Abstract
Serotype changes among natural isolates of Streptococcus pneumoniae are well documented and occur by recombinational exchanges at the capsular biosynthetic locus. However, the frequency with which this phenomenon occurs within the nasopharynx of children is not clear and is likely to be highest in the nasopharynx of children, who have high rates of pneumococcal carriage. A birth cohort of 100 infants was studied, and pneumococci were recovered from nasopharyngeal samples taken at monthly intervals during the first 6 months of life and then at 2-monthly intervals until the age of 2 years. Among the 1,353 nasopharyngeal samples were 523 that contained presumptive pneumococci, and three colonies from each were serotyped. A total of 333 isolates, including all isolates of differing serotypes from the same child, were characterized by multilocus sequence typing. Sixty-eight children carried multiple serotypes during the first 2 years of life. Two children carried a typeable and a nonserotypeable pneumococcus of identical genotype, and five children carried genetically indistinguishable isolates of serotypes 15B and 15C. These isolates were considered, respectively, to be due to loss of capsule expression and the known ability of serotype 15B and 15C pneumococci to interconvert by loss or gain of an acetyl group on the capsular polysaccharide. In all other cases, isolates from the same children that differed in serotype also differed in genotype, indicating the acquisition of a different pneumococcal strain rather than a change in capsular type. There was therefore no evidence in this study for any change of serotype due to recombinational replacements at the capsular locus among the pneumococci carried within the nasopharynges of the children.
Streptococcus pneumoniae causes diseases that range in severity from acute otitis media and sinusitis to pneumonia, septicemia, and meningitis (1, 14). Pneumococcal disease is considered to occur subsequent to nasopharyngeal colonization, which typically occurs soon after birth (10, 14). Carriage is a highly dynamic process, with pneumococci being acquired, carried for a period of weeks or months, and then lost (10). A subset of the >90 pneumococcal serotypes are commonly carried by children, and these isolates also are the major causes of disease in children (11). Multivalent conjugate capsular polysaccharide vaccines have been developed to protect infants from pneumococcal disease caused by these common childhood serotypes, and in clinical trials these have proved effective in preventing invasive disease by the vaccine serotypes (3).
In almost all of the clinical trials a reduction in carriage of the vaccine serotypes and an increase in carriage of nonvaccine serotypes have been observed among vaccinated children (7, 12, 14). The extent, duration, and clinical significance of this serotype replacement are unclear (12, 16a). Isolates of the nonvaccine serotypes are considered to be less virulent than those of the vaccine serotypes, and thus even if vaccine serotypes are replaced with a new set of nonvaccine serotypes after mass vaccination, it is considered likely that a large reduction in the prevalence of invasive pneumococcal disease will be maintained (12, 16a). The effect of replacement may be more significant in acute otitis media since in the Finnish efficacy trials of seven-valent conjugate pneumococcal vaccines, serotype replacement led to a significant increase in otitis media caused by nonvaccine serotypes (9).
One consequence of serotype replacement following mass vaccination is the increased selective pressure for emergence of penicillin resistance and multiple antibiotic resistance among those nonvaccine serotypes that become more prevalent in the nasopharynges of children (16a). At present almost all of the major antibiotic-resistant clones of S. pneumoniae are of vaccine serotypes or serotypes for which the vaccines should provide cross-protection—notably, serotypes 6B, 6A, 9V, 14, 19F, 19A, and 23F (12). Resistance to penicillin is difficult to achieve de novo, but resistance to penicillin and to other classes of antibiotics can occur in a new serotype, in a single step, by a change of serotype within one of the highly successful international multiply antibiotic-resistant clones (16a). Evidence that pneumococci can change their serotype in vivo was first obtained from the analysis of populations of antibiotic-resistant pneumococci, in which isolates were identified that were indistinguishable in genotype but that differed in serotype (4). Subsequently, there have been numerous reports of serotype variants of the major penicillin-resistant and multiply antibiotic-resistant clones (13), and molecular studies have shown that these serotype changes occur by recombinational events that replace the capsular biosynthetic genes of a recipient pneumococcus with the corresponding genes from a donor pneumococcus of a different serotype (5, 6). Almost all reported examples of serotype changes have been from one vaccine serotype to another vaccine serotype, presumably because these are the serotypes most commonly present to act as donors and recipients of capsular genes within the nasopharynx of children. Serotype replacement after mass vaccination would lead to more donors of capsular genes of nonvaccine serotypes in the nasopharynx, which, combined with selection favoring both the emergence of antibiotic resistance and of nonvaccine serotypes, could lead to the appearance of variants of the successful major antibiotic-resistant clones with nonvaccine serotypes (16a).
Although there is ample evidence for the occurrence of serotype changes, there is very little information about the frequency of these events during colonization of the nasopharynx. In this study, nasopharyngeal swabs were taken at 15 sampling points, between 1 month and 2 years of age, from a birth cohort of 100 children. Children who carried at least two pneumococci of differing serotype during the first 2 years of life were identified, and molecular characterization of 333 isolates from these children was carried out using multilocus sequence typing (MLST) (8). There was no evidence in these children for any changes of serotype mediated by recombinational replacements at the capsular locus during nasopharyngeal carriage.
MATERIALS AND METHODS
Identification and microbiological characterization of pneumococcal isolates.
Study isolates were obtained from a longitudinal pneumococcal carriage study performed by the Oxford Vaccine Group (University of Oxford) during the period from 1999 to 2001. Children were enrolled in this study at birth and lived throughout the city of Oxford, United Kingdom, or in the surroundings of Oxford. Samples of the nasopharyngeal flora were obtained from 100 children using cotton-tipped swabs, which were streaked onto sheep blood-gentamicin agar. Three alpha-hemolytic colonies from the primary agar plate were isolated in an attempt to recognize some of the children who were colonized with multiple strains of pneumococci. Identification of organisms as S. pneumoniae was performed using standard microbiological methods: colony morphology, bile solubility, and Optochin susceptibility. Serotyping was performed using the Quellung reaction with sera purchased from the Statens Serum Institut, Copenhagen, Denmark. Isolates that were nonserotypeable but Optochin susceptible and bile soluble were considered to be presumptive pneumococci. The presence of the pneumolysin gene in these isolates was examined using PCR (15). As described below, MLST resolved these isolates into those that appeared to be pneumococci that failed to express a capsule, and into isolates that were similar to, but distinct from, pneumococci. A similar phylogenetic distinction has been made by Whatmore et al., who showed that both of these groups of isolates possess the pneumolysin gene (18).
Molecular characterization of pneumococcal isolates.
Pneumococcal isolates, and the nonserotypeable presumptive pneumococci, were unambiguously characterized by MLST as described by Enright and Spratt (8). The sequences (alleles) at each locus were compared to those at the MLST website (www.mlst.net) and were assigned allele numbers if they corresponded to sequences already submitted to the pneumococcal MLST database; novel sequences were assigned new allele numbers and were deposited in the database. The allelic profiles of isolates (the allele numbers at the seven loci) were compared to those at the MLST website, and sequence types (STs) were assigned. Allelic profiles that were not represented in the MLST database were assigned new ST numbers and were deposited in the database. The similarities between the STs were shown by cluster analysis, using the matrix of pairwise differences between the allelic profiles of all isolates, and the unweighted pair-group method with arithmetic averages.
RESULTS
Sampling and selection of carried pneumococci for molecular characterization.
The carriage study was designed to sample the nasopharynges of 100 children, living in Oxford or the surrounding region, at 15 sampling points from 1 month to 2 years of age. Of these, 21 children were not sampled for the whole 2-year period, and for 2 others 1 of the 15 samples was not obtained, but 1,353 of the anticipated 1,500 swabs were available for analysis. Almost all children were colonized during the first 2 years of life. Of the 100 children, there were only 8 who appeared not to be colonized by a pneumococcus during this period, but 7 of these were children who withdrew from the study before the end of the 2 years. Presumptive pneumococci were present in 523 of the 1,353 nasopharyngeal samples from the 92 children in which colonization was detected, and three colonies from each positive nasopharyngeal sample were serotyped. Pneumococci of only a single serotype were identified in 18 of these children during the 2-year period. For the other 74 colonized children, two or more pneumococcal isolates of different serotypes (or a serotypeable pneumococcus and a nonserotypeable presumptive pneumococcus) were obtained from the nasopharyngeal samples.
A total of 333 carried isolates from the 74 children were characterized by MLST. Isolates were selected for characterization by MLST if they were of a serotype different from that of any of the isolates obtained in the previous positive sample from the child (even if isolates of that serotype had been previously recovered in an earlier sample), or where there were isolates of differing serotype in the same sample. Table 1 shows the pneumococci isolated from two of the children and those selected for characterization by MLST using the above criteria.
TABLE 1.
Selection of isolates for molecular characterization
| Mo of sampling | Child 2
|
Child 28
|
||
|---|---|---|---|---|
| Serotype | Serotype selected for MLST | Serotype | Serotype selected for MLST | |
| 1 | 21 | 19A | ||
| 2 | 21 | 19A | ||
| 3 | 21 | |||
| 4 | 21 | 19A | 19A | |
| 5 | 21 | 11A | 11A | |
| 6 | 21 | |||
| 8 | 21 | 21 | 14 | 14 |
| 10 | 19F | 19F | 21 | 21 |
| 12 | 8 + 14 | 8 + 14 | 6A | 6A |
| 14 | 22F | 22F | 6B | 6B |
| 16 | 6B | |||
| 18 | ||||
| 20 | 6A | 6A | ||
| 22 | 6A | |||
| 24 | 23A | 23A | 6A | 6A |
Characterization of nonserotypeable isolates.
There were 31 nonserotypeable presumptive pneumococci among the 333 isolates, all of which tested positive for the pneumolysin gene using PCR. Four of these possessed alleles at six or all seven of the MLST loci that were found in serotypeable pneumococci within the MLST database. Two of these four isolates were identical in ST to serotypeable isolates from the same children, and isolates with an identical ST and serotype were also present within the MLST database. The other two isolates each had six alleles that were found in serotypeable pneumococci within the MLST database, although their allelic profiles were not closely similar to any serotypeable isolates in the MLST database. These four isolates were therefore assigned as pneumococci that were not expressing capsular polysaccharide. The other 27 nonserotypeable isolates had alleles at six or all seven loci that were not found in any serotypeable pneumococci. Furthermore, as these novel alleles were several percent divergent from the alleles found in serotypeable pneumococci, they were not considered to be pneumococci and were not used in the further analyses.
Characterization of pneumococci from longitudinal carriage.
The remaining 306 pneumococcal isolates were resolved by MLST into 101 STs. Children often carried the same ST over several sampling points, and 235 pneumococcal isolates remained when multiple isolates of the same strain from the same child were removed. The serotype distribution and genetic relatedness of these 235 isolates were studied, as this subset indicates the prevalence of the serotypes and STs carried by the children. Serotypes 6B, 19F, 23F, 14, and 6A were the five most commonly carried serotypes, together representing 56% of all carried isolates (Table 2).
TABLE 2.
Serotype distribution and diversity of carried pneumococcal isolates
| Serotype(s) | No. of isolates | No. of STsa |
|---|---|---|
| 6B | 44 | 11 |
| 19F | 30 | 16 |
| 23F | 26 | 6 |
| 14 | 17 | 5 |
| 6A | 14 | 7 |
| 15B, 15Cb | 12 | 4 |
| 19A | 9 | 6 |
| 9V | 8 | 4 |
| 18C | 7 | 2 |
| 3 | 6 | 1 |
| 21 | 5 | 5 |
| 9N | 5 | 2 |
| 11A | 5 | 2 |
| 23A | 4 | 4 |
| 8 | 4 | 2 |
| 22F | 4 | 2 |
| 33F | 4 | 2 |
| 17F | 4 | 1 |
| 16F | 3 | 3 |
| 20 | 3 | 2 |
| 38 | 3 | 2 |
| 15A | 2 | 2 |
| 10A | 2 | 1 |
| 12F | 2 | 1 |
| 24F | 2 | 1 |
| 4 | 1 | 1 |
| 7F | 1 | 1 |
| 13 | 1 | 1 |
| 23B | 1 | 1 |
| 27 | 1 | 1 |
| 31 | 1 | 1 |
| 35F | 1 | 1 |
| 37 | 1 | 1 |
| NTc | 2 | 2 |
The total number of STs is greater than 101 as isolates of some STs were of differing serotype.
Isolates of serotypes 15B and 15C were considered together (see text).
NT, nonserotypeable.
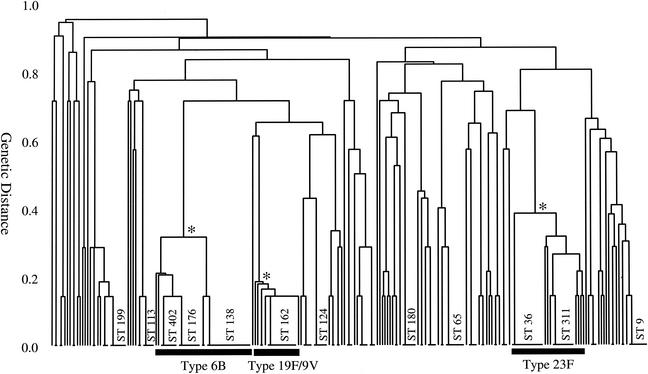
The genotypes of the carried isolates were very diverse and 64 of the 101 STs were recovered from only a single child; the other 37 STs colonized between 2 and 18 children. Table 3 shows the properties of the pneumococcal STs that were recovered from more than one child. Figure 1 displays the similarities between the genotypes of the 235 isolates as a dendrogram which shows there were three major clusters of closely related STs (clonal complexes). Together these three clonal complexes included 81 of the 235 carriage isolates (35%).
TABLE 3.
Properties of pneumococci carried by more than one child
| ST | Serotype(s)a | No. of children | Allele at MLST locusb:
|
Comment | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | ||||
| 138 | 6A(1)/6B | 18 | 7 | 5 | 8 | 5 | 10 | 6 | 14 | |
| 36 | 23F | 13 | 1 | 8 | 4 | 1 | 1 | 4 | 6 | |
| 162 | 9V(5)/19F | 12 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | |
| 176 | 6B | 9 | 7 | 13 | 8 | 6 | 10 | 6 | 14 | |
| 311 | 23F | 9 | 1 | 8 | 9 | 1 | 6 | 4 | 6 | |
| 65 | 6A | 8 | 2 | 7 | 4 | 10 | 10 | 1 | 27 | |
| 9 | 14 | 8 | 1 | 5 | 4 | 5 | 5 | 1 | 8 | England14-9 clone |
| 402 | 6B | 7 | 7 | 1 | 8 | 6 | 10 | 6 | 14 | |
| 199 | 15B(3)/15C | 7 | 8 | 13 | 14 | 4 | 17 | 4 | 14 | |
| 180 | 3 | 6 | 7 | 15 | 2 | 10 | 6 | 1 | 22 | |
| 124 | 14 | 6 | 7 | 5 | 1 | 8 | 14 | 11 | 14 | |
| 113 | 18C | 5 | 7 | 2 | 1 | 1 | 10 | 1 | 21 | |
| 66 | 9N | 4 | 2 | 8 | 2 | 4 | 6 | 1 | 1 | |
| 62 | 11A | 4 | 2 | 5 | 29 | 12 | 16 | 3 | 14 | |
| 392 | 17F | 4 | 7 | 5 | 1 | 1 | 6 | 31 | 14 | |
| 422 | 19F | 4 | 18 | 9 | 4 | 18 | 15 | 1 | 14 | |
| 53 | 8 | 3 | 2 | 5 | 1 | 11 | 16 | 3 | 14 | |
| 411 | 15B(1)/15C | 3 | 2 | 13 | 14 | 4 | 17 | 4 | 14 | |
| 415 | 19A | 3 | 1 | 5 | 12 | 5 | 14 | 15 | 31 | |
| 433 | 22F | 3 | 1 | 1 | 4 | 1 | 18 | 58 | 17 | |
| 60 | 33F | 3 | 2 | 5 | 23 | 18 | 10 | 3 | 1 | |
| 146 | 6B | 2 | 7 | 6 | 1 | 2 | 6 | 15 | 14 | |
| 273 | 6B | 2 | 5 | 6 | 1 | 2 | 6 | 1 | 14 | |
| 400 | 6B | 2 | 7 | 5 | 8 | 5 | 57 | 6 | 14 | |
| 156 | 9V/14 | 2 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | Spain9V-3 clone |
| 97 | 10A | 2 | 5 | 7 | 4 | 2 | 10 | 1 | 27 | |
| 218 | 12F | 2 | 10 | 20 | 14 | 1 | 6 | 1 | 29 | |
| 121 | 18C | 2 | 7 | 2 | 25 | 1 | 10 | 1 | 21 | |
| 416 | 19A | 2 | 1 | 13 | 14 | 4 | 17 | 51 | 14 | |
| 420 | 19F | 2 | 15 | 38 | 19 | 5 | 36 | 20 | 6 | |
| 421 | 19F | 2 | 3 | 10 | 2 | 5 | 9 | 48 | 6 | |
| 423 | 19F | 2 | 1 | 5 | 4 | 12 | 5 | 3 | 8 | |
| 424 | 19F | 2 | 1 | 5 | 4 | 5 | 9 | 3 | 8 | |
| 177 | 19F | 2 | 7 | 14 | 4 | 12 | 1 | 1 | 14 | |
| 235 | 20 | 2 | 15 | 8 | 8 | 18 | 15 | 1 | 31 | |
| 72 | 24F | 2 | 2 | 13 | 2 | 4 | 9 | 4 | 1 | |
| 393 | 38 | 2 | 10 | 43 | 41 | 18 | 13 | 49 | 6 | |
Number of isolates of the minority serotype in parentheses.
aroE, gene encoding shikimate dehydrogenase; gdh, gene encoding glucose-6-phosphate dehydrogenase; gki, gene encoding glucose kinase; spi, gene encoding signal peptidase I; xpt, gene encoding xanthine phosphotransferase; ddl, gene encoding d-ala-d-ala ligase.
FIG. 1.
Relatedness of isolates from the nasopharynges of children. The 235 isolates that represent the diverse strains recovered from the children were analyzed by MLST, and their relatedness was displayed as a dendrogram. ST numbers are shown for those STs recovered from more than four children. The node that defines each of three major clonal complexes within the carriage population is indicated with an asterisk. The serotype 6B clonal complex included 36 isolates of type 6B, 1 isolate of type 6A, and 1 isolate of type 23F. The serotype 23F clonal complex included 25 isolates, all of type 23F, and the 19F/9V complex included 10 isolates of serotype 19F, 7 isolates of type 9V, and 1 isolate of type 14.
Following the removal of the 27 nonserotypeable isolates that were not considered to be pneumococci, there were 68 children who carried at least two pneumococci of differing serotype, and 229 isolates from these children remained when multiple isolates of the same strain from the same child were removed. Examination of the pneumococci from the 68 children showed that, in almost all cases, isolates of different serotypes from the same child were also different in ST. Table 4 provides details of the isolates from the 14 children who over the 2 years carried isolates of multiple serotypes and from which at least seven pneumococci were selected for characterization by MLST, according to the criteria described above. Details of the pneumococci carried by all of the 68 children are available from the authors on request.
TABLE 4.
Isolates from children carrying more than six pneumococcal isolates
| Isolatea | Sero- type | Allele at MLST locusb:
|
ST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | |||
| 2-8.21 | 21 | 10 | 8 | 4 | 35 | 6 | 1 | 18 | 429 |
| 2-10.19F | 19F | 15 | 38 | 19 | 5 | 36 | 20 | 6 | 420 |
| 2-12.8 | 8 | 2 | 5 | 1 | 11 | 16 | 3 | 14 | 53 |
| 2-12.14 | 14 | 1 | 5 | 4 | 5 | 5 | 1 | 8 | 9 |
| 2-14.22F | 22F | 1 | 1 | 4 | 1 | 18 | 58 | 17 | 433 |
| 2-20.6A | 6A | 1 | 5 | 7 | 12 | 10 | 1 | 14 | 327 |
| 2-24.23A | 23A | 1 | 51 | 9 | 2 | 6 | 4 | 6 | 435 |
| 5-6.6B | 6B | 5 | 6 | 1 | 2 | 6 | 1 | 14 | 273 |
| 5-10.23F | 23F | 1 | 8 | 9 | 1 | 6 | 4 | 6 | 311 |
| 5-12.18C | 18C | 7 | 2 | 1 | 1 | 10 | 1 | 21 | 113 |
| 5-12.23F | 23F | 1 | 8 | 9 | 1 | 6 | 4 | 6 | 311 |
| 5-16.19F | 19F | 3 | 10 | 2 | 5 | 9 | 48 | 6 | 421 |
| 5-18.6B | 6B | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 90 |
| 5-20.19F | 19F | 3 | 10 | 2 | 5 | 9 | 48 | 6 | 421 |
| 5-20.6B | 6B | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 90 |
| 5-24.3 | 3 | 7 | 15 | 2 | 10 | 6 | 1 | 22 | 180 |
| 5-24.6B | 6B | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 90 |
| 5-24.19F | 19F | 3 | 10 | 2 | 5 | 9 | 48 | 6 | 421 |
| 25-6.NT | NTc | 8 | 5 | 2 | 27 | 2 | 11 | 71 | 448 |
| 25-12.15B | 15B | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 199 |
| 25-12.15C | 15C | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 199 |
| 25-14.15B | 15B | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 199 |
| 25-14.15C | 15C | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 199 |
| 25-14.NT | NT | 8 | 37 | 9 | 29 | 2 | 47 | 5 | 449 |
| 25-16.19A | 19A | 1 | 5 | 12 | 5 | 14 | 15 | 31 | 415 |
| 28-4.19A | 19A | 10 | 13 | 14 | 4 | 17 | 95 | 14 | 450 |
| 28-5.11A | 11A | 2 | 5 | 29 | 12 | 16 | 3 | 14 | 62 |
| 28-8.14 | 14 | 8 | 5 | 4 | 5 | 1 | 3 | 8 | 409 |
| 28-10.21 | 21 | 10 | 8 | 4 | 35 | 6 | 1 | 14 | 432 |
| 28-12.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 28-14.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 28-24.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 32-5.19F | 19F | 7 | 11 | 10 | 6 | 6 | 8 | 14 | 165 |
| 32-6.19F | 19F | 7 | 11 | 10 | 6 | 6 | 8 | 14 | 165 |
| 32-8.14 | 14 | 1 | 5 | 4 | 5 | 5 | 1 | 8 | 9 |
| 32-10.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 32-12.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 32-12.19F | 19F | 7 | 11 | 10 | 6 | 6 | 8 | 14 | 165 |
| 32-14.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 32-16.19F | 19F | 7 | 11 | 10 | 6 | 6 | 8 | 14 | 165 |
| 32-20.11A | 11A | 2 | 5 | 29 | 12 | 16 | 3 | 14 | 62 |
| 32-22.11A | 11A | 2 | 5 | 29 | 12 | 16 | 3 | 14 | 62 |
| 32-24.23F | 23F | 1 | 8 | 4 | 1 | 1 | 4 | 6 | 36 |
| 38-10.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 38-14.23F | 23F | 7 | 5 | 1 | 1 | 13 | 31 | 14 | 440 |
| 38-20.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 38-20.23F | 23F | 7 | 13 | 8 | 6 | 10 | 6 | 69 | 441 |
| 38-22.23F | 23F | 1 | 8 | 4 | 1 | 1 | 4 | 6 | 36 |
| 38-24.3 | 3 | 7 | 15 | 2 | 10 | 6 | 1 | 22 | 180 |
| 38-24.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 44-4.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 44-6.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 44-12.23B | 23B | 1 | 8 | 9 | 2 | 6 | 4 | 6 | 439 |
| 44-14.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 44-14.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 44-16.6B | 6B | 7 | 5 | 8 | 5 | 57 | 6 | 14 | 400 |
| 44-22.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 44-24.6B | 6B | 7 | 6 | 1 | 2 | 6 | 71 | 14 | 401 |
| 47-5.38 | 38 | 1 | 43 | 41 | 18 | 13 | 49 | 6 | 310 |
| 47-6.18C | 18C | 7 | 2 | 1 | 1 | 10 | 1 | 21 | 113 |
| 47-8.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 47-10.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 47-12.18C | 18C | 7 | 2 | 1 | 1 | 10 | 1 | 21 | 113 |
| 47-14.9N | 9N | 2 | 8 | 2 | 4 | 6 | 1 | 1 | 66 |
| 47-24.6B | 6B | 7 | 5 | 8 | 5 | 57 | 6 | 14 | 400 |
| 52-4.19F | 19F | 1 | 5 | 4 | 12 | 5 | 3 | 8 | 423 |
| 52-5.19F | 19F | 1 | 5 | 4 | 12 | 5 | 3 | 8 | 423 |
| 52-6.19F | 19F | 1 | 5 | 4 | 12 | 5 | 3 | 8 | 423 |
| 52-10.14 | 14 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 156 |
| 52-10.19F | 19F | 1 | 5 | 4 | 12 | 5 | 3 | 8 | 423 |
| 52-12.14 | 14 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 156 |
| 52-14.14 | 14 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 156 |
| 52-18.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 52-22.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 53-10.6B | 6B | 7 | 6 | 1 | 2 | 6 | 15 | 14 | 146 |
| 53-14.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 53-16.15B | 15B | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 199 |
| 53-18.19A | 19A | 1 | 5 | 12 | 5 | 14 | 15 | 31 | 415 |
| 53-22.9V | 9V | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 162 |
| 53-22.22F | 22F | 1 | 1 | 4 | 1 | 18 | 58 | 17 | 433 |
| 53-24.33F | 33F | 2 | 5 | 29 | 5 | 42 | 3 | 18 | 445 |
| 58-6.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 58-8.14 | 14 | 1 | 5 | 4 | 5 | 5 | 1 | 8 | 9 |
| 58-10.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 58-12.6B | 6B | 7 | 1 | 8 | 6 | 10 | 6 | 14 | 402 |
| 58-12.19F | 19F | 18 | 9 | 4 | 18 | 15 | 1 | 14 | 422 |
| 58-16.6B | 6B | 7 | 13 | 8 | 6 | 10 | 54 | 14 | 403 |
| 58-18.6B | 6B | 7 | 13 | 8 | 6 | 10 | 54 | 14 | 403 |
| 58-22.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 58-24.6A | 6A | 2 | 7 | 4 | 10 | 10 | 1 | 27 | 65 |
| 58-24.6B | 6B | 7 | 13 | 8 | 6 | 10 | 54 | 14 | 403 |
| 74-6.3 | 3 | 7 | 15 | 2 | 10 | 6 | 1 | 22 | 180 |
| 74-12.9N | 9N | 10 | 20 | 2 | 1 | 6 | 1 | 29 | 405 |
| 74-14.9N | 9N | 10 | 20 | 2 | 1 | 6 | 1 | 29 | 405 |
| 74-14.27 | 27 | 8 | 17 | 46 | 24 | 9 | 55 | 14 | 443 |
| 74-16.21 | 21 | 8 | 10 | 2 | 16 | 1 | 26 | 1 | 193 |
| 74-18.15B | 15B | 2 | 13 | 14 | 4 | 17 | 4 | 14 | 411 |
| 74-20.19A | 19A | 1 | 5 | 12 | 5 | 14 | 15 | 32 | 418 |
| 81-4.23A | 23A | 1 | 5 | 9 | 9 | 6 | 4 | 6 | 438 |
| 81-6.19F | 19F | 1 | 10 | 4 | 1 | 9 | 3 | 8 | 43 |
| 81-10.23A | 23A | 1 | 5 | 9 | 9 | 6 | 4 | 6 | 438 |
| 81-12.23A | 23A | 1 | 5 | 9 | 9 | 6 | 4 | 6 | 438 |
| 81-16.19F | 19F | 1 | 10 | 4 | 1 | 9 | 3 | 8 | 43 |
| 81-18.19F | 19F | 1 | 10 | 4 | 1 | 9 | 3 | 8 | 43 |
| 81-18.6B | 6B | 5 | 6 | 1 | 2 | 6 | 1 | 14 | 273 |
| 81-22.6B | 6B | 5 | 6 | 1 | 2 | 6 | 1 | 14 | 273 |
| 98-12.15A | 15A | 8 | 10 | 2 | 16 | 7 | 26 | 1 | 410 |
| 98-16.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 98-20.6B | 6B | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 138 |
| 98-20.19F | 19F | 1 | 5 | 1 | 15 | 9 | 28 | 8 | 427 |
| 98-22.9V | 9V | 7 | 11 | 10 | 1 | 6 | 73 | 14 | 407 |
| 98-22.19F | 19F | 1 | 5 | 1 | 15 | 9 | 28 | 8 | 427 |
| 98-24.19F | 19F | 1 | 5 | 1 | 15 | 9 | 28 | 8 | 427 |
Isolates are named as follows: child identification number-month sampled after birth.serotype.
See footnote b of Table 3 for explanation of genes.
NT, nonserotypeable.
There were a few examples of isolates of the same ST, from the same child, that differed in serotype. Five children carried isolates of the same ST that were either serotype 15B or 15C (e.g., child 25 [Table 4]), and two children carried isolates of the same ST that were serotypeable and nonserotypeable. In one of these children, serotype 18C and nonserotypeable isolates with the same allelic profile (ST113) were recovered in the 2-month sample, and an identical serotype 18C isolate was also recovered at the 5-month sampling. In the other child a serotype 14 isolate (ST9) was present at the 12-month sampling, and an identical isolate that was nonserotypeable was recovered at the 16-month sampling.
DISCUSSION
A total of 333 serotypeable or nonserotypeable presumptive pneumococci were characterized by MLST. Thirty-one of the isolates were nonserotypeable. The large majority (87%) of these nonserotypeable presumptive pneumococci were not considered to be pneumococci, whereas others were assigned as pneumococci that did not express capsular polysaccharide. MLST provides a clear way of distinguishing these two classes of nonserotypeable isolates. Isolates with alleles at all or most loci that are found in serotypeable pneumococci, or that have unique alleles which differ at only two or three nucleotides from a known pneumococcal allele, are assigned as pneumococci. Those that have alleles at all or most loci that are not found in serotypeable pneumococci, and which are several percent diverged in sequence from pneumococcal alleles, are assigned as isolates that are similar to, but distinct from, pneumococci. The identification among nonserotypeable presumptive pneumococci of both pneumococci that do not express a capsule and isolates very closely related to, but genetically distinct from, pneumococci has also been shown using molecular techniques by Whatmore et al. (18). As found by these authors, the latter class of isolates all possessed the pneumolysin gene, which is not a totally reliable indicator that an isolate is a pneumococcus, as besides being present in the latter isolates, the gene is present (and expressed) in some more distantly related isolates that are more closely allied to Streptococcus mitis than to S. pneumoniae (18).
The isolates carried by the children were of 33 different serotypes, although isolates of the common childhood serotypes predominated. As found previously for pneumococci from invasive disease (8), multiple STs are represented within most serotypes of carried pneumococci. The STs that were commonly carried typically corresponded to STs that have also been recovered from cases of invasive disease. Isolates from invasive disease were present in the MLST database for 12 of the 16 STs carried by at least four children. This report focuses on serotype changes during carriage, and a comparison of the carried and invasive isolates from age-matched children in the Oxford region will be reported elsewhere.
The vast majority of isolates with different serotypes obtained from the same child had different STs and were therefore genetically distinct. In most cases, the appearance of an isolate with a different serotype from that which was previously carried was thus due to the acquisition of a new pneumococcal strain, rather than to a change in the serotype of the resident strain. There were a few exceptions which need to be considered. Five children carried isolates of serotypes 15B and 15C that had the same ST, and two children carried a serotypeable and a nonserotypeable isolate of the same ST. The capsular polysaccharides of serotypes 15B and 15C differ only in the presence or absence of an acetyl group, and these serotypes are known to interconvert in vitro and in vivo, by an unknown molecular mechanism, at a low frequency (17). The isolation of genetically indistinguishable serotype 15B and 15C pneumococci from the same children is therefore considered to be a reflection of this interconversion process rather than a change of capsular serotype mediated by recombinational replacements at the capsular locus. Similarly, the interconversion from serotypeable to nonserotypeable presumably reflects a loss of capsular expression rather than a true change of serotype.
There were therefore no examples of changes of serotype mediated by recombinational events at the capsular locus among the pneumococcal isolates recovered from sequential samplings of the nasopharynx in this cohort of children. These results put some limits on the frequency of serotype changes during nasopharyngeal carriage and suggest that the phenomenon is relatively rare. Single examples of putative serotype changes during nasopharyngeal colonization have been reported in two previous studies. One involved a child in a day care center who initially carried a multiresistant serotype 23F isolate and subsequently carried an isolate of serotype 14 that was shown to be similar by pulsed-field gel electrophoresis (2). The other was a longitudinal study of carriage in a birth cohort of 19 children (16). In the latter study there were only 10 children who carried pneumococci of more than one serotype, and using the selection criteria used here (Table 1) there were 26 isolates of differing serotype obtained from these children, compared with 68 children and 229 isolates of differing serotype in our study. It is not clear why serotype changes were apparently found in these previous studies but were not found in our considerably larger study.
Serotype changes are well documented from the presence among pneumococcal populations of isolates of the same ST with different serotypes, and although there were no examples of serotype changes in individual children, there were several examples among the population of carried pneumococci studied here. Excluding the two STs that included isolates of both serotypes 15B and 15C, which are not considered to be due to a recombinational change of serotype, there were three STs that included isolates of two different serotypes (Table 3). One of these was ST156, a widely disseminated penicillin-resistant clone (Spain9V-3 [13]), isolates of which are usually serotype 9V, but (as in this study) serotype 14 isolates also are encountered (6).
The sequences of the capsular genes of different serotype 14 variants of the Spain9V-3 clone, and of serotype 19A and 19F variants of the multiresistant Spain23F-1 clone, have demonstrated that each of these three classes of serotype variants has arisen on multiple occasions by recombinational events at the capsular locus (5, 6; T. J. Coffey and B. G. Spratt, unpublished results). Although serotype changes appear to be relatively common, on the basis of both the analysis of pneumococcal populations and the multiple origins of the serotype variants of the major antibiotic-resistant clones, these events are not so common that they can readily be observed by monitoring the pneumococci carried over time by a cohort of children. This does not imply that these events are insignificant in the evolution of the pneumococcus, since isolates that do undergo a change of serotype may on occasion increase in frequency within the nasopharynx, either by chance or by selection favoring the variant serotype, providing the possibility of transmission to new hosts. Whether serotype changes are of any great consequence is unclear, but the possibility of selection (for both antibiotic resistance and the nonvaccine serotype) leading to the emergence in vaccinated populations of nonvaccine serotype variants of the successful antibiotic-resistant clones needs to be monitored, as transmission of these among children could limit the expected reduction in antibiotic resistance among isolates causing pneumococcal disease.
Acknowledgments
This work was funded by the Wellcome Trust and the Oxford Vaccine Group. M.C.E. received funding from a Royal Society University Research Fellowship. B.G.S. received funding from a Wellcome Trust Principal Research Fellowship.
REFERENCES
- 1.Austrian, R. 1997. The enduring pneumococcus: unfinished business and opportunities for the future. Microb. Drug Resist. 3:111-115. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, D. M., S. Whittier, P. H. Gilligan, S. Soares, A. Tomasz, and F. W. Henderson. 1995. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J. Infect. Dis. 171:890-896. [DOI] [PubMed] [Google Scholar]
- 3.Black, S. B., H. R. Shinefield, J. Hansen, L. Elvin, D. Laufer, and F. Malinoski. 2001. Postlicensure evaluation of the effectiveness of seven valent pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 20:1105-1107. [DOI] [PubMed] [Google Scholar]
- 4.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 6.Coffey, T. J., M. Daniels, M. C. Enright, and B. G. Spratt. 1999. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology 145:2023-2031. [DOI] [PubMed]
- 7.Dagan, R., N. Givon-Lavi, O. Zamir, M. Sikuler-Cohen, L. Guy, J. Janco, P. Yagupsky, and D. Fraser. 2002. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J. Infect. Dis. 185:927-936. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 9.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, P. H. Makela, and The Finnish Otitis Media Study Group. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. 2001. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 10.Gray, B. M., G. M. Converse, and H. C. Dillon. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142:923-933. [DOI] [PubMed] [Google Scholar]
- 11.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 12.Klugman, K. P. 2001. Efficacy of pneumococcal conjugate vaccines and their effect on carriage and antimicrobial resistance. Lancet Infect. Dis. 1:85-91. [DOI] [PubMed] [Google Scholar]
- 13.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J-C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network (PMEN). J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obaro, S., and R. Adegbola. 2002. The pneumococcus: carriage, disease and conjugate vaccines. J. Med. Microbiol. 51:98-104. [DOI] [PubMed] [Google Scholar]
- 15.Salo, P., A. Ortqvist, and M. Leinonen. 1995. Diagnosis of bacteremic pneumococcal pneumoniae by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 171:479-482. [DOI] [PubMed] [Google Scholar]
- 16.Sluijter, M., H. Faden, R. de Groot, N. Lemmens, W. H. F. Goessens, A. van Belkum, and P. W. M. Hermans. 1998. Molecular characterization of pneumococcal nasopharynx isolates collected from children during the first 2 years of life. J. Clin. Microbiol. 36:2248-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Spratt, B. G., and B. M. Greenwood. 2000. Prevention of disease by vaccination: does serotype replacement matter? Lancet 356:1210-1211. [DOI] [PubMed] [Google Scholar]
- 17.Venkateswaran, P. S., N. Stanton, and R. Austrian. 1983. Type variation of strains of Streptococcus pneumoniae in capsular serogroup 15. J. Infect. Dis. 147:1041-1054. [DOI] [PubMed] [Google Scholar]
- 18.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “Atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]