Abstract
Salmonella enterica subsp. enterica serotype Enteritidis is not readily subtyped beyond the level of phage type (PT). A recently developed method for ribotyping of this organism, which uses a mixture of PstI and SphI (PS) for restriction of DNA (PS ribotyping), has proved useful for further subtyping of a number of PTs of this organism, including PT 4. However, it has not been extensively tested with PT 8. In the present study the PS ribotyping method was used to investigate outbreaks of both S. enterica serotype Enteritidis PT 4 and PT 8 and provided subtyping data that were consistent with information obtained from epidemiologic investigations. The method proved to be more discriminatory than phage typing and pulsed-field gel electrophoresis (PFGE) combined and was useful for investigating a pseudo-outbreak involving isolates that had identical PTs and PFGE types but that could not be linked epidemiologically. Several PS ribotypes were found within the cluster of isolates indistinguishable by other subtyping methods, confirming the epidemiologic findings. Although the PS ribotyping method proved to have a superior discriminatory ability in resolving clusters, it did not have high enough throughput for use in outbreak investigations. This method has therefore been adapted for use in automated ribotyping with a RiboPrinter, and the results were compared with those obtained by manual ribotyping. Both methods produce equivalent results and are useful for obtaining epidemiologically relevant subtyping data for S. enterica serotype Enteritidis, including PT 8 strains not extensively tested previously.
Salmonella enterica subsp. enterica serotype Enteritidis isolates have proved difficult to subtype. While biotyping and serotyping are traditionally the first steps in the characterization process, the technique of phage typing has been applied to these enteric pathogens with a high degree of success and has proved invaluable for epidemiologic investigations of outbreaks of human enteritis. However, this procedure is not routinely performed with all isolates across North America. By these subtyping methods, a worldwide increase in the incidence of human salmonellosis, in particular, those cases caused by S. enterica serotype Enteritidis, has been detected in industrialized countries (6). Indeed, intensive surveillance over the last two decades has revealed that S. enterica serotype Enteritidis has emerged as the second most common Salmonella serotype causing enteric disease in Canada (4). Although the proportion of S. enterica serotype Enteritidis phage type (PT) 4 isolates increased through 1992 (8), PT 8 has until very recently comprised the predominant PT isolated from humans in Canada.
Pulsed-field gel electrophoresis (PFGE) has been used to define a limited diversity within several established PTs of S. enterica serotype Enteritidis (14, 21); however, use of this technique has often resulted in failure to distinguish epidemiologically unrelated strains. In the absence of phage typing data, PFGE has not been effective for distinguishing S. enterica serotype Enteritidis PT 1 isolates from serotype Enteritidis PT 4 isolates due to pattern similarities (16). Randomly amplified polymorphic DNA (RAPD) analysis, PCR ribotyping, and typing by PCR for repetitive elements (M13 and the enterobacterial repetitive intergenic consensus sequence [ERIC]) have low indices of discrimination (9, 11, 12, 15). This can create problems when clusters of similar isolates are detected within a short period of time and can result in extensive and costly epidemiologic investigations that are ultimately unproductive. Most ribotyping methods do not discriminate well among S. enterica serotype Enteritidis strains (21); and those developed to date have proved too cumbersome, too expensive, too difficult to standardize, or too time-consuming to be effective when used with the large numbers of isolates that form the basis of molecular typing networks such as PulseNet (22) in the United States and PulseNet Canada (24).
Initial investigations on the application of ribotyping used SphI as the digestion enzyme. The data thus generated by this procedure suggested that use of this enzyme may serve to discriminate among many S. enterica serotype Enteritidis PT 8 strains. However, some strains remain untypeable because their DNA was not restricted (7). Recently, Landeras and colleagues (10, 12, 13) developed a method for ribotyping of S. enterica serotype Enteritidis isolates using a mixture of restriction enzymes, PstI and SphI (PS; PS ribotyping), and this appeared to be more discriminatory than other methods (10, 11, 14). However, the efficacy of the PS ribotyping method for the differentiation of S. enterica serotype Enteritidis PT 8 strains has been tested with only a relatively few isolates.
The aim of the present work was to determine the utility of the method for the differentiation of S. enterica serotype Enteritidis isolates and, in particular, PT 8 isolates that, by PFGE or other fingerprinting methods, appear to be identical or very similar. A further goal was to adapt the method for automated ribotyping, thereby making it suitable for use with a large number of specimens and hence applicable for use in the PulseNet and PulseNet Canada identification schemes. PS ribotyping was found to be very effective for the subtyping of S. enterica serotype Enteritidis PT 8 strains with identical PTs and PFGE patterns and can be adapted for use in automated ribotyping with the RiboPrinter (RP).
MATERIALS AND METHODS
Strains.
The isolates used in this study were from the National Laboratory for Enteric Pathogens (NLEP) culture collection, to which Canadian provincial public health laboratories submit isolates for further characterization. The isolates selected for this study were from confirmed outbreaks of human disease due to S. enterica serotype Enteritidis, as well as from clusters of cases resembling outbreaks and sporadic cases of human illness. Bacteria were maintained on slants of IP maintenance medium (10 g of Difco Laboratories [Detroit, Mich.] peptone per liter, 5 g of Difco beef extract per liter, 3 g of NaCl, 2 g of Na2HPO4 · 12H2O per liter, 8 g of Difco granulated agarose per liter [pH 7.4]) at room temperature in the dark for long-term storage. Subcultures, frozen at −80°C in brain heart infusion broth (Difco) containing 15% glycerol, were used to prepare working cultures. The methods used for serotyping and phage typing were as described previously (19, 23).
Preparation of riboprobe.
The riboprobe, which contained the entire 7.5-kb Escherichia coli rrnB rRNA operon (16S, 23S, and 5S rRNA genes), was amplified from recombinant plasmid pKK3535 (2) with primers rrnB F2 (5′-TGG ATC CGC CTA CCT TTC ACG AGT-3′) and rrnB R3 (5′-CTT TTG GCA GAC GCA GAC CTA CG-3′). PCR amplification was done with the Expand High Fidelity PCR system (Roche Molecular Biochemicals, Laval, Quebec, Canada) according to the instructions of the manufacturer. The protocol consisted of 30 cycles of the following steps: denaturation at 94°C for 1 min, annealing at 64°C for 1.5 min, and extension at 68°C for 6 min.
Manual ribotyping.
Ribotyping was performed with bacteria embedded in agarose plugs (“bugs in plugs” method). Bacterial cultures were grown overnight at 37°C on nutrient agar containing 1.5% NaCl. High-molecular-weight genomic DNA was prepared in 1.2% Seakem Gold agarose (Mandel Scientific Co. Inc., Guelph, Ontario, Canada) plugs by the standardized protocol of the Centers for Disease Control and Prevention (3). Digestion of the bacteria embedded in agarose plugs was done by the method of Nair et al. (17) with the enzyme system developed by Landeras and colleagues (10, 11, 12). After the plugs were cut out of the agarose, three-quarters of each original plug was equilibrated for 1 h with 1× buffer H (Roche Molecular Biochemicals). Digestion of DNA was accomplished by adding 40 U of PstI, 40 U of SphI, and 1.0 μl of a 0.5-mg/ml RNase solution (Roche Molecular Biochemicals) in a total volume of 100 μl, followed by incubation for 4 h at 37°C. Finally, the intact plugs containing the digested DNA were equilibrated with 0.5 ml of 0.5× TBE (Tris-borate-EDTA; 10× TBE was obtained from Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) for 15 min at room temperature and placed into a 1% agarose gel. The samples were electrophoresed for 18 h at 60 V in 0.5× TBE. The molecular size marker used was a 1-kb DNA extension ladder (Gibco Life Technologies, Burlington, Ontario, Canada). The gels were stained with ethidium bromide, photographed, and blotted with a Vacugene XL blotting apparatus (Amersham Pharmacia Biotech Ltd., Baie d'Urfé, Quebec, Canada). Blotting was performed by protocol 1 of the manufacturer's instructions. Blots were cross-linked with UV irradiation and probed with 500 ng of labeled riboprobe and 100 ng of labeled DNA ladder.
All subsequent procedures were done by the protocol for hybridization in tubes supplied with the Enhanced Chemiluminescent (ECL) Direct Nucleic Acid Labeling and Detection kit (Amersham Life Science Inc., Oakville, Ontario, Canada). The rrnB rRNA operon of E. coli originally from plasmid pKK3535 (2) was used as a probe for the rRNA genes, and the labeled size standard was used to visualize standard lanes. Developed blots were exposed to Hyperfilm-MP photographic film (Amersham Life Science Inc.).
Automated ribotyping.
Ribotyping was performed with the RP microbial characterization system (Qualicon, Inc., DuPont, Wilmington, Del.). The software included with the system compared single patterns of the strains or composite patterns in RiboGroups (21). Briefly, colonies were picked from individual culture plates, placed in tubes containing lysis buffer, and loaded into the RP unit. Within the unit, bacterial DNA digestion was accomplished with 27 μl of PstI at 40 U/μl (Qualicon) and 27 μl of SphI at 40 U/μl (Roche Molecular Biochemicals). The substitute restriction enzyme protocol in which digestion takes place at 37°C for 2 h was used. The RiboPrint pattern for each isolate was then compared to the patterns generated for the other isolates. As no database was available for comparison of isolates in assays with the restriction enzymes combined, characterization was critical. Automated ribotyping and manual ribotyping were performed with distinct, but overlapping, groups of isolates in order to facilitate comparison of the two methods and to gather more extensive data on isolates of interest.
PFGE.
PFGE was done by previously described protocols (3). Electrophoresis was performed with a CHEF DR III unit (Bio-Rad Ltd., Mississauga, Ontario, Canada) in 1% PFGE agarose at 14°C. Initial and final switch times were 2.2 and 63.8 s, respectively, and the total run time was 22 h. Following electrophoresis the gels were stained with ethidium bromide (0.5 μg/ml) and imaged with an Alpha Imager 2000 (Canberra Packard Canada, Mississauga, Ontario, Canada).
PCR.
ERIC PCR was performed as described by López-Molina et al. (15) with primers ERIC 1R (5′-ATG TAA GCT CCT GGG GAT TCA C-3′) and ERIC 2 (5′-AAG TAA GTG ACT GGG GTG AGC G-3′), synthesized at the University of Manitoba DNA Laboratory (Winnipeg, Manitoba, Canada). Amplifications were done in a Perkin-Elmer 9600 thermal cycler. Ten microliters of each of the amplified products was run on 1% DNA typing-grade agarose gels (Gibco BRL Life Technologies) in 0.5× TBE for 105 min at 120 V. A 100-bp ladder (Gibco BRL Life Technologies) was used as a marker. After electrophoresis the gels were stained in a solution of 0.5 μg of ethidium bromide (Gibco BRL Life Technologies) per ml and photographed under UV irradiation.
Analysis of data.
Interpretation of the PFGE and ribotype patterns was aided by use of Bionumerics (version 1.50) software (Applied Maths, Kortrijk, Belgium). All associations obtained with this software package were checked visually by at least two laboratorians. For the isolates analyzed in this study, each unique pattern was given a separate number. Similarity coefficients were obtained by calculating Dice coefficients. Automated cluster analysis was performed by using the unweighted pair group method with arithmetic averages within Bionumerics software. Dendrograms were constructed with Bionumerics software. Band position tolerances and optimization values of 1% were used for all analyses.
RESULTS
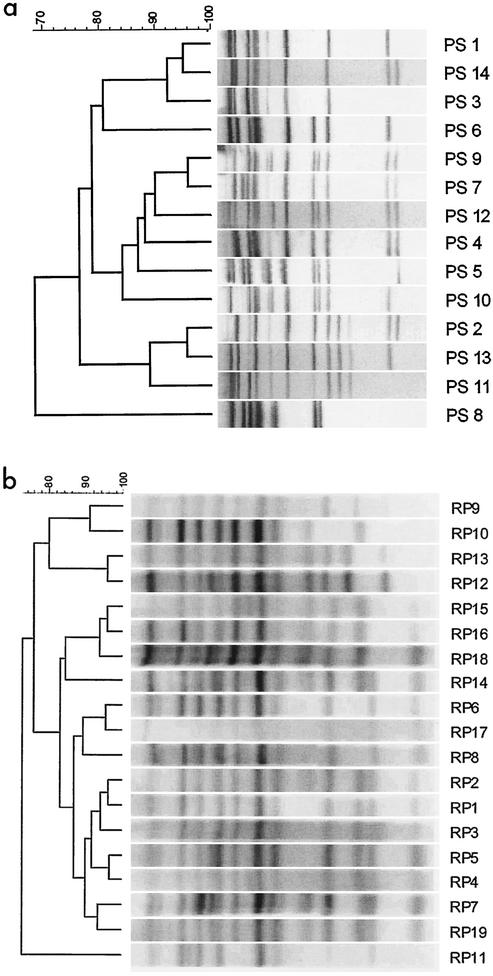
The ribotypes obtained by the manual and automated methods are summarized in Fig. 1. Dendrograms were constructed to aid in the visual comparison of ribotype patterns and were used to obtain an estimate of pattern relatedness only; they were not used to estimate the phylogenetic relationships among strains. All ribotype patterns occupying different branches of the dendrograms had clear differences, confirming that both ribotyping methods were capable of discriminating among strains not subtyped by other methods.
FIG. 1.
Results from ribotyping of S. enterica serotype Enteritidis strains by two methods. (a) Dendrogram of PS ribotype patterns obtained by manual ribotyping. (b) Dendrogram of ribotype patterns obtained by automated ribotyping with the RP unit. The ribotype designations are given at the right in both panels.
Comparison of methods for epidemiologic differentiation of isolates.
A major Canada-wide outbreak (outbreak 98002) of S. enterica serotype Enteritidis associated with contaminated cheese in a commercial product occurred in March and April 1998 (20). Phage typing and PFGE linked the clinical and cheese isolates of serotype Enteritidis PT 8 but failed to differentiate outbreak from nonoutbreak strains (1). Although all the isolates from humans and contaminated cheese found in food products obtained from different provinces across Canada proved to be PT 8 and PFGE XbaI pattern 3, three PS ribotype patterns, PS 4, PS 7, and PS 12, were detected (Fig. 1a and Table 1) along with two profiles detected with the RP unit, defined as RP 4 and RP 5 (Fig. 1b and Table 2). Two of the PS types, PS 4 and PS 12, were found in both humans and cheese, while PS 7 was found only in humans. RP 5 was also common to isolates from humans and cheese. ERIC PCR further differentiated three isolates of PS 4 by PS ribotyping. Automated ribotyping of a distinct, but overlapping, group of strains was capable of distinguishing between sporadic isolates and outbreak isolates with identical PTs (Table 2). One S. enterica serotype Enteritidis strain isolated as part of environmental investigations of the plant producing the contaminated product had a different PT, PFGE type, and PS ribotype but had ERIC PCR, repetitive extragenic palindromic sequence (REP), and RAPD types that were in common with the outbreak strain (data not shown). This isolate could therefore be eliminated as an outbreak strain on the basis of the PT, PS ribotype, and PFGE type, but not by ERIC PCR, REP, and RAPD typing.
TABLE 1.
Manual ribotyping of S. enterica serotype Enteritidis strains associated with an outbreak in 1998 and a pseudo-outbreak in 1999a
| Yr | Outbreak source | PT | Pattern by PFGE with XbaI | PS ribotype | ERIC PCR type | No. of isolates |
|---|---|---|---|---|---|---|
| 1998 | 98002/human, ON | 8 | 3 | 4 | 1 | 2 |
| 1998 | 98002/cheese, NF, NS | 8 | 3 | 4 | 1 | 6 |
| 1998 | 98002/cheese, ON | 8 | 3 | 4 | 2 | 3 |
| 1998 | 98002/human, NF, ON | 8 | 3 | 7 | 1 | 3 |
| 1998 | 98002/human, ON | 8 | 3 | 12 | 2 | 1 |
| 1998 | 98002/cheese, ON | 8 | 3 | 12 | 2 | 1 |
| 1999 | 99031/human, ON | 8 | 3 | 4 | 2 | 5 |
| 1999 | 99031/human, ON | 8 | 3 | 9 | 2 | 5 |
| 1999 | 99031/human, ON | 8 | 3 | 10 | 2 | 3 |
| 1999 | 99031/human, ON | 8 | 3 | 6 | 2 | 1 |
| 1999 | 99031/human, ON | 8 | 3 | 13 | 2 | 1 |
Abbreviations for Canadian provinces: AB, Alberta; BC, British Columbia; MB, Manitoba; NB, New Brunswick; NS, Nova Scotia; ON, Ontario; PEI, Prince Edward Island; QC, Québec; NF, Newfoundland.
TABLE 2.
Results of automated and manual ribotyping for selected S. enterica serotype Enteritidis strainsa
| Yr | Outbreak/source | PT | RP ribotype | PS ribotype | No. of isolates |
|---|---|---|---|---|---|
| 1998 | 98002/human, AB, MB, QC, NS, NB, NF | 8 | 5 | ND | 9 |
| 1998 | 98002/cheese, NS | 8 | 5 | 4 | 1 |
| 1998 | 98002/NS | 8 | 4 | ND | 1 |
| 1998 | Sporadic/Latin America | 8 | 14 | 12 | 1 |
| 1998 | Sporadic/AB, MB | 8 | 19 | 10 | 2 |
| 1999 | 99031/ON | 8 | 1 | 4 | 4 |
| 1999 | 99031/ON | 11b | 1 | 9 | 1 |
| 1999 | 99031/BC | Atypical | 1 | ND | 1 |
| 1999 | 99031/ON | 8 | 3 | 13 | 1 |
| 1999 | Sporadic/ON | 8 | 2 | ND | 2 |
| 1999 | Sporadic/PEI | 4 | 6 | 1 | 1 |
| 1999 | Sporadic/QC | 6a | 6 | 1 | 1 |
| 1999 | Sporadic/ON | 4a | 10 | 8 | 1 |
| 1999 | Sporadic/ON | 21 | 8 | ND | 1 |
| 1999 | Sporadic/ON | 21 | 17 | 14 | 1 |
| 2000 | 00031/BC | 8, atypical | 12 | ND | 5 |
| 2000 | 00031/BC | 8 | 13 | ND | 1 |
| 2000 | 00031/BC | 8 | 15 | ND | 1 |
| 2000 | 00031/BC | 8 | 16 | ND | 1 |
| 2000 | 00031/BC | 8 | 18 | ND | 1 |
| 2000 | Sporadic/QC | 8 | 2 | ND | 1 |
| 2000 | Sporadic/BC | 4 | 6 | 1 | 1 |
| 2000 | Sporadic/BC | 4 | 9 | ND | 1 |
| 2000 | Sporadic/PEI | 4 | 11 | 3 | 1 |
| 2000 | Sporadic/AB | 2 | 7 | ND | 2 |
ND, not determined; see footnote a of Table 1 for province name abbreviations.
In 1999 there was an increased incidence in southern Ontario of S. enterica serotype Enteritidis PT 8 isolations, investigated as outbreak 99031. While the submission to NLEP of a relatively large number of isolates within a fairly short span of time suggested the possibility of an outbreak, epidemiologists indicated that they could find no epidemiologic evidence that an outbreak was indeed occurring. PFGE with XbaI indicated that all isolates tested produced an identical PFGE pattern indistinguishable from the one associated with the cheese outbreak in 1998. However, manual ribotyping detected five different groups of strains (Table 1). One group of five isolates generated a PS ribotype (PS 4) identical to that of strains involved in the cheese outbreak. However, another group of five isolates produced the PS 9 ribotype, three produced PS 10, and the other two strains produced unique ribotypes. Automated ribotyping of selected isolates supported this analysis (Table 2).
An increase in travel-related S. enterica serotype Enteritidis cases was noticed from January through April 2000 (5). Phage typing, PFGE with two enzymes, and PS ribotyping grouped many of these PT 4 strains into a single group that included strains from patients who had traveled to Thailand and the Dominican Republic (Table 3). Two strains isolated from patients who had traveled to the Dominican Republic differed only by their PFGE patterns, while another two differed only by their PS ribotype patterns. The single characteristic that differentiated the pattern for an isolate from a sporadic case in Ontario in 1998 from the predominant pattern for isolates from patients with travel-associated disease was the PS ribotype, with the sporadic isolate being PS 6 and the travel-associated isolates being PS 1 and PS 3. In contrast, S. enterica serotype Enteritidis isolates from patients who had traveled to Jamaica and Cuba had different PTs, PFGE patterns, and PS ribotypes (Table 3).
TABLE 3.
Manual ribotyping of S. enterica serotype Enteritidis isolates for comparison of isolates from sporadic cases with strains associated with travel in 1999
| Sample sourcea | Travel destination(s) (yr) | PT | Pattern by PFGE with:
|
PS (RP) ribotype | ERIC PCR type | No. of isolates | |
|---|---|---|---|---|---|---|---|
| XbaI | SmaI | ||||||
| NS | Thailand | 4 | 1 | 1 | 1 (6) | 1 | 1 |
| NS, ON | Dominican Republic | 4 | 1 | 1 | 1 | 1 | 3 |
| NS | Dominican Republic | 4 | 2 | 2 | 1 | 1 | 2 |
| PEI | Dominican Republic | 4 | 1 | 1 | 3 (11) | 1 | 2 |
| PEI | Sporadicb | 4 | 1 | NDc | 1 | 1 | 1 |
| BC, ON | Sporadicb | 4 | 1 | 1 | 1 | 1 | 4 |
| ON | Sporadicb (1998) | 4 | 1 | ND | 6 | 1 | 1 |
| NS | Jamaica, Cuba | 5a | 12 | 8 | 8 | 1 | 2 |
See footnote a of Table 1 for province name abbreviations.
These isolates were not associated with travel outside Canada.
ND, not determined.
An outbreak of S. enterica serotype Enteritidis PT 8 (outbreak 00031) that occurred in 2000 was analyzed immediately by phage typing and retrospectively by automated ribotyping. Of the eight PT 8 isolates and one PT atypical isolate, five were RP 12 and four exhibited unique RP patterns (Table 2). While the RP 13 pattern was very similar to the RP 12 pattern, the other three RP patterns differed markedly (Fig. 1b).
S. enterica serotype Enteritidis isolates from sporadic cases of human disease could generally be distinguished from each other and from outbreak strains by phage typing and/or ribotyping (Table 2). Two isolates of PT 4, PS 1, and RP 6 were detected in Prince Edward Island in 1999 and in British Columbia in 2000, respectively, and were therefore considered epidemiologically unrelated. In other cases more than one isolate with a unique combination of types was isolated during the same year within the same province. Although no epidemiologic evidence was available to link the individuals from whom the isolates were obtained, there was also little evidence to prove that no link existed.
ERIC PCR was capable of differentiating isolates not distinguished by other methods (Table 1). REP and RAPD PCRs did not differentiate PT 4, PT 5a, and PT 8 strains (data not shown), confirming the very low discriminatory powers of these assays for S. enterica serotype Enteritidis.
Although PS ribotyping and automated ribotyping were capable of differentiating among the different PTs, there was not a direct relationship between ribotype and PT (Table 4). PS 6 was detected among three PTs types, while PS 1, PS 8, PS 9, and PS 10 were each found to be associated with two PTs. Five PTs had more than one PS type, with PT 8 producing six PS types. In a different group of isolates, RP 1 was found to be distributed among three PTs and RP 6 was found to be distributed into two PTs, with PT 8 divided among 12 RP types.
TABLE 4.
Ribotypes associated with specific PTs
| PT | PS ribotype(s) | RP ribotype(s) |
|---|---|---|
| 2 | NDa | 7 |
| 4 | 1,b 3, 6 | 6, 9, 11 |
| 4a | 8, 11 | 10 |
| 5a | 8 | ND |
| 6a | ND | 6 |
| 8 | 2, 4, 6, 9, 10, 13 | 1, 2, 3, 4, 5, 12, 13, 14, 15, 16, 18, 19 |
| 11b | 9 | 1 |
| 13 | 7 | |
| 13a | 6, 10 | |
| 21 | 1, 14 | 8, 17 |
| 30 | 5 | |
| Atypical | ND | 1 |
ND, not determined.
Ribotypes in bold indicate those shared by more than one PT.
Comparison of ribotyping methods.
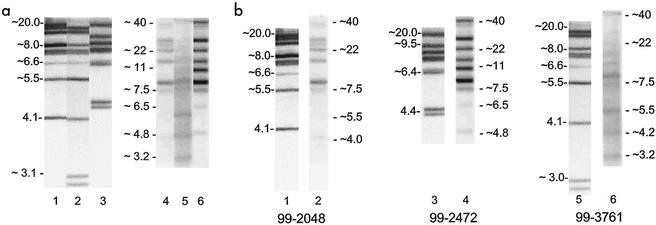
Manual and automated ribotyping produced patterns that were difficult to compare directly due to differences in the separation on gels; indeed, gels run by the two methods gave profiles that differed markedly in appearance. Differences in gel composition and run conditions caused substantial differences in the positions of bands on the gels and in the estimated fragment sizes, although differentiation of unrelated strains was still possible. Manual ribotyping separated smaller fragments with greater resolution but did not separate larger fragments well (Fig. 2a). Ribotyping with the RP unit separated fragments larger than 6 kb better than manual ribotyping but did not have as good a resolution for smaller bands. This can be seen by the lack of separation of two bands of approximately 4.4 kb for isolate 99-2472 by ribotyping with the RP unit that were clearly separated by the manual method (Fig. 2b, lanes 3 and 4). A similar lack of separation is seen for strain 99-3761, in which manual ribotyping detected two bands smaller than 3 kb, while automated ribotyping only detected a single band of about 3.2 kb (Fig. 2b, lanes 5 and 6). Several bands larger than 10 kb were not well separated by manual ribotyping but were much better separated with the RP unit. Two bands near 20 kb in the PS pattern of isolate 99-2048 were resolved into three bands larger than 20 kb in the corresponding RP pattern (Fig. 2b, lanes 1 and 2), and similar differences can be seen in comparisons between the remaining two isolates. In all cases, the bands obtained following analysis with the RP unit were not as sharp as those obtained by manual ribotyping, making it much more difficult to determine whether one or two fragments were present in a given band. The patterns derived by manual and automated ribotyping of each isolate were clearly consistent with each other, despite these differences.
FIG. 2.
Comparison of ribotype patterns obtained by manual ribotyping with DNA restricted in plugs and automated ribotyping with the RPunit. (a) Variation among patterns within each method. Lanes 1 to 3 were generated by manual ribotyping, lanes 4 to 6 were produced with the RP unit. Lanes 1 and 4, strain 99-2048, PS 3, RP 11; lanes 2 and 5, strain 99-3761, PS 14, RP 17; lanes 3 and 6, strain 99-2472, PS 8, RP 10. Approximate DNA fragment sizes (in kilobases) are shown at the left of each set of three lanes. (b) Comparison of manual and automated ribotype patterns for each strain. The strains in each lane were the same as those in panel a. Lanes 1, 3, and 5 were generated by manual ribotyping, while lanes 2, 4, and 6 were generated by automated ribotyping. Approximate DNA fragment sizes (in kilobases) are shown for each lane.
A set of nine isolates was analyzed by manual and automated ribotyping to facilitate comparison of the two methods. Both were capable of discriminating among different S. enterica serotype Enteritidis strains. In only one instance did the two techniques not differentiate the same isolate. Isolates 99-3769 (PS 4, RP 1) and 99-3772 (PS 9, RP 1) were separated by manual ribotyping on the basis of clear differences in banding patterns that were not seen when automated ribotyping was used. A pair of bands located at 4.4 kb in the PS pattern of isolate 99-3772 was not resolved in the RP pattern (see the PS 9 and RP 1 patterns in Fig. 1). Thus, while the discriminatory powers of the two assays were equivalent, each method subtyped the S. enterica serotype Enteritidis population under study in a slightly different way.
The patterns obtained by both manual and automated ribotyping were reproducible when the analysis was repeated with freshly prepared bacteria. The only difference in the banding pattern was seen with isolate EN5472, in which doublets at approximately 6.5 and 17 kb were not seen in the initial analysis but were seen in subsequent analyses (data not shown). Differences in pattern intensities suggest that slight differences in the cell concentration used for the analysis may be responsible for the pattern differences seen, making band designation more difficult. Similar pattern intensity differences were seen in the RP patterns for strains EN5580 and 99-3761. While these differences did not appear to interfere with the appropriate band designation for isolate EN5580, they may have led to the introduction of a separate RP designation for isolate 99-3761, which shared a common PT and similar RP pattern with isolate 99-3763. In contrast, the intensities of the PS patterns obtained by manual ribotyping with the bugs in plugs method were remarkably reproducible within and between blots.
The isolates were characterized and assigned to a specific RiboGroup that defined their genetic relatedness by using the software provided with the RP system. Visual examination of the patterns was obtained, and a degree of similarity equal to or higher than 0.95 allowed the samples in the RiboGroups to be merged. The 46 isolates analyzed were clustered into eight RiboGroups, S-1 to S-8 (Table 5). A few of these groups, for example, S-3 and S-6, incorporated a number of different RP types.
TABLE 5.
Comparison of RP types with RiboGroups created with RP system software
| RiboGroup | RP type | PT | No. of strains | % of strains |
|---|---|---|---|---|
| S-1 | 2 | 8 | 3 | 6.38 |
| S-2 | 14 | ND | 1 | |
| 19 | 8 | 2 | 6.38 | |
| S-3 | 8 | 21 | 1 | |
| 17 | 21 | 1 | ||
| 3 | 8 | 1 | ||
| 6 | 4 | 2 | ||
| 9 | 4 | 1 | ||
| 4 | 8 | 1 | ||
| 12 | 8 | 4 | ||
| 12 | Atypical | 1 | ||
| 18 | 8 | 1 | 27.65 | |
| S-4 | 11 | 4 | 1 | 2.12 |
| S-5 | 10 | 4a | 1 | |
| 13 | 8 | 1 | ||
| 16 | 8 | 1 | 6.38 | |
| S-6 | 1 | 8 | 3 | |
| 1 | 11b | 1 | ||
| 1 | Atypical | 1 | ||
| 5 | 8 | 12 | ||
| 6 | 6a | 1 | 40.42 | |
| S-7 | 1 | 8 | 1 | |
| 15 | 8 | 1 | 4.25 | |
| S-8 | 2 | 8 | 1 | |
| 7 | 2 | 2 | 6.38 |
DISCUSSION
PS ribotyping provided additional strain discrimination that proved effective for the description of outbreak strains as well as for the investigation of clusters of human disease caused by S. enterica serotype Enteritidis. The use of both manual and automated ribotyping with a mixture of PstI and SphI was capable of subtyping serotype Enteritidis PTs in a manner that provided results consistent with epidemiologic data. Isolates from three confirmed outbreaks of serotype Enteritidis PT 8 were grouped by both ribotyping methods, while epidemiologically unrelated isolates revealed different ribotype patterns. Retrospective PS ribotyping analysis differentiated strains that were responsible for a cluster resembling an outbreak in Ontario in 1999 and supported the epidemiologic investigations that found no links among infected individuals. Had the data from PS ribotyping been available at the time of the outbreak investigation, it would have been more helpful to apply epidemiologic methods to the smaller clusters that were found. Strains phage typed as PT 8 and PT 4 can be subtyped by PFGE. Manual PS ribotyping and automated ribotyping were highly effective for subtyping of these PTs. The PS ribotype patterns obtained here were similar to those obtained by Landeras and Mendoza (12), although they could not be compared directly because of differences in run conditions.
The diversity of patterns associated with each PT is not known and may be different for different PTs. There appeared to be a relatively high degree of diversity of ribotypes for PT 4 and PT 8. In contrast, all isolates of S. enterica serotype Enteritidis PT 30 from patients and from almonds associated with an international outbreak of enteric disease due to contaminated almonds had a single unique pattern by PFGE with XbaI (data not shown) and PS 5 by ribotyping. In the absence of epidemiologic evidence, it would be difficult to determine whether new patient isolates were associated with the outbreak. With further data collection, the predictive power of the ribotyping method(s) would improve as the sizes of the databases increase. Some ribotypes were found among multiple PTs, suggesting that the two methods measured independent characteristics.
The results of manual and automated ribotyping were not easily subject to direct comparison, although the two methods did appear to provide comparable strain discrimination. Both methods were reproducible. Previous work reported a 96% overall reproducibility for automated ribotyping (18). The minor differences between the two methods appear to be related to the fact that each procedure exhibits differences in resolution for different ranges of DNA fragment sizes. During the development of the automated ribotyping method, it was noted that differences in the intensities of RP patterns that affect pattern interpretation could arise quite readily if the input bacterial cell concentrations were not precisely standardized. While the stability of the ribotype patterns was not probed extensively here, repeat analysis of each isolate gave identical results in all but one instance. Similarly, Landeras and Mendoza (12) found that only a single band of about 7 kb, similar to the weak band that migrated at about 6.6 kb in this study, was not always reproducible and, hence, did not include that band in pattern analysis.
Use of the RP program to merge groups resulted in a loss of discriminatory power for automated ribotyping compared with that obtained by use of Bionumerics software to score every band. Comparison of the data from Table 5 and Fig. 1b indicated that in some cases the pattern differences were subtle (e.g., RP 2 versus RP 7 in S-8), although in other cases clear multiple band differences were seen (e.g., RP 4 and RP 12 in S-3). The interpretive criteria used for manual ribotyping generally assume that a single band difference is significant, allowing greater differentiation among related strains. Since this was the purpose for developing this assay, we propose that the same criteria be adopted for the interpretation of automated ribotyping of S. enterica serotype Enteritidis isolates by using PstI and SphI. A practical consequence of this recommendation is that pattern analysis would have to be carried out with a software package such as Bionumerics to allow scoring of individual bands, database construction, and information exchange. This would increase the time to completion for the testing of isolates.
Since both manual and automated ribotyping can subtype S. enterica serotype Enteritidis strains well, the choice of method to be used may depend on considerations other than the discriminatory power of the assay. Automated ribotyping has a much higher throughput and a shorter turnaround time than manual ribotyping and could be readily standardized among different laboratories. Standardization of the cell concentration used appears to be critical for the reproducible designation of RP band patterns produced by automated ribotyping. Manual ribotyping has proved successful for the subtyping of strains in ways that are epidemiologically meaningful, but it may be more difficult to standardize among different laboratories and also requires greater preparation time. Relatively few laboratories have access to RPs, which are relatively expensive, although some laboratories are now beginning to offer automated ribotyping on a fee-for-service basis. It is anticipated that the public health benefits resulting from adoption and use of either method will justify further development and use of ribotyping with a combination of the restriction enzymes PstI and SphI, especially for investigations of outbreaks of enteric disease due to S. enterica serotype Enteritidis.
Acknowledgments
We thank David Woodward and Helen Tabor for serotyping all Salmonella isolates; Walter Demczuk for phage typing; Dave Spreitzer, Russell Easy, Jennifer Campbell, and Shelley Johnson for technical assistance; and the directors of Canadian provincial public health laboratories for provision of strains.
REFERENCES
- 1.Ahmed, R., G. Soule, W. H. Demczuk, C. Clark, R. Khakhria, S. Ratnam, S. Marshall, L.-K. Ng, D. L. Woodward, W. M. Johnson, and F. G. Rodgers. 2000. Epidemiologic typing of Salmonella enterica serotype Enteritidis in a Canada-wide outbreak of gastroenteritis due to contaminated cheese. J. Clin. Microbiol. 38:2403-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112-118. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1998. One-day (24-48 h) standardized laboratory protocol for molecular subtyping of non-typhoidal Salmonella by pulsed field gel electrophoresis (PFGE), standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis (manual). Centers for Disease Control and Prevention, Atlanta, Ga.
- 4.Demczuk, W., R. Ahmed, D. Woodward, C. Clark, and F. Rodgers. 2001. Laboratory surveillance data for enteric pathogens in Canada: 2000 annual summary. Minister of Public Works and Government Services Canada, Ottawa, Ontario, Canada.
- 5.Division of Enteric, Foodborne and Waterborne Diseases, Bureau of Infectious Diseases, CIDPC, and the National Laboratory for Enteric Pathogens, Bureau of Microbiology, PPHB, Health Canada. 2001. Risk of enteric illness associated with travel: a case review of gastroenteritis among Canadian travellers: January to April 2000. Can. Commun. Dis. Rep. 27(6):45-49. [PubMed] [Google Scholar]
- 6.Gomez, T. M., Y. Motarjemi, S. Miyagawa, F. K. Käferstein, and K. Stöhr. 1997. Foodborne salmonellosis. World Health Statist. Q. 50:81-88. [PubMed] [Google Scholar]
- 7.Gruner, E., G. M. Lucchini, R. K. Hoop, and M. Altwegg. 1994. Molecular epidemiology of Salmonella enteritidis. Eur. J. Epidemiol. 10:85-89. [DOI] [PubMed] [Google Scholar]
- 8.Khakhria, R., D. Woodward, W. M. Johnson, and C. Poppe. 1997. Salmonella isolated from humans, animals, and other sources in Canada, 1983-92. Epidemiol. Infect. 119:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagatolla, C., L. Dolzani, E. Tonin, A. Lavenia, M. Di Michele, T. Tommasini, and C. Monti-Bragadin. 1996. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J. Clin. Microbiol. 34:2440-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landeras, E., M. A. González-Hevia, R. Alzugaray, and M. C. Mendoza. 1996. Epidemiological differentiation of pathogenic strains of Salmonella enteritidis by ribotyping. J. Clin. Microbiol. 34:2294-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landeras, E., M. A. González-Hevia, R. Alzugaray, and M. C. Mendoza. 1998. Molecular epidemiology of Salmonella serotype Enteritidis. Relationships between food, water, and pathogenic strains. Int. J. Food Microbiol. 43:81-90. [DOI] [PubMed] [Google Scholar]
- 12.Landeras, E., and M. C. Mendoza. 1998. Evaluation of PCR-based methods and ribotyping performed with a mixture of Pst I and Sph I to differentiate strains of Salmonella serotype Enteritidis. J. Med. Microbiol. 47:427-434. [DOI] [PubMed] [Google Scholar]
- 13.Landeras, E., M. A. Usera, C. Calderón, and M. C. Mendoza. 1997. Usefulness of phage typing and “two-way ribotyping” to differentiate Salmonella enteritidis strains. Microbiol. Semin. 13:471-480. [PubMed] [Google Scholar]
- 14.Liebana, E., L. Garcia-Migure, M. F. Breslin, R. H. Davies, and M. Woodward. 2001. Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J. Clin. Microbiol. 39:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Molina, N., I. Laconcha, A. Rementería, A. Audicana, I. Perales, and J. Garaizar. 1998. Typing of Salmonella enteritidis of different phage types of PCR fingerprinting. J. Appl. Microbiol. 84:877-882. [DOI] [PubMed] [Google Scholar]
- 16.Lukinmaa, S., R. Schildt, T. Rinttilä, and A. Siitonen. 1999. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J. Clin. Microbiol. 37:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair, S., R. Karim, M. J. Cardosa, G. Ismail, and T. Pang. 1999. Convenient and versatile DNA extraction using agarose plugs for ribotyping of problematic bacterial species. J. Microbiol. Methods 38:63-67. [DOI] [PubMed] [Google Scholar]
- 18.Pfaller, M. A., C. Wendt, R. Hollis, R. Wenzel, S. J. Fritschel, J. Neubauer, and L. Herwaldt. 1996. Comparative evaluation of an automated ribotyping system versus PFGE for epidemiological typing of clinical isolates of Escherichia coli and Pseudomonas aeruginosa from patients with recurrent gram-negative bacteremia. Diagn. Microbiol. Infect. Dis. 25:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Popoff, M. Y., and L. LeMinor. 1997. Antigenic formulas of the Salmonella serovars, 7th ed. World Health Organization Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 20.Ratnam, S., F. Stratton, C. O'Keefe, A. Roberts, R. Coates, M. Yetman, S. Squires, R. Khakhria, and J. Hockin. 1999. Salmonella Enteritidis outbreak due to contaminated cheese—Newfoundland. Can. Commun. Dis. Rep. 25:17-21. [PubMed] [Google Scholar]
- 21.Ridley, A. M., E. J. Threlfall, and B. Rowe. 1998. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC PulseNet Task Force. 2001. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward, L. R., J. D. H. De Sa, and B. Rowe. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodward, D. W., C. G. Clark, R. A. Caldeira, R. Ahmed, and F. G. Rodgers. Verotoxigenic Escherichia coli (VTEC): a major public health threat in Canada. Can. J. Infect. Dis. 13:321-330. [DOI] [PMC free article] [PubMed]