UTI strain and CFT-073 as combined drivers resulted in 40 tester-specific sPCR fragments ranging in size from 300 to 700 bp. Twenty-two fragments were found to hybridize strongly to the sequenced K-12 strain and were not used for subsequent analysis.
GenBank searches were conducted on the remaining tester-specific sPCR fragments, one of which had no homology to currently sequenced genes. Four of the remaining 18 fragments had open reading frames (ORFs), which showed similarity to cryptic phage-related proteins found on the E. coli O157:H7 strain (Table 1). We also identified sPCR fragments containing ORFs with homology to several proteins from the E. coli O157:H7 strain and the E. coli neonatal meningitis strain. Among these the initial distribution of S2T2-31 (homologous to putative cryptic prophage integrase CP933U from E. coli O157:H7) looked interesting; however, an accurate estimate of its distribution proved to be difficult, possibly due to its shared homology with other prophage integrase genes.
TABLE 1.
Initial screen of sPCR fragments with first-UTI (n = 88), pyelonephritis (n = 88), and rectal (n = 88) E. coli strains
| sPCR fragment | % UTI | % Pyelonephritis | % Rectal | Potential function (% amino acid identity) |
|---|---|---|---|---|
| S2T2-6 | 52 | 65 | 22 | Heat-resistant agglutinin (Hra) protein E. coli (91) |
| S2T1-91 | 34 | 44 | 15 | Putative transposase in Y. pestis (51) |
| S2T1-115 | 6 | 15 | 3 | No match |
| S2T2-9 | 15 | 20 | 11 | lifA gene (90) |
| S2T1-43 | 36 | 32 | 28 | Ribokinase RbsK from lactobacillus (38) |
| S2T1-54 | 14 | 14 | 7 | Hypothetical protein E. coli O157:H7 (25) |
| S2T1-93 | 15 | 13 | 6 | sugR ATP binding protein S. enterica serovar Typhimurium (86) |
| S2T2-11 | 81 | 48 | 67 | No match |
| S2T1-135 | 15 | 15 | 14 | Hypothetical lipoprotein H. pylori (50) |
| S2T1-48 | 17 | 22 | 5 | yscA protein B. subtilis (40) |
| S2T1-116 | 2 | 14 | 25 | Hypothetical protein in V. cholera (27) |
| S2T1-30 | 20 | 13 | 14 | Hyaluronan synthetase P. multicoda (66) |
| S2T1-86 | 83 | 61 | 82 | Hypothetical type II secretion protein E. coli EPEC strain (92) |
| S2T0-25 | 0 | 8 | 3 | Predicted protein L. lactis (50) |
| S2T2-31 | —a | — | — | Putative integrase CP933U E. coli O157:H7 (97) |
| S2T1-89 | 20 | 11 | 14 | Unknown protein CP933M E. coli O157:H7 (51) |
| S2T2-48 | 13 | 9 | 6 | Putative tail component CP933P E. coli O157:H7 (52) |
| S2T2-37 | 19 | 7 | 7 | Putative integrase prophage CP933C E. coli O157:H7 (90) |
—, probe showed high degree of cross hybridization, possibly due to presence of related prophage sequences in most E. coli strains.
Several other sPCR fragments like S2T2-48 (homology match to the tail component of putative cryptic prophage CP933P in E. coli O157:H7), S2T2-37 (homology to putative integrase of CP933C in E. coli O157:H7), and S2T2-9 (homology match to lifA gene from E. coli O157:H7) were interesting. These were twofold more prevalent in first-UTI and pyelonephritis strains than in the rectal strains (Table 1) and their distribution in UTI strains (15 to 20%) is in the same range as of the Dr family of adhesins (13). However, a 400-bp sPCR fragment, S2T2-6, with a 96% homology to heat-resistant agglutinin (hra), was present in over half the UTI strains and showed the highest preferential distribution in UTI versus non-UTI strains (52% of first-UTI strains and 65% of pyelonephritis strains versus 22% of rectal strains). This hra homolog was tested further (below).
Distribution of hra in UTI and non-UTI isolates.
To confirm and extend the observations of hra gene distributions, we hybridized the hra probe to an additional 621 E. coli strains (total of 885). The results of this probing are shown in Table 2. hra occurred in 43%-66% of the UTI causing E. coli isolates from different collections but in only 28% of rectal strains. Overall, hra was found in 55% of 486 UTI strains compared to 28% of 165 rectal strains (P = 0.001). The relative prevalence ranged from 1.5 to 2.1 depending upon the collection (Table 2). hra also occurred 1.4 times more frequently in vaginal and periurethral strains than in rectal strains. The prevalence of hra in UTI strains was not significantly different from those of periurethral and vaginal strains. The prevalence of hra among vaginal and periurethral strains though higher than among rectal strains is not significant. Periurethral and vaginal strains are known to consist of a mix of UTI and non-UTI strains and this may explain the observed overlapping confidence intervals (Table 2).
TABLE 2.
Distribution of hra by E. coli collection, and prevalence ratio and confidence intervals for the prevalence of hra in UTI collections compared to that in rectal isolates
| E. coli collection (n = 885) | Total no. of isolates (%) | Prevalence ratio | 95% confidence interval |
|---|---|---|---|
| UTI | |||
| First UTI (Michigan) | 96 (54) | 1.9 | 1.4-2.6 |
| First UTI (Texas) | 91 (43) | 1.5 | 1.1-2.1 |
| Pyelonephritis | 148 (66) | 2.4 | 1.8-3.1 |
| UTI 40-65 | 90 (58) | 2.1 | 1.5-2.8 |
| Recurring UTI | 61 (56) | 2.0 | 1.4-2.8 |
| Non-UTI | |||
| Rectal | 165 (28) | 1.0 | Reference |
| Periurethral | 79 (40) | 1.4 | 1.1-2.0 |
| Vaginal | 155 (39) | 1.4 | 1.0-1.9 |
Associations of hra with known uropathogenic virulence factors.
We performed pairwise comparisons (Table 3) between hra and each of the previously known virulence factors, aerobactin (aer), group II capsule (kpsMT), cytotoxic necrotizing factor (cnf), Dr family of adhesins (drb), hemolysin (hly), outer membrane protease (ompT), three classes of P-pili (papGJ96 [class I], papGAD [class II], and prsGJ96 [class III]), S-fimbrial adhesin (sfa), and the newly discovered putative uropathogenic factors iha, usp, and iroNEcoli (2, 10). hra was positively associated with genes fim, ppili, capII, hly, cnf, papGAD, usp, iroNEcoli, and prsGJ96 (Table 3). This suggested that the observed association of hra with UTI might be an artifact; that is, the observed association with UTI occurs only indirectly because of the association with known uropathogenic virulence factors. To test this hypothesis, we examined the association of hra with UTI E. coli strains among strains positive for hly and those negative for hly. Shown in Table 4 is the analysis stratifying by the presence of hly. In two collections, first-UTI (Michigan) and UTI 40-65 strains, when hly is absent, we see a strong association between hra and UTI, but essentially no association in UTI strains containing hly. The odds ratios were significantly different by strata using the Breslow Day test. A similar effect was seen on stratification with cnf and prsGJ96 (data not shown). We saw a similar but not significant trend for pyelonephritis and for all UTI collections combined (all UTI). For the first-UTI (Texas) and recurrent-UTI group the sample size is small and gave unstable results with overlapping confidence intervals. Thus, we rejected the hypothesis that the association of hra with UTI was due solely to the association with other known uropathogenic factors.
TABLE 3.
Associations of hra with nine known virulence genes in 885 uropathogenic and commensal E. coli isolatesa
| Virulence factor (gene name) | OR | 95% confidence interval |
|---|---|---|
| Aerobactin (aer) | 0.9 | 0.7-1.2 |
| Capsule, group II (capII) | 2.5 | 1.7-3.6 |
| Capsule, group III (capIII) | 1.1 | 0.6-2.1 |
| Catechole siderophore receptor homolog (iroNEcoli) | 3.2 | 2.4-4.4 |
| Cytotoxic necrotizing factor (cnf) | 16.8 | 10.2-27.2 |
| Dr family of adhesins (drb) | 0.6 | 0.4-0.9 |
| Hemolysin (hly) | 6.6 | 4.7-9.8 |
| Nonhemagglutinating adhesin (iha) | 0.9 | 0.6-1.2 |
| OmpT (ompT) | 3.8 | 2.4-5.9 |
| P pilus family (pff) | 5.8 | 4.2-7.9 |
| papGAD (class II) | 2.1 | 1.5-2.8 |
| prsGJ96 (class III) | 14.5 | 8.1-25 |
| papGJ96 (class I) | 2.1 | 0.5-8.5 |
| Type I pilus (fim) | 6.2 | 4.2-9.1 |
| Uropathogenic specific protein (usp) | 2.7 | 1.9-3.7 |
Odds ratios (OR) and 95% confidence intervals for pairwise comparisons are shown. Statistically significant odd ratios are shown in boldface type (P < 0.05).
TABLE 4.
Association of hra among 496 UPEC and 165 rectal isolates in the presence or absence of hly
| UTI collection | ORa (95% confidence interval)
|
Breslow Day P value | ||
|---|---|---|---|---|
| Crude | hly absent | hly present | ||
| All UTI (n = 496) | 3.3 (2.2-4.9) | 2.9 (1.8-4.5) | 1.2 (0.4-3.5) | 0.13 |
| First UTI (Michigan) (n = 96) | 2.9 (1.7-5.0) | 3.5 (1.9-6.6) | 0.7 (0.1-2.3) | 0.02 |
| UTI 40-65 (n = 90) | 3.5 (2.0-6.0) | 4.4 (2.4-8.5) | 0.7 (0.2-2.4) | 0.01 |
| Pyelonephritis (n = 148) | 5.0 (3.1-8.2) | 4.2 (2.3-7.6) | 1.8 (0.6-5.8) | 0.20 |
| Recurring UTIb (n = 61) | 3.2 (1.7-6.0) | 1.3 (0.5-3.0) | 3.2 (0.7-13.2) | 0.30 |
| First UTI (Texas)b (n = 91) | 1.9 (1.1-3.3) | 1.0 (0.5-2.1) | 1.0 (0.3-3.3) | 0.98 |
OR, odds ratio.
Low number of hly-positive and hra-negative strains (10 of 47 strains in recurring-UTI isolates and 19 of 58 strains in first-UTI [Texas] isolates were hly+ hra−).
Cloning of hra.
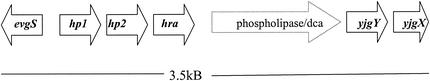
The heat-resistant agglutinin gene in 1128-11 was cloned and sequenced and determined to be a 710-bp gene with a 93% DNA homology to the sequenced hra in an enteric E. coli strain (12) and 91% homology to the E. coli meningitis strain RS218 (http://www.genome.wisc.edu). The regions of 1128-11 adjoining hra were cloned to determine the genetic location of hra in 1128-11 in comparison to the sequenced K-12 genome. Figure 1 describes the alignment of different sequences from cloning the genetic regions adjacent to hra in 1128-11. Two IS600-related elements of unknown function, hp1 and hp2, and the sensor gene evgS for a two-component regulatory system (evgS/evgA) found in E. coli K-12 at 53.4 min, are present immediately adjacent to hra in 1128-11. Only a fragment of IS element hp1 is found next to hra and the evgS gene in 1128-11 is missing 300 bp at the N terminus. On the other side, hra is flanked by a gene fragment of unknown function, yjgY, present at 96.9 min on the K-12 genome map, and a fragment of another gene of unknown function, yjgX.
FIG. 1.
Alignment of the hra-flanking regions in 1128-11. Solid arrows indicate regions with >90% DNA sequence homology to known gene sequences. The amino-terminal region of evgS (∼200 bp) is not present in the cloned region near hra in 1128-11. A partial segment of IS elements hp1 and hp2 found in CFT073 flank hra. yjgY and yjgX are hypothetical genes present in strain K-12. The cloned region contains a 200-bp internal region of the yjgX gene. The dashed arrow indicates a region of low homology (<30% at the translated-nucleotide level) between the 1128-11 sequence and published sequences.
DISCUSSION
We report the discovery in E. coli strain 1128-11 of a heat-resistant agglutinin (hra) gene that is widely distributed among UTI E. coli isolates and occurs twice as frequently among UPEC strains as it does among rectal E. coli strains. hra occurred less frequently in periurethral and vaginal strains than in UTI strains, although the confidence intervals for the prevalence ratio of hra in UTI strains overlaps with those of the vaginal and periurethral strains. Since periurethral E. coli strains are somewhat different from uropathogens, they may cause UTI only if something mechanical, like bladder catheterization, facilitates their movement (13). This and subsequent data analyses suggesting that hra in 1128-11 is independent of pathogenicity island (PAI) factors hly, cnf, and prsGJ96 imply that hra has a high potential to be a gene important in UTI virulence, although the functional role of this putative virulence determinant in UTI has not been established.
The hra gene in 1128-11 was cloned, sequenced, and found to have 91% homology to the hra gene from E. coli meningitis strain RS218 (http://www.genome.wisc.edu) and C5 (3). hra also shows significant homology to the 756-bp tia loci in the enterotoxigenic E. coli (ETEC) strain H10407 (7) and Salmonella enterica serovar Typhimurium (5). The hra gene in ETEC consists of a 792-bp ORF coding for a putative protein of 29 kDa with a predicted N-terminal secretory signal sequence. An hra-like gene fragment has also been found on PAI V of E. coli J96 (16) at 94 min on the K-12 map at tRNA leuX.
Colonization of host tissues is usually mediated by adhesins, which recognize and bind to specific receptor moieties of host cells (15). While the functional role of hra in UTI is not known, hra in an ETEC O9:H10:K99 strain was determined to be a mannose-resistant hemagglutinating protein (12). The ETEC Hra functions as an outer membrane protein that acts as a nonfimbrial adhesin and promotes agglutination of human and animal erythrocytes and human colonic cells (8). The E. coli strain C1212 isolated from UTI was found to adhere to urinary epithelial cells in a mannose resistant manner (14).
The region of 1128-11 genome containing hra appears to be relatively plastic. In addition to hra, it contains part of the IS600-related sequences hp1 (found in CFT073) and hp2 (found on CFT073 and homologous to b4285 on K-12) and a region of transposase for IS600 sequence (Fig. 1). A gene encoding a putative membrane protein of unknown function, yjgY, present at 96.9 min on the K-12 map, is present adjacent to hra in 1128-11. Insertion sequences are known to be responsible for the integration of foreign DNA into E. coli genomes. The presence of an IS600 element and transposase suggests that hra could have integrated into the E. coli genome of uropathogenic 1128-11 as part of a mobile genetic element.
hra was positively associated with genes fim, pff, capII, hly, cnf, papGAD, usp, iroNEcoli, and prsGJ96. This association could arise either from genetic or functional linkage. hra can be present in UTI strains with hly/cnf/prsgj96, e.g., on the same PAI, PAI V, as in the uropathogenic strain J96 (16). We think functional linkage with PAI V factors is unlikely, since in our collection of 486 UTI strains, hra occurred with equal frequency with hly and without hly (137 strains or 28% had both hra and hly and 135 strains [27%] had hra without hly). Further, hra in strain 1128-11 does not appear to be in a region similar to PAI V.
The N-terminal sequence of evgS is not found in the region flanking hra in 1128-11, although it is present in the 1128-11 genome (results not shown), suggesting that at the very least the insertion of hra has resulted in the disruption of this gene. It is interesting to speculate that hra in 1128-11 is on a novel PAI. The determination of the genetic location of the N-terminal end of evgS and adjacent genes will help determine the exact nature (PAI- versus IS-related) of the hra insertion in 1128-11. Further studies to determine the distribution of genomic positions of hra in UTI strains will also help in our understanding of the different modes of acquisition and transfer of hra among UTI-causing E. coli.
In summary, hra is present in more than half the UTI-causing E. coli strains and only about a quarter of rectal strains. This implies that hra may be important in UTI virulence. Functional studies will be needed to further establish the definitive role of Hra and relative importance of mannose resistant adhesin like Hra among the already known adhesins in their contribution to UTI pathogenesis.
Acknowledgments
We thank Lixin Zhang for his technical help and useful discussions and Pat Tallman for isolation and purification of E. coli strains. We also thank Guy Plunkett, University of Wisconsin—Madison, for his help with the E. coli K1 sequence information.
This work was supported by an award from the National Institutes of Health (grant RO1 DK55496 to C.F.M.).
REFERENCES
- 1.Arthur, M., C. E. Johnson, R. H. Rubin, R. D. Arbeit, C. Campanelli, C. Kim, S. Steinbach, M. Agarwal, R. Wilkinson, and R. Goldstein. 1989. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect. Immun. 57:303-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, R. J., L. Zhang, B. Foxman, A. Siitonen, M. E. Jantunen, H. Saxem, and C. F. Marrs. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection—usp, iha, and iroNE. coli. J. Infect. Dis. 185:1521-1524. [DOI] [PubMed] [Google Scholar]
- 3.Bonacorsi, S. P., O. Clermont, C. Tinsley, I. Le Gall, J. C. Beaudoin, J. Elion, X. Nassif, and E. Bingen. 2000. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 68:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbonetti, N. H., S. Boonchai, S. H. Parry, V. Vaisanen-Rhen, T. K. Korhonen, and P. H. Williams. 1986. Aerobactin-mediated iron uptake by Escherichia coli isolates from human extraintestinal infections. Infect. Immun. 51:966-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 95:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diatchenko, L., Y. F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleckenstein, J. M., L. E. Lindler, E. A. Elsinghorst, and J. B. Dale. 2000. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foxman, B., L. Zhang, K. Palin, P. Tallman, and C. F. Marrs. 1995. Bacterial virulence characteristics of Escherichia coli isolates from first-time urinary tract infection. J. Infect. Dis. 171:1514-1521. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunin, C. M. 1997. Urinary tract infections: detection, prevention and management, 5th ed. Williams and Wilkins, Baltimore, Md.
- 12.Lutwyche, P., R. Rupps, J. Cavanagh, R. A. Warren, and D. E. Brooks. 1994. Cloning, sequencing, and viscometric adhesion analysis of heat-resistant agglutinin 1, an integral membrane hemagglutinin from Escherichia coli O9:H10:K99. Infect. Immun. 62:5020-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marrs, C. F., L. Zhang, P. Tallman, S. D. Manning, P. Somsel, P. Raz, R. Colodner, M. E. Jantunen, A. Siitonen, H. Saxen, and B. Foxman. 2002. Variations in 10 putative uropathogen virulence genes among urinary, faecal and peri-urethral Escherichia coli. J. Med. Microbiol. 51:138-142. [DOI] [PubMed] [Google Scholar]
- 14.Ørskov, I., F. Ørskov, and A. Birch-Andersen. 1980. Comparison of Escherichia coli fimbrial antigen F7 with type 1 fimbriae. Infect. Immun. 27:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, L., B. Foxman, S. D. Manning, P. Tallman, and C. F. Marrs. 2000. Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect. Immun. 68:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44:225-233. [DOI] [PubMed] [Google Scholar]